Exploring Different Roles of StWRKY4 and StWRKY56 in Transgenic Potato Against Salt Stress
Abstract
1. Introduction
2. Methodology
2.1. Plant Material and Growth Conditions
2.2. Cloning of a Unique Sequence of StWRKY4 and StWRKY56 into RNAi Vector for Transformation into Agrobacterium Tumefaciens
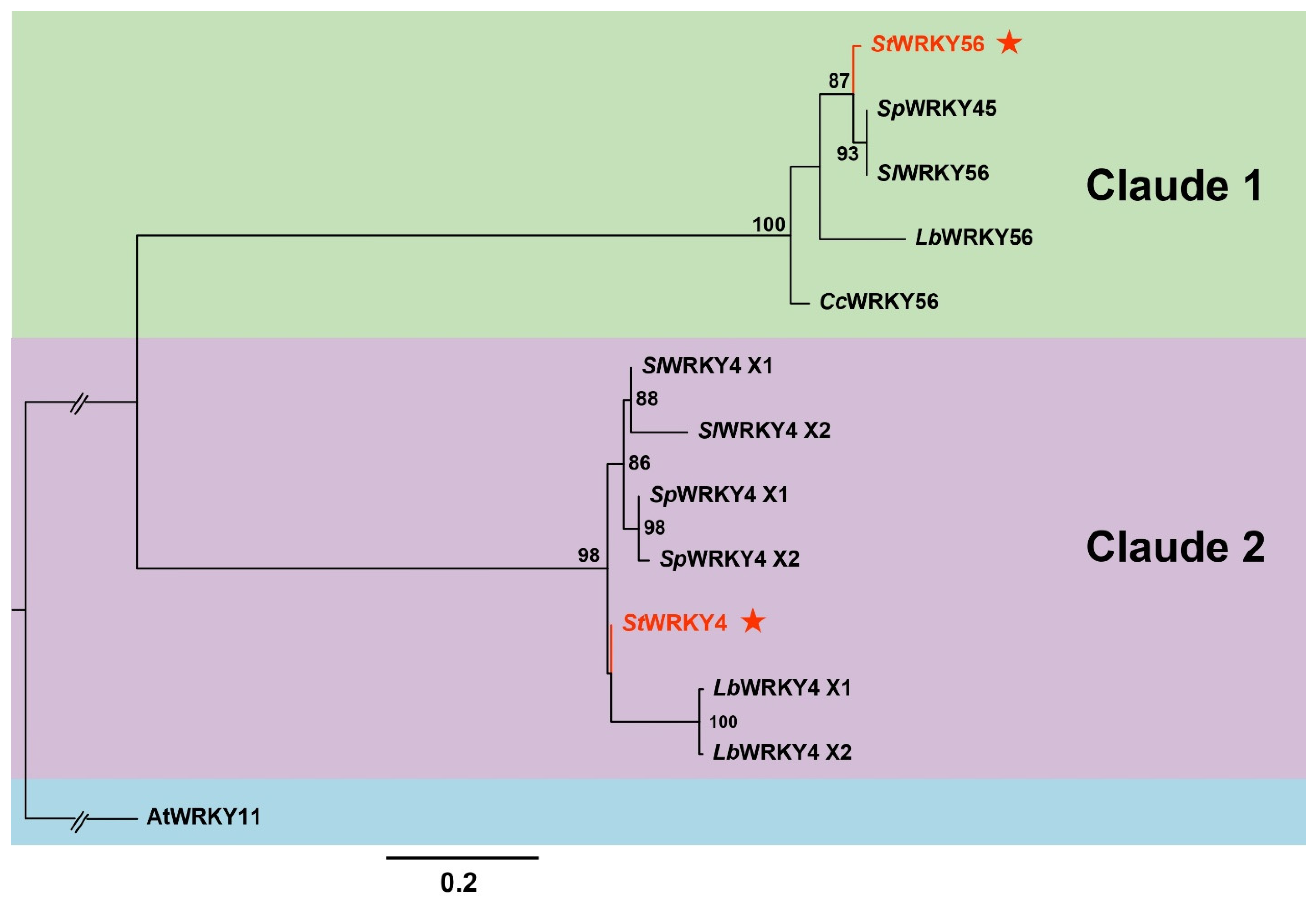
2.3. Phylogenetic Tree Construction for StWRKY4 and StWRKY56
2.4. Development of StWRKY4 and StWRKY56 Transgenic Lines
2.5. Salt Stress Analysis
2.6. Pigment Analysis for StWRKY4 and StWRKY56
2.7. Proline Content
2.8. Sodium and Potassium Ratio Determination
2.9. RNA Extraction Quantification and Complementary DNA (cDNA) Synthesis
2.10. Quantitative Real-Time qPCR
2.11. Statistical Analysis
3. Results
3.1. Development of Transgenic Potato StWRKY4 and StWRKY56
3.2. Phylogenetic Tree for StWRKY4 and StWRKY56
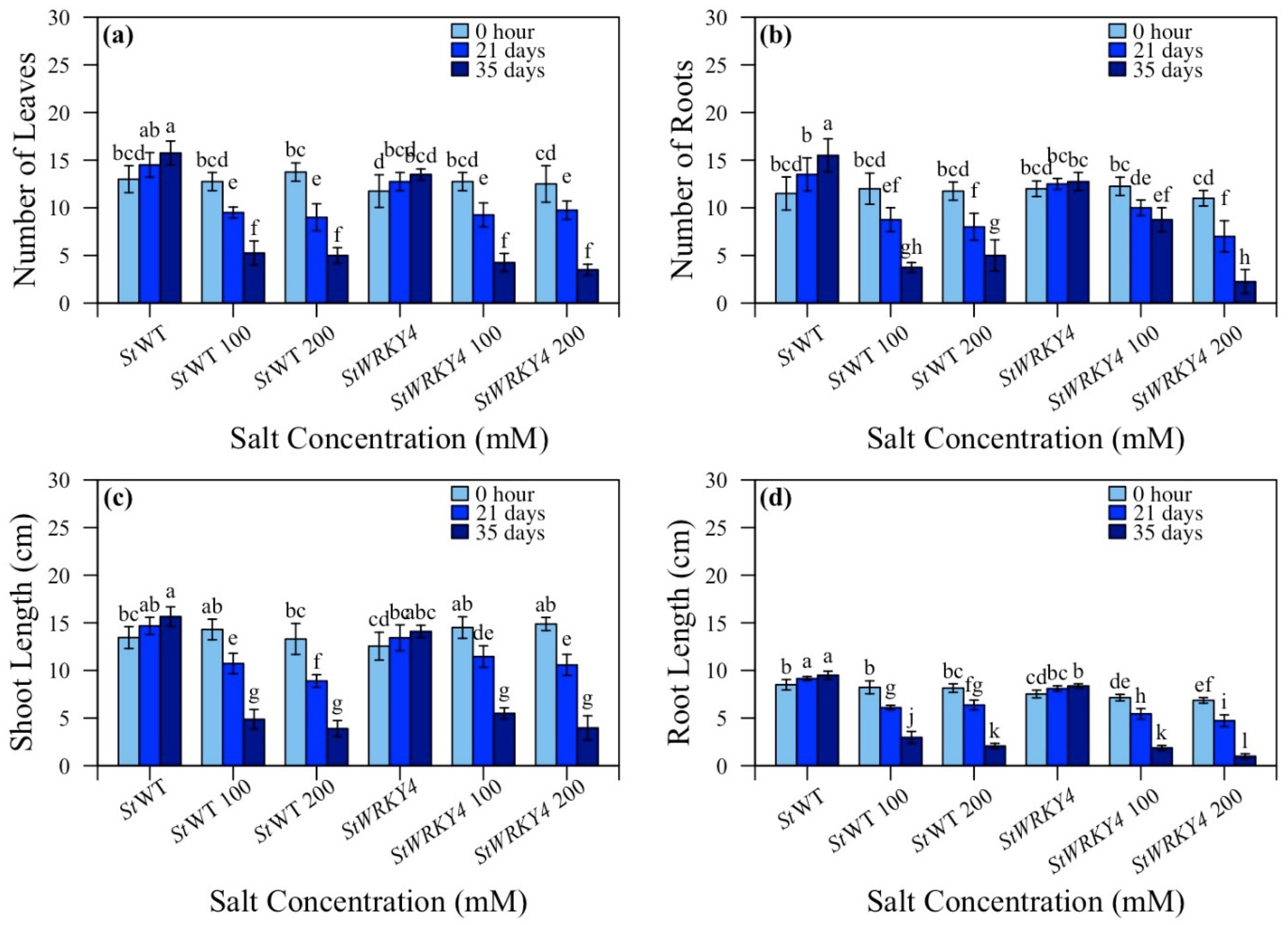
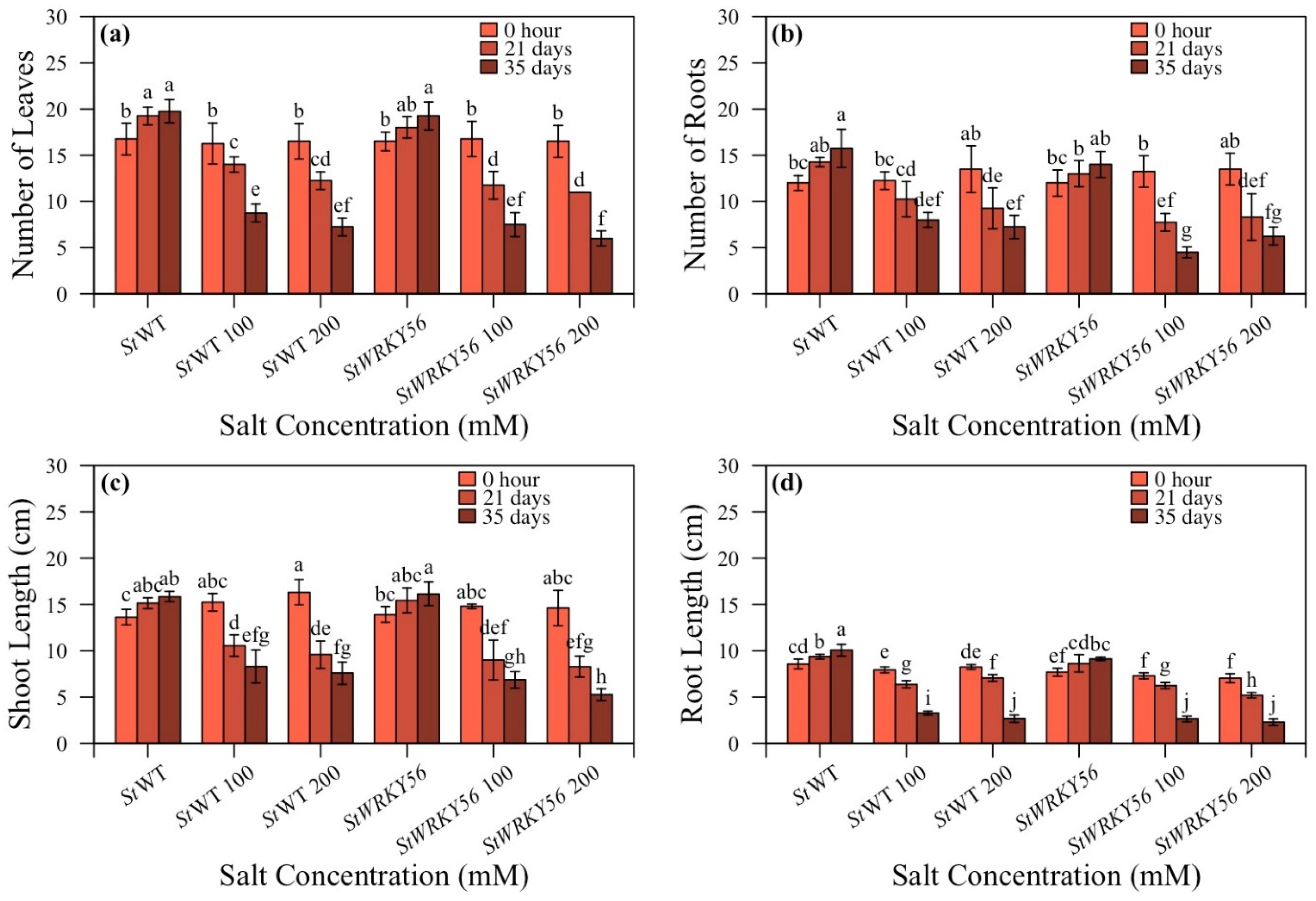
3.3. Number of Leaves, Number of Roots, Shoot Length and Root Length
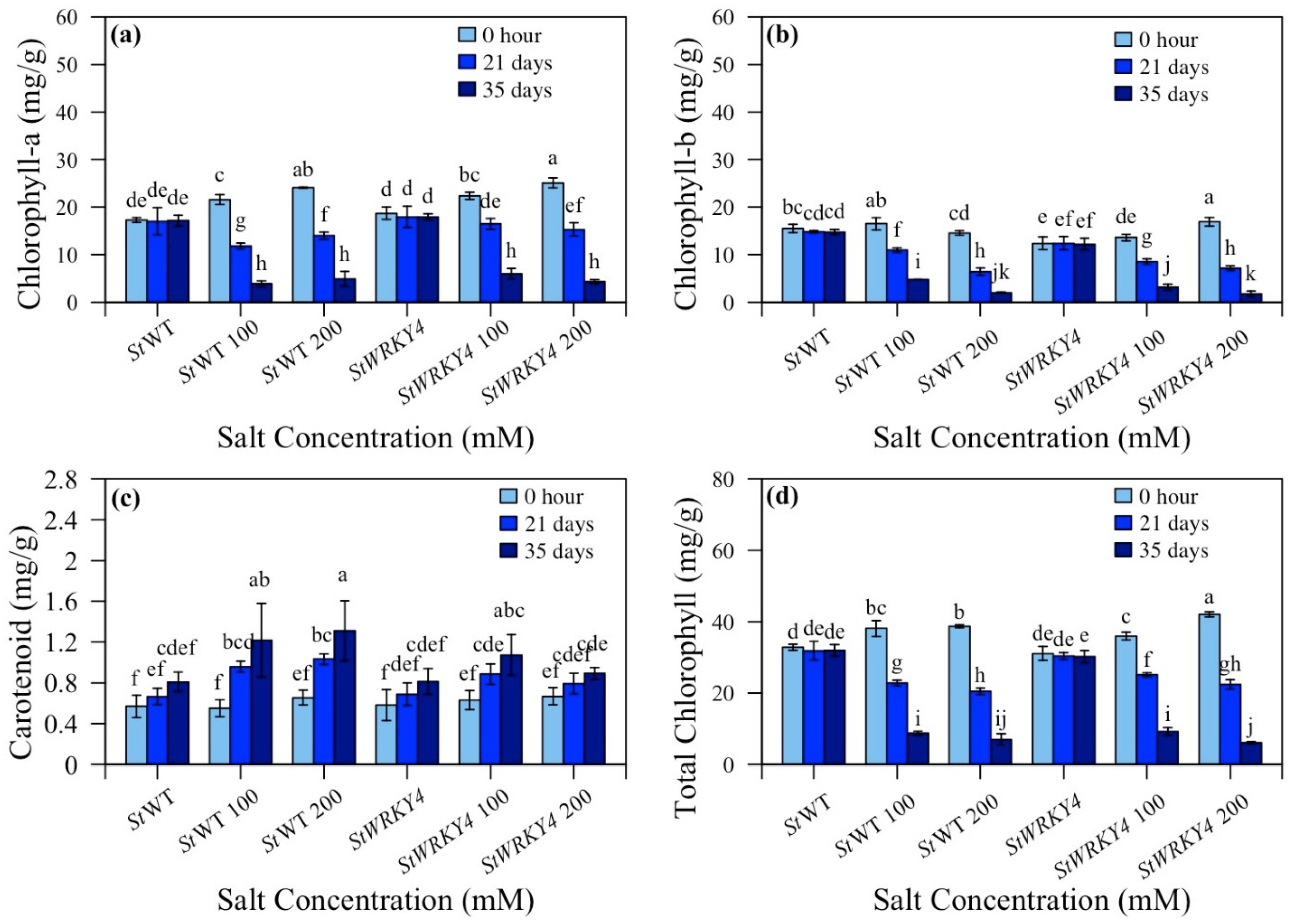
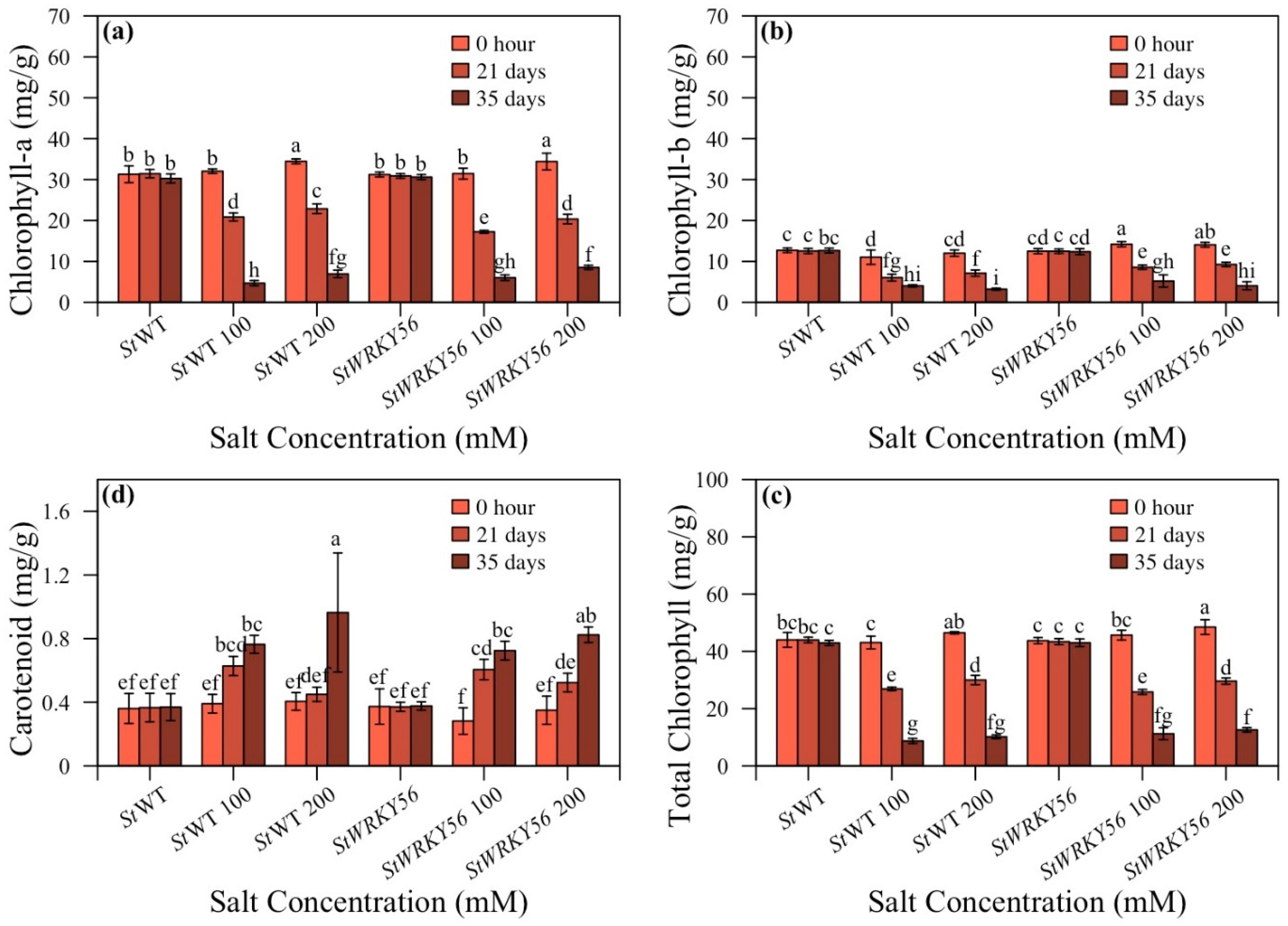
3.4. Chlorophyll and Carotenoid Content
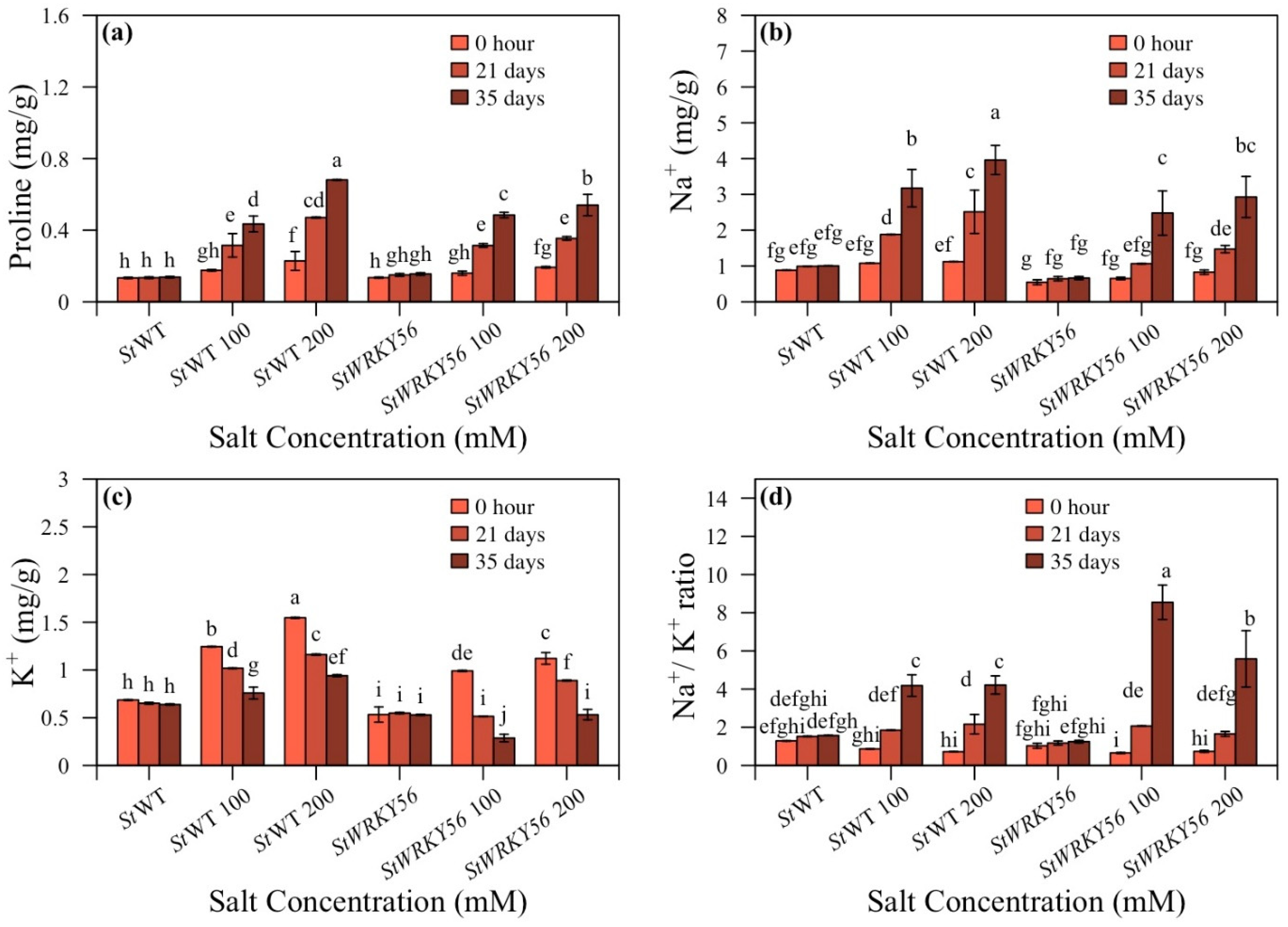
3.5. Proline Content and Na+/K+ Ratio
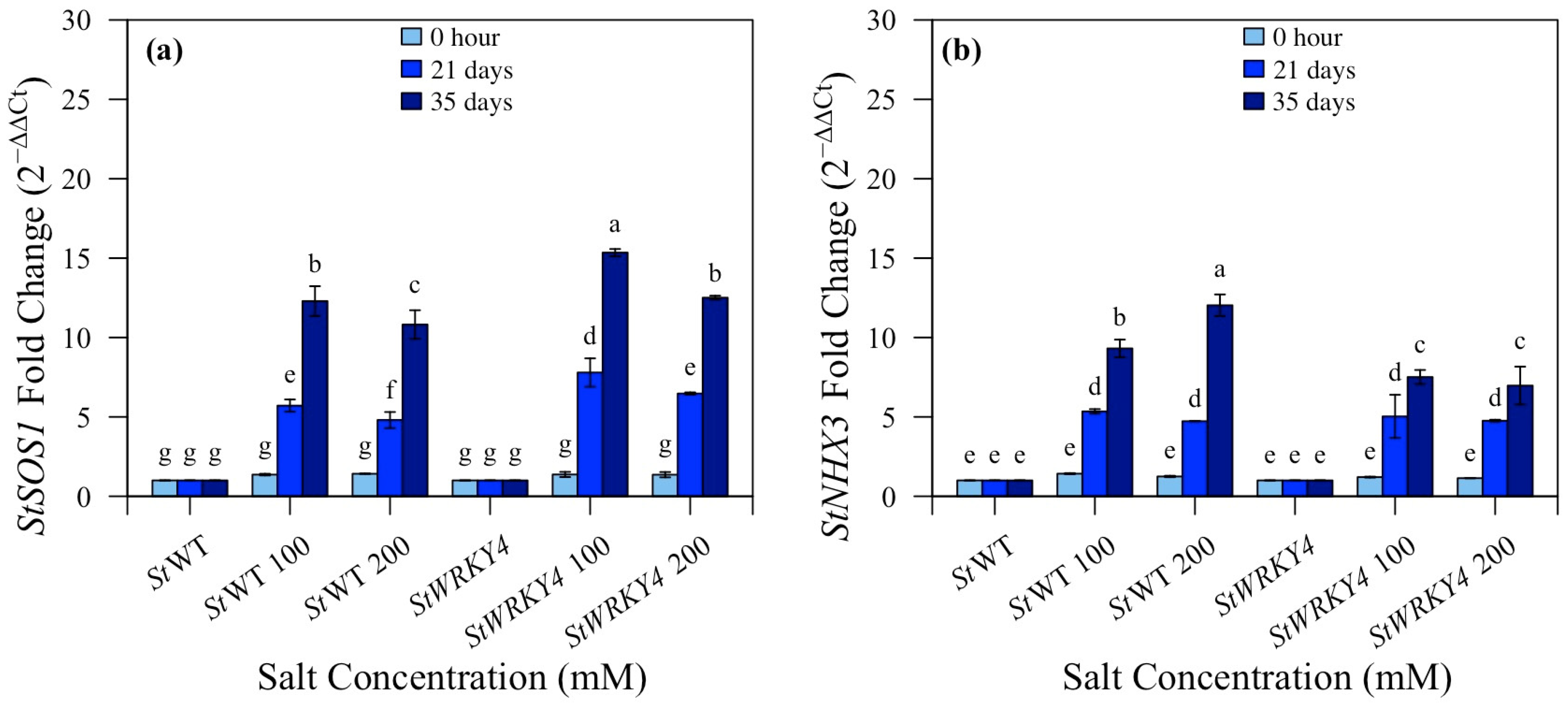
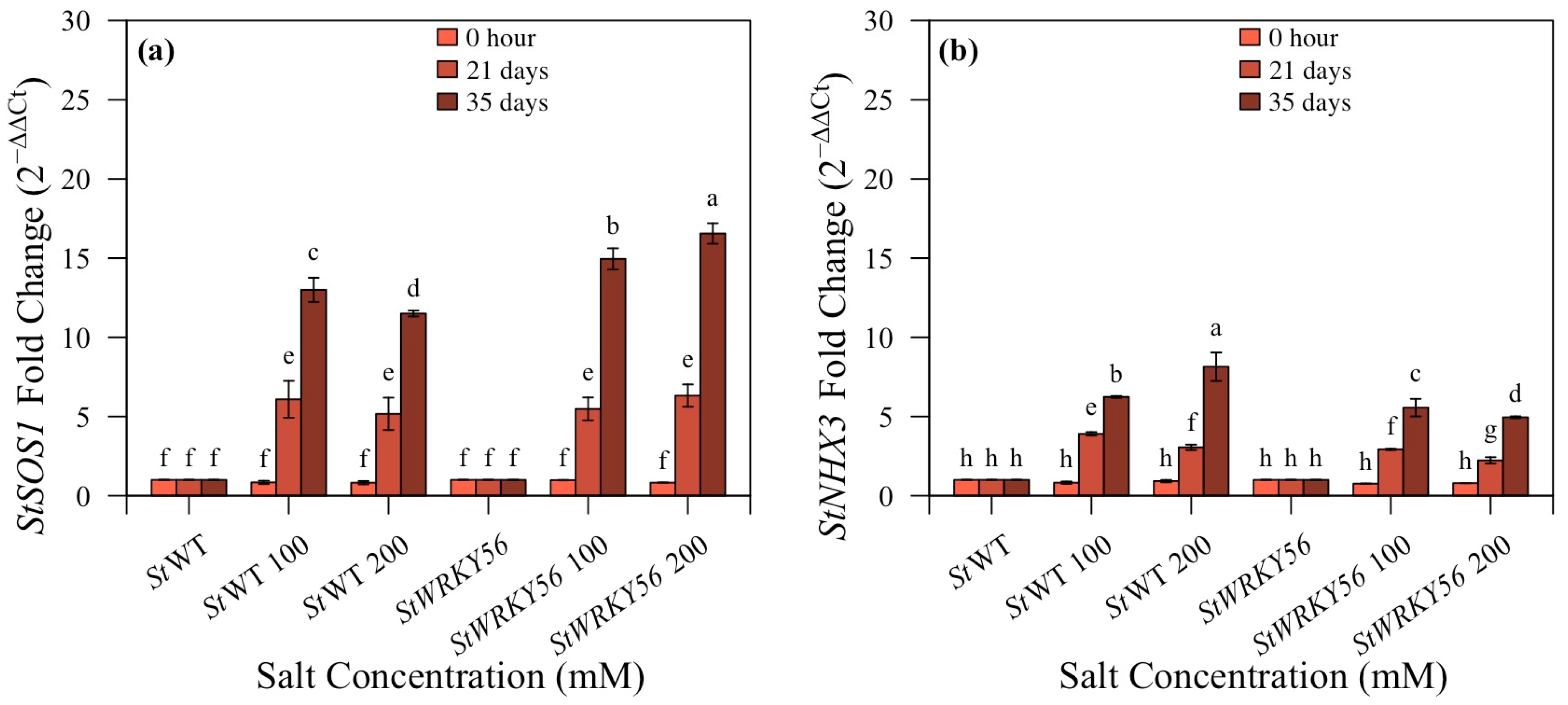
3.6. Quantitative Analysis of StSOS1 and StNHX3
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Çalışkan, M.E.; Yousaf, M.F.; Yavuz, C.; Zia, M.A.B.; Çalışkan, S. History, production, current trends, and future prospects. In Potato Production Worldwide; Academic Press: Cambridge, MA, USA, 2023; pp. 1–18. [Google Scholar]
- Devaux, A.; Goffart, J.P.; Kromann, P.; Andrade-Piedra, J.; Polar, V.; Hareau, G. The potato of the future, opportunities and challenges in sustainable agri-food systems. Potato Res. 2021, 64, 681–720. [Google Scholar] [CrossRef]
- Mir, B.A.; John, A.; Rahayu, F.; Martasari, C.; Husni, A.; Sukmadjaja, D.; Wani, A.K. Potato stress resilience: Unraveling the role of signalling molecules and phytohormones. Plant Gene 2024, 38, 100456. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, B. Protection of halophytes and their uses for cultivation of saline-alkali soil in China. Biology 2021, 10, 353. [Google Scholar] [CrossRef] [PubMed]
- Food and Agricultural Organization of the United Nations. FAOSTAT. 2024. Available online: https://www.fao.org/faostat/en/#home (accessed on 11 March 2025).
- Yokoi, S.; Quintero, F.J.; Cubero, B.; Ruiz, M.T.; Bressan, R.A.; Hasegawa, P.M.; Pardo, J.M. Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 2002, 30, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, A.; Huynh, H.D.; Endo, T.; Watanabe, K. Review of recent transgenic studies on abiotic stress tolerance and future molecular breeding in potato. Breed. Sci. 2015, 65, 85–102. [Google Scholar] [CrossRef]
- Qu, L.; Huang, X.; Su, X.; Zhu, G.; Zheng, L.; Lin, J.; Xue, H. Potato: From functional genomics to genetic improvement. Mol. Hortic. 2024, 4, 34. [Google Scholar] [CrossRef]
- Dzinyela, R.; Alhassan, A.R.; Suglo, P.; Movahedi, A. Advanced study of functional proteins involved in salt stress regulatory pathways in plants. S. Afr. J. Bot. 2023, 159, 425–438. [Google Scholar] [CrossRef]
- Chi, Y.; Yang, Y.; Zhou, Y.; Zhou, J.; Fan, B.; Yu, J.Q.; Chen, Z. Protein–protein interactions in the regulation of WRKY transcription factors. Mol. Plant 2013, 6, 287–300. [Google Scholar] [CrossRef]
- Villano, C.; Esposito, S.; D’Amelia, V.; Garramone, R.; Alioto, D.; Zoina, A.; Carputo, D. WRKY genes family study reveals tissue-specific and stress-responsive TFs in wild potato species. Sci. Rep. 2020, 10, 7196. [Google Scholar] [CrossRef]
- Shahzad, R.; Harlina, P.W.; Cong-hua, X.; Ewas, M.; Nishawy, E.; Zhenyuan, P.; Foly, M.M. Overexpression of potato transcription factor (StWRKY1) conferred resistance to Phytophthora infestans and improved tolerance to water stress. Plant Omics 2016, 9, 149–158. [Google Scholar] [CrossRef]
- Shahzad, R.; Ewas, M.; Harlina, P.W.; Nishawy, E.; Ayaad, M.; Manan, A.; Khames, E. Multiple stress responsive WRKY transcription factor, StWRKY2, enhances drought and late blight resistance in transgenic potato. Int. J. Agric. Biol. 2020, 24, 154–164. [Google Scholar]
- Dai, W.; Wang, M.; Gong, X.; Liu, J.H. The transcription factor FcWRKY 40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs. New Phytol. 2018, 219, 972–989. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Song, A.; Gao, C.; Wang, L.; Wang, Y.; Sun, J.; Chen, S. Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Rep. 2015, 34, 1365–1378. [Google Scholar] [CrossRef]
- Gao, H.; Wang, Y.; Xu, P.; Zhang, Z. Overexpression of a WRKY transcription factor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Front. Plant Sci. 2018, 9, 997. [Google Scholar] [CrossRef]
- Qin, Y.; Tian, Y.; Liu, X. A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2015, 464, 428–433. [Google Scholar] [CrossRef]
- Yu, Y.; He, L.; Wu, Y. Wheat WRKY transcription factor TaWRKY24 confers drought and salt tolerance in transgenic plants. Plant Physiol. Biochem. 2023, 205, 108137. [Google Scholar] [CrossRef]
- Zheng, L.; Liu, G.; Meng, X.; Liu, Y.; Ji, X.; Li, Y.; Wang, Y. A WRKY gene from Tamarix hispida, ThWRKY4, mediates abiotic stress responses by modulating reactive oxygen species and expression of stress-responsive genes. Plant Mol. Biol. 2013, 82, 303–320. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, Y. Overexpression of GmWRKY17, a class IIb WRKY transcription factor from Glycine max, enhances drought tolerance in Arabidopsis plants. Plant Growth Regul. 2024, 104, 253–265. [Google Scholar] [CrossRef]
- Duan, D.; Yi, R.; Ma, Y.; Dong, Q.; Mao, K.; Ma, F. Apple WRKY transcription factor MdWRKY56 positively modulates drought stress tolerance. Environ. Exp. Bot. 2023, 212, 105400. [Google Scholar] [CrossRef]
- Zhao, K.; Zhang, D.; Lv, K.; Zhang, X.; Cheng, Z.; Li, R.; Jiang, T. Functional characterization of popular WRKY75 in salt and osmotic tolerance. Plant Sci. 2019, 289, 110259. [Google Scholar] [CrossRef]
- Waqas, M.; Azhar, M.T.; Rana, I.A.; Azeem, F.; Ali, M.A.; Nawaz, M.A.; Atif, R.M. Genome-wide identification and expression analyses of WRKY transcription factor family members from chickpea (Cicer arietinum L.) reveal their role in abiotic stress-responses. Genes Genom. 2019, 41, 467–481. [Google Scholar] [CrossRef]
- Khan, I.; Lubna, B.S.; Abdelbacki, A.M.; Kang, S.M.; AL-Harrasi, A.; Lee, I.J. Genome-wide and transcriptome analysis of PdWRKY transcription factors in date palm (Phoenix dactylifera) revealing insights into heat and drought stress tolerance. BMC Genom. 2025, 26, 589. [Google Scholar] [CrossRef]
- Jiang, D.; Huang, W.; Liu, J. Phylogenetic relationship of WRKY transcription factors in Solanum and potato genes in response to hormonal and biotic stresses. Plant Signal. Behav. 2025, 20, e249146. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Mehboob, I.; Baig, S.; Siddique, M.; Shan, X.; Baig, A.; Shah, M.M.; Khalid, S. Deciphering the role of SlWRKY36 and SlWRKY51 in salt stress tolerance via modulating ion homeostasis and proline biosynthesis. Curr. Plant Biol. 2024, 39, 100380. [Google Scholar] [CrossRef]
- Camacho, C.; Boratyn, G.M.; Joukov, V.; Vera Alvarez, R.; Madden, T.L. Elastic BLAST: Accelerating sequence search via cloud computing. BMC Bioinform. 2023, 24, 117. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Rashid, S.; Saad, M.; Baig, S.; Shah, M.M.; Nawaz, I.; Baig, A. Effective regeneration and transformation of potato variety Desiree with nucleotide diphosphate kinase 2 using IAA and BAP. Pak. J. Bot. 2021, 53, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.A.; Erban, A.; Kopka, J.; Zörb, C. Metabolic contribution to salt stress in two maize hybrids with contrasting resistance. Plant Sci. 2015, 233, 107–115. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Buschmann, C. Extraction of photosynthetic tissues: Chlorophylls and Carotenoids. Curr. Protoc. Food Anal. Chem. 2001, 1, F4.2.1–F4.2.6. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.A.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Ali, S.G.; Rab, A.; Khan, N.U.; Nawab, K. Enhanced proline synthesis may determine resistance to salt stress in tomato cultivars. Pak. J. Bot. 2011, 43, 2707–2710. [Google Scholar]
- Qadir, A.; Khan, S.A.; Ahmad, R.; Masood, S.; Irshad, M.; Kaleem, F.; Shahzad, M. Exogenous Ca2SiO4 enrichment reduces the leaf apoplastic Na+ and increases the growth of okra (Abelmoschus esculentus L.) under salt stress. Sci. Hortic. 2017, 214, 1–8. [Google Scholar] [CrossRef]
- Barbier, F.F.; Chabikwa, T.G.; Ahsan, M.U.; Cook, S.E.; Powell, R.; Tanurdzic, M.; Beveridge, C.A. A phenol/chloroform-free method to extract nucleic acids from recalcitrant, woody tropical species for gene expression and sequencing. Plant Methods 2019, 15, 62. [Google Scholar] [CrossRef]
- Liang, L.; Guo, L.; Zhai, Y.; Hou, Z.; Wu, W.; Zhang, X.; Liu, W. Genome-wide characterization of SOS1 gene family in potato (Solanum tuberosum) and expression analyses under salt and hormone stress. Front. Plant Sci. 2023, 14, 1201730. [Google Scholar] [CrossRef]
- Jaarsma, R.; De Boer, A.H. Salinity tolerance of two potato cultivars (Solanum tuberosum) correlates with differences in vacuolar transport activity. Front. Plant Sci. 2018, 9, 737. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2025. [Google Scholar]
- Agarwal, P.; Reddy, M.P.; Chikara, J. WRKY: Its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol. Biol. Rep. 2011, 38, 3883–3896. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wang, P.; Hou, L.; Zhao, S.; Zhao, C.; Xia, H.; Wang, X. Global analysis of WRKY genes and their response to dehydration and salt stress in soybean. Front. Plant Sci. 2016, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Song, Y.; Xing, F.; Wang, N.; Wen, F.; Zhu, C. GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana benthamiana. Protoplasma 2016, 253, 1265–1281. [Google Scholar] [CrossRef]
- Zhao, X.; Qi, G.; Liu, J.; Chen, K.; Miao, X.; Hussain, J.; Ren, H. Genome-wide identification of WRKY transcription factors in Casuarina equisetifolia and the function analysis of CeqWRKY11 in response to NaCl/NaHCO3 stresses. BMC Plant Biol. 2024, 24, 376. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, N.N.; Gong, S.Y.; Lu, R.; Li, Y.; Li, X.B. Overexpression of a cotton (Gossypium hirsutum) WRKY gene, GhWRKY34, in Arabidopsis enhances salt-tolerance of the transgenic plants. Plant Physiol. Biochem. 2015, 96, 311–320. [Google Scholar] [CrossRef]
- Huang, J.; Liu, F.; Chao, D.; Xin, B.; Liu, K.; Cao, S.; Xia, J. The WRKY transcription factor OsWRKY54 is involved in salt tolerance in rice. Int. J. Mol. Sci. 2022, 23, 11999. [Google Scholar] [CrossRef]
- Hichri, I.; Muhovski, Y.; Žižková, E.; Dobrev, P.I.; Gharbi, E.; Franco-Zorrilla, J.M.; Lutts, S. The Solanum lycopersicum WRKY3 transcription factor SlWRKY3 is involved in salt stress tolerance in tomato. Front. Plant Sci. 2017, 8, 1343. [Google Scholar] [CrossRef]
- Liang, Q.Y.; Wu, Y.H.; Wang, K.; Bai, Z.Y.; Liu, Q.L.; Pan, Y.Z.; Jiang, B.B. Chrysanthemum WRKY gene DgWRKY5 enhances tolerance to salt stress in transgenic chrysanthemum. Sci. Rep. 2017, 7, 4799. [Google Scholar] [CrossRef]
- Yuan, Y.; Ren, S.; Liu, X.; Su, L.; Wu, Y.; Zhang, W.; Zhang, Y. SlWRKY35 positively regulates carotenoid biosynthesis by activating the MEP pathway in tomato fruit. New Phytol. 2022, 234, 164–178. [Google Scholar] [CrossRef]
- Ruiz-Sola, M.Á.; Arbona, V.; Gomez-Cadenas, A.; Rodriguez-Concepcion, M.; Rodriguez-Villalon, A. A root specific induction of carotenoid biosynthesis contributes to ABA production upon salt stress in Arabidopsis. PLoS ONE 2014, 9, e90765. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, M.; Bouma, J.; Venema, J.H.; van der Schoot, H.; Verstappen, F.; de Zeeuw, T.; Karlova, R. Potato cultivars use distinct mechanisms for salt stress acclimation. Plant Stress 2025, 15, 100798. [Google Scholar] [CrossRef]
- Mansour, M.M.F.; Ali, E.F. Evaluation of proline functions in saline conditions. Phytochemistry 2017, 140, 52–68. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Chen, S.; Chen, R.; Li, X.; Wu, J.; Zheng, Y.; Zhan, Y. Okra WRKY transcription factors AeWRKY32 and AeWRKY70 are involved in salt stress response. Int. J. Mol. Sci. 2024, 25, 12820. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.Y.; Du, Y.T.; Ma, J.; Min, D.H.; Jin, L.G.; Chen, J.; Zhang, X.H. The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in soybeans. Int. J. Mol. Sci. 2018, 19, 4087. [Google Scholar] [CrossRef]
- Ahammed, G.J.; Li, X.; Wan, H.; Zhou, G.; Cheng, Y. SlWRKY81 reduces drought tolerance by attenuating proline biosynthesis in tomato. Sci. Hortic. 2020, 270, 109444. [Google Scholar] [CrossRef]
- Ma, L.; Li, X.; Zhang, J.; Yi, D.; Li, F.; Wen, H.; Wang, X. MsWRKY33 increases alfalfa (Medicago sativa L.) salt stress tolerance through altering the ROS scavenger via activating MsERF5 transcription. Plant Cell Environ. 2023, 46, 3887–3901. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, Z.; Huang, Y.; He, D.; Lu, L.; Wei, M.; Jiang, Z. WRKY45 positively regulates salinity and osmotic stress responses in Arabidopsis. Plant Physiol. Biochem. 2025, 219, 109408. [Google Scholar] [CrossRef]
- Chen, H.; Lai, Z.; Shi, J.; Xiao, Y.; Chen, Z.; Xu, X. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 2010, 10, 281. [Google Scholar] [CrossRef]
- Li, C.; Guan, J.; Liang, W.H.; Yao, S.; He, L.; Wei, X.D.; Zhao, L.; Zhuo, L.H.; Zhao, C.F.; Zhao, Q.Y.; et al. OsWRKY72 enhances salt tolerance in rice via SKC1-mediated Na+ regulation. Plant Stress 2025, 17, 100962. [Google Scholar] [CrossRef]
- Lin, L.; Yuan, K.; Huang, Y.; Dong, H.; Qiao, Q.; Xing, C.; Zhang, S. A WRKY transcription factor PbWRKY40 from Pyrus betulaefolia functions positively in salt tolerance and modulating organic acid accumulation by regulating PbVHA-B1 expression. Environ. Exp. Bot. 2022, 196, 104782. [Google Scholar] [CrossRef]
- Yu, J.; Zhu, C.; Xuan, W.; An, H.; Tian, Y.; Wang, B.; Wan, J. Genome-wide association studies identify OsWRKY53 as a key regulator of salt tolerance in rice. Nat. Commun. 2023, 14, 3550. [Google Scholar] [CrossRef]


| Primers | Forward Sequence | Reverse Sequence |
|---|---|---|
| StWRKY4 | 5′ GCTCCACCAACTCTACATTCCC 3′ | 5′ CAGAATGAGCAACAAGAGCCCC 3′ |
| StWRKY56 | 5′ CCCTTGTGAAAAGCTAATGGAG 3′ | 5′ GCATGTGTGATGTGTACATCG 3′ |
| StNHX3 | 5′ TTGGCACAGACGTGAACCTA 3′ | 5′ GTGGCTTCTGACCAGTGACA 3′ |
| StSOS1 | 5′ TCCTGGAGACGGTAGCCAAA 3′ | 5′ ATTCCACCAATGGCAGCAGA 3′ |
| StActin | 5′ ATGAAGCTGTCCTTTTCACTTGTTTT 3′ | 5′ CTACATAGTATGCATGTCCGTATTT 3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gul, N.; Baig, S.; Shan, X.; Shahzadi, I.; Siddique, M.; Zhao, H.; Ahmad, R.; Hussain, J.; Khalid, S.; Baig, A. Exploring Different Roles of StWRKY4 and StWRKY56 in Transgenic Potato Against Salt Stress. Life 2025, 15, 1389. https://doi.org/10.3390/life15091389
Gul N, Baig S, Shan X, Shahzadi I, Siddique M, Zhao H, Ahmad R, Hussain J, Khalid S, Baig A. Exploring Different Roles of StWRKY4 and StWRKY56 in Transgenic Potato Against Salt Stress. Life. 2025; 15(9):1389. https://doi.org/10.3390/life15091389
Chicago/Turabian StyleGul, Nadia, Sofia Baig, Xiaoliang Shan, Irum Shahzadi, Maria Siddique, Hongwei Zhao, Raza Ahmad, Jamshaid Hussain, Samina Khalid, and Ayesha Baig. 2025. "Exploring Different Roles of StWRKY4 and StWRKY56 in Transgenic Potato Against Salt Stress" Life 15, no. 9: 1389. https://doi.org/10.3390/life15091389
APA StyleGul, N., Baig, S., Shan, X., Shahzadi, I., Siddique, M., Zhao, H., Ahmad, R., Hussain, J., Khalid, S., & Baig, A. (2025). Exploring Different Roles of StWRKY4 and StWRKY56 in Transgenic Potato Against Salt Stress. Life, 15(9), 1389. https://doi.org/10.3390/life15091389





