Molecular Dynamics Simulation Analysis of JAK1 Initial Activation: Phosphorylation-Induced Conformational Dynamics and Domain Interactions
Abstract
1. Introduction
2. Method
2.1. JAK1 Modeling
2.2. Molecular Dynamics Simulation
2.3. Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuation (RMSF)
2.4. Hydrogen Bonding
2.5. Electrostatic Potential Study
2.6. Comparison of Conformational Differences Between Phosphorylated and Non-Phosphorylated JAK1
3. Results
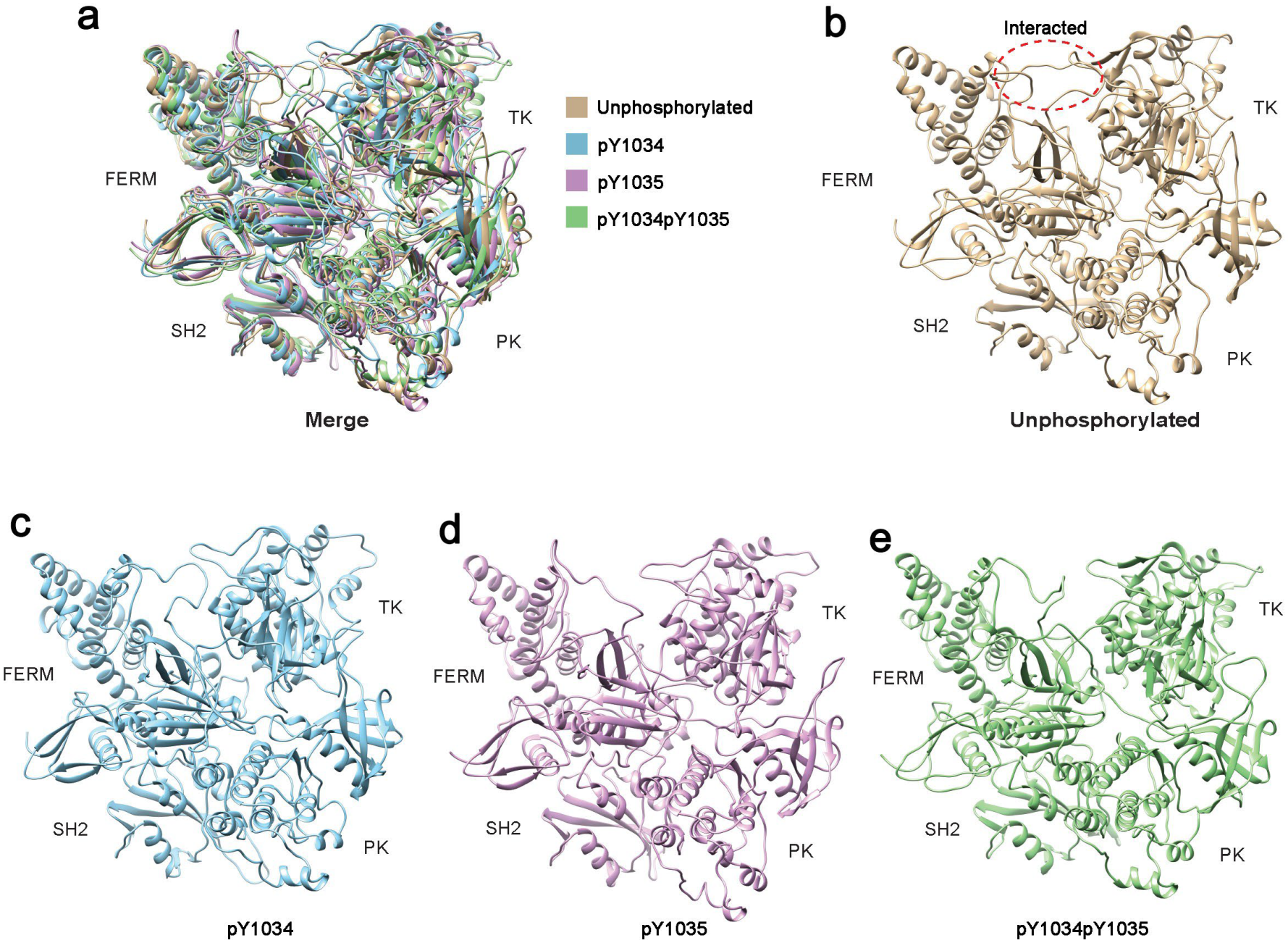
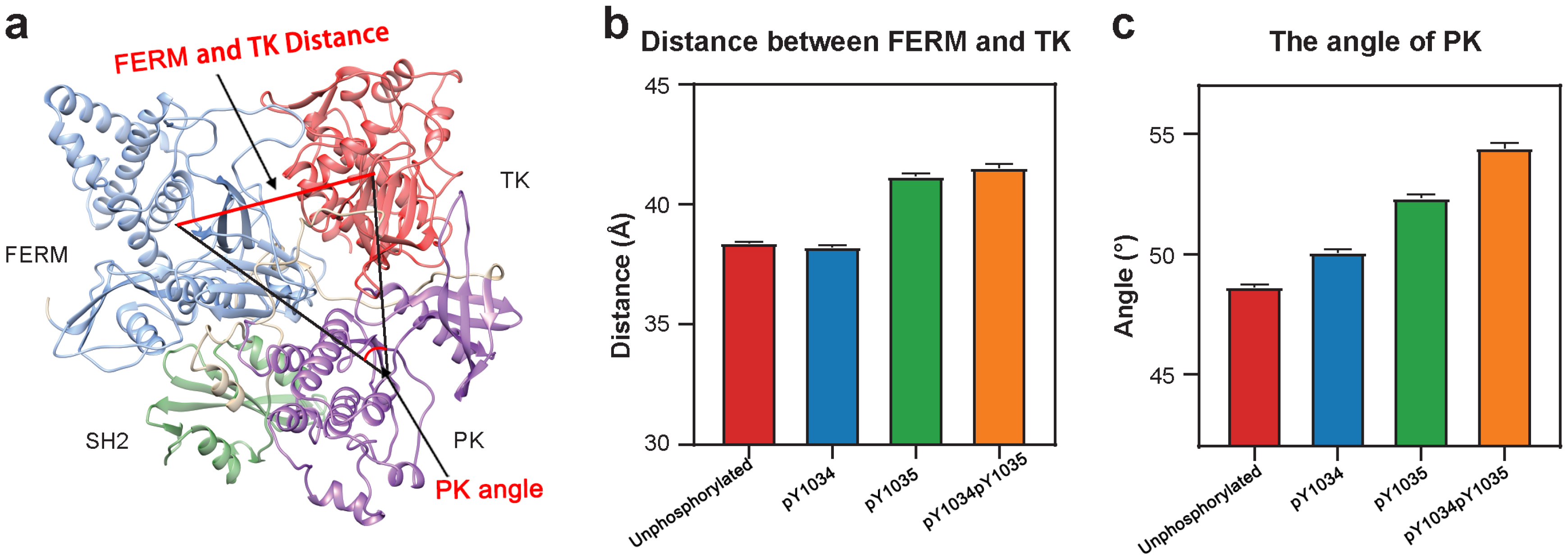
3.1. Tyr Phosphorylation in Activation Segment Has an Effect on the Conformation of JAK1
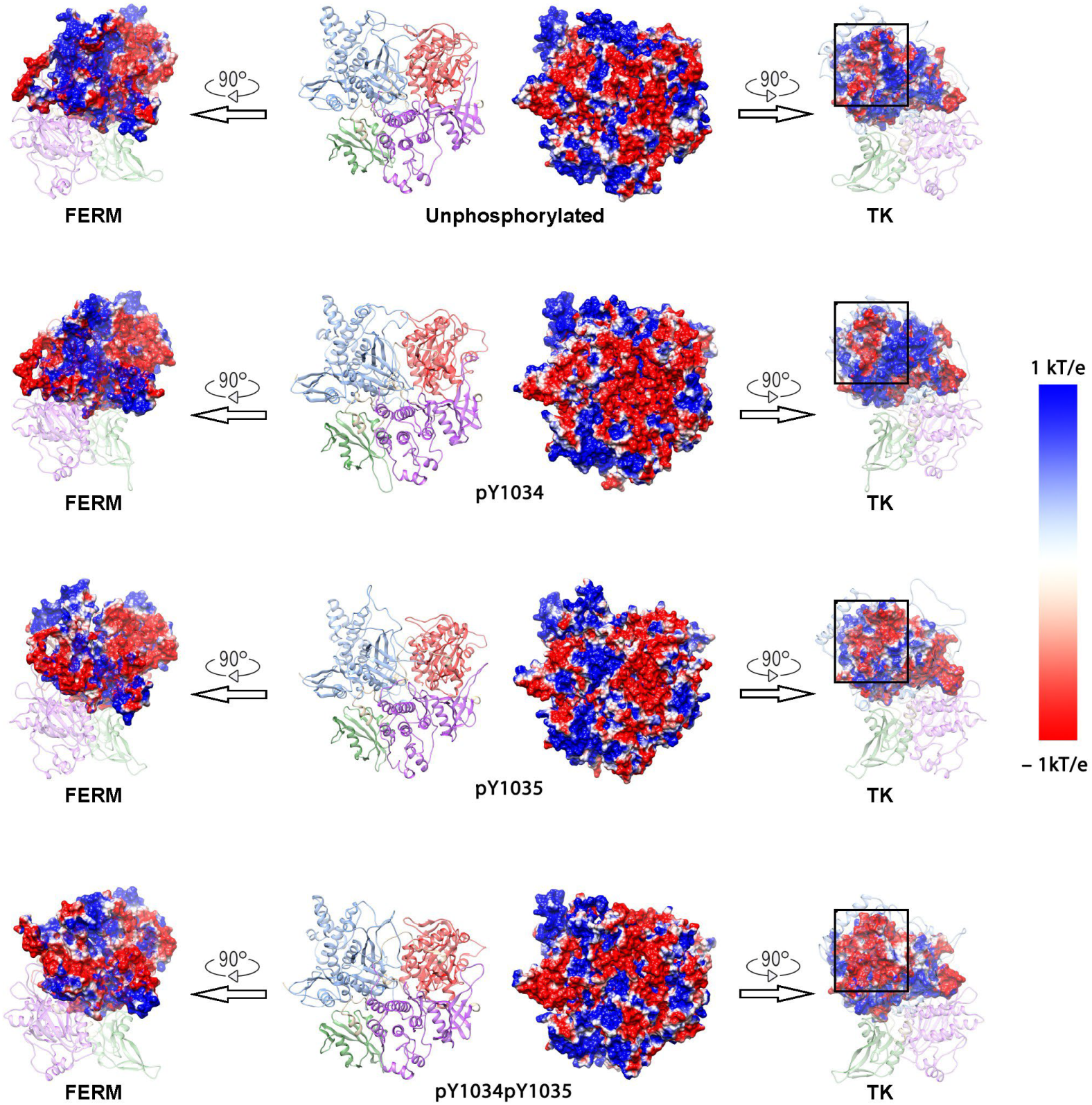
3.2. Surface Negative Charge of TK Domain Increased After Phosphorylation in Y1034/Y1035
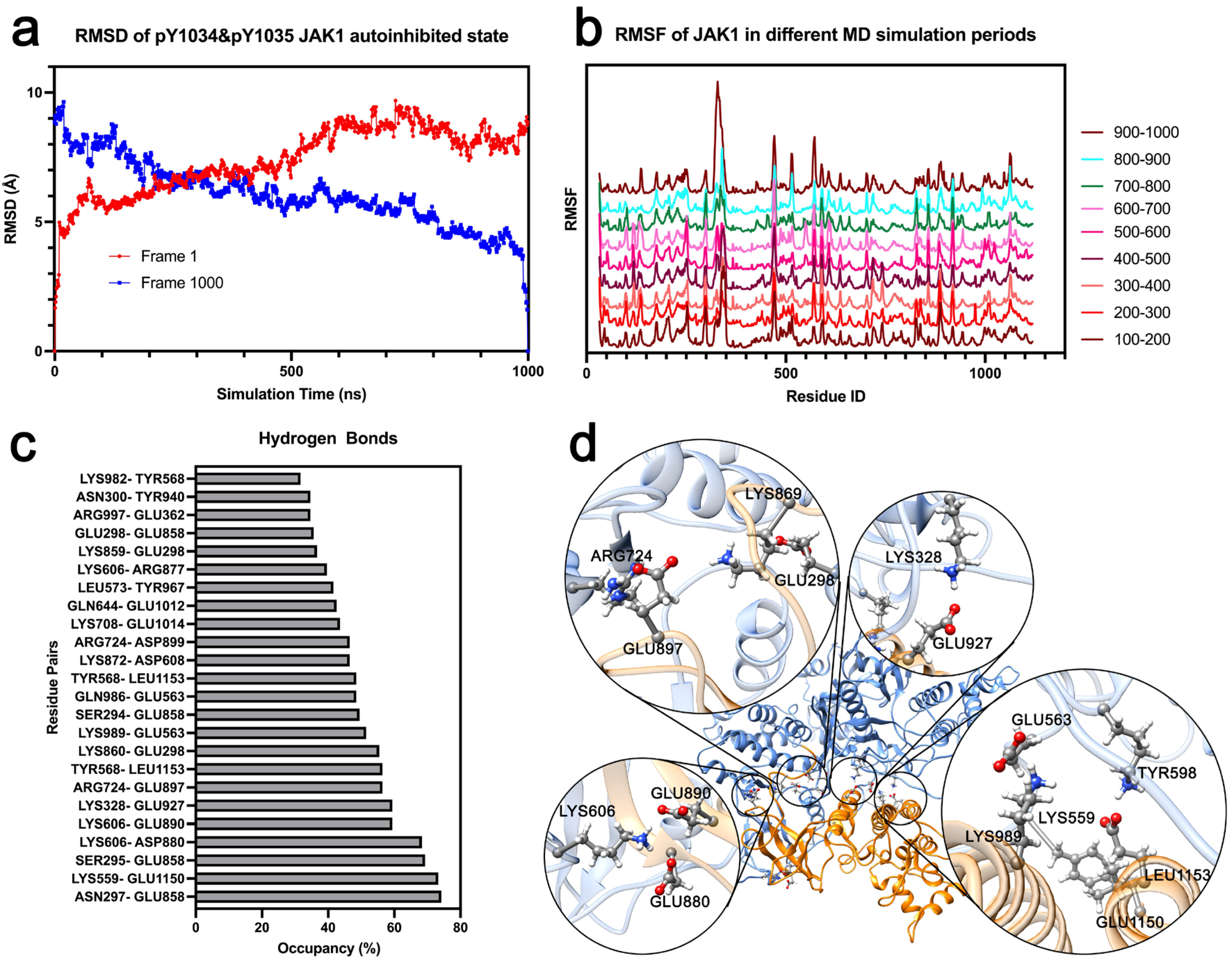
3.3. Phosphorylation in Y1034/Y1035 Leads to Weakening of Hydrogen Bonding Between TK and FERM
3.4. Phosphorylation in Y1034 and Y1035 Changed JAK1 Structural Stability in Partial Amino Residue
3.5. MD Simulation Lasting for 1.0 µs Further Provides the Stuck Activation of JAK1 from the Autoinhibited State
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bousoik, E.; Montazeri Aliabadi, H. “Do We Know Jack” About JAK? A Closer Look at JAK/STAT Signaling Pathway. Front. Oncol. 2018, 8, 287. [Google Scholar] [CrossRef]
- Stark, G.R.; Darnell, J.E. The JAK-STAT pathway at twenty. Immunity 2012, 36, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Schindler, C.; Levy, D.E.; Decker, T. JAK-STAT signaling: From interferons to cytokines. J. Biol. Chem. 2007, 282, 20059–20063. [Google Scholar] [CrossRef] [PubMed]
- Watowich, S.S.; Yoshimura, A.; Longmore, G.D.; Hilton, D.J.; Yoshimura, Y.; Lodish, H.F. Homodimerization and constitutive activation of the erythropoietin receptor. Proc. Natl. Acad. Sci. USA 1992, 89, 2140–2144. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, S.; Hafer, M.; Vuorio, J.; Tucker, J.A.; Winkelmann, H.; Löchte, S.; Stanly, T.A.; Pulgar Prieto, K.D.; Poojari, C.; Sharma, V.; et al. Mechanism of homodimeric cytokine receptor activation and dysregulation by oncogenic mutations. Science 2020, 367, 643–652. [Google Scholar] [CrossRef]
- Hubbard, S.R. Mechanistic Insights into Regulation of JAK2 Tyrosine Kinase. Front. Endocrinol. 2017, 8, 361. [Google Scholar] [CrossRef]
- Xin, P.; Xu, X.; Deng, C.; Liu, S.; Wang, Y.; Zhou, X.; Ma, H.; Wei, D.; Sun, S. The role of JAK/STAT signaling pathway and its inhibitors in diseases. Int. Immunopharmacol. 2020, 80, 106210. [Google Scholar] [CrossRef]
- Xue, C.; Yao, Q.; Gu, X.; Shi, Q.; Yuan, X.; Chu, Q.; Bao, Z.; Lu, J.; Li, L. Evolving cognition of the JAK-STAT signaling pathway: Autoimmune disorders and cancer. Signal Transduct. Target. Ther. 2023, 8, 204. [Google Scholar] [CrossRef]
- Raivola, J.; Haikarainen, T.; Abraham, B.G.; Silvennoinen, O. Janus Kinases in Leukemia. Cancers 2021, 13, 800. [Google Scholar] [CrossRef]
- Hammarén, H.M.; Ungureanu, D.; Grisouard, J.; Skoda, R.C.; Hubbard, S.R.; Silvennoinen, O. ATP binding to the pseudokinase domain of JAK2 is critical for pathogenic activation. Proc. Natl. Acad. Sci. USA 2015, 112, 4642–4647. [Google Scholar] [CrossRef]
- Hammarén, H.M.; Virtanen, A.T.; Raivola, J.; Silvennoinen, O. The regulation of JAKs in cytokine signaling and its breakdown in disease. Cytokine 2019, 118, 48–63. [Google Scholar] [CrossRef]
- Valentino, L.; Pierre, J. JAK/STAT signal transduction: Regulators and implication in hematological malignancies. Biochem. Pharmacol. 2006, 71, 713–721. [Google Scholar] [CrossRef]
- Saharinen, P.; Silvennoinen, O. The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J. Biol. Chem. 2002, 277, 47954–47963. [Google Scholar] [CrossRef] [PubMed]
- Saharinen, P.; Takaluoma, K.; Silvennoinen, O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol. Cell Biol. 2000, 20, 3387–3395. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lupardus, P.J.; Ultsch, M.; Wallweber, H.; Bir Kohli, P.; Johnson, A.R.; Eigenbrot, C. Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 8025–8030. [Google Scholar] [CrossRef]
- Meharena, H.S.; Chang, P.; Keshwani, M.M.; Oruganty, K.; Nene, A.K.; Kannan, N.; Taylor, S.S.; Kornev, A.P. Deciphering the structural basis of eukaryotic protein kinase regulation. PLoS Biol. 2013, 11, e1001680. [Google Scholar] [CrossRef]
- Nolen, B.; Taylor, S.; Ghosh, G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 2004, 15, 661–675. [Google Scholar] [CrossRef]
- Roskoski, R. Janus kinase (JAK) inhibitors in the treatment of neoplastic and inflammatory disorders. Pharmacol. Res. 2022, 183, 106362. [Google Scholar] [CrossRef]
- Liu, K.D.; Gaffen, S.L.; Goldsmith, M.A.; Greene, W.C. Janus kinases in interleukin-2-mediated signaling: JAK1 and JAK3 are differentially regulated by tyrosine phosphorylation. Curr. Biol. 1997, 7, 817–826. [Google Scholar] [CrossRef]
- Feng, J.; Witthuhn, B.A.; Matsuda, T.; Kohlhuber, F.; Kerr, I.M.; Ihle, J.N. Activation of Jak2 catalytic activity requires phosphorylation of Y1007 in the kinase activation loop. Mol. Cell. Biol. 1997, 17, 2497–2501. [Google Scholar] [CrossRef]
- Zhou, Y.J.; Hanson, E.P.; Chen, Y.Q.; Magnuson, K.; Chen, M.; Swann, P.G.; Wange, R.L.; Changelian, P.S.; O’Shea, J.J. Distinct tyrosine phosphorylation sites in JAK3 kinase domain positively and negatively regulate its enzymatic activity. Proc. Natl. Acad. Sci. USA 1997, 94, 13850–13855. [Google Scholar] [CrossRef] [PubMed]
- Gauzzi, M.C.; Velazquez, L.; McKendry, R.; Mogensen, K.E.; Fellous, M.; Pellegrini, S. Interferon-α-dependent activation of Tyk2 requires phosphorylation of positive regulatory tyrosines by another kinase. J. Biol. Chem. 1996, 271, 20494–20500. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, S.R.; Wei, L.; Hendrickson, W.A. Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature 1994, 372, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Hubbard, S.R.; Hendrickson, W.A.; Ellis, L. Expression, Characterization, and Crystallization of the Catalytic Core of the Human Insulin Receptor Protein-tyrosine Kinase Domain (∗). J. Biol. Chem. 1995, 270, 8122–8130. [Google Scholar] [CrossRef]
- Glassman, C.R.; Tsutsumi, N.; Saxton, R.A.; Lupardus, P.J.; Jude, K.M.; Garcia, K.C. Structure of a Janus kinase cytokine receptor complex reveals the basis for dimeric activation. Science 2022, 376, 163–169. [Google Scholar] [CrossRef]
- Lupardus, P.J.; Skiniotis, G.; Rice, A.J.; Thomas, C.; Fischer, S.; Walz, T.; Garcia, K.C. Structural snapshots of full-length Jak1, a transmembrane gp130/IL-6/IL-6Rα cytokine receptor complex, and the receptor-Jak1 holocomplex. Structure 2011, 19, 45–55. [Google Scholar] [CrossRef]
- Sun, S.; Rodriguez, G.; Xie, Y.; Guo, W.; Hernandez, A.E.L.; Sanchez, J.E.; Kirken, R.A.; Li, L. Phosphorylation of tyrosine 841 plays a significant role in JAK3 activation. Life 2023, 13, 981. [Google Scholar] [CrossRef]
- Sun, S.; Rodriguez, G.; Zhao, G.; Sanchez, J.E.; Guo, W.; Du, D.; Rodriguez Moncivais, O.J.; Hu, D.; Liu, J.; Kirken, R.A. A novel approach to study multi-domain motions in JAK1’s activation mechanism based on energy landscape. Brief. Bioinform. 2024, 25, bbae079. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Price, D.J.; Brooks, C.L., III. A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 2004, 121, 10096–10103. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [PubMed]
- York, D.M.; Darden, T.A.; Pedersen, L.G. The effect of long-range electrostatic interactions in simulations of macromolecular crystals: A comparison of the Ewald and truncated list methods. J. Chem. Phys. 1993, 99, 8345–8348. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Li, L.; Li, C.; Sarkar, S.; Zhang, J.; Witham, S.; Zhang, Z.; Wang, L.; Smith, N.; Petukh, M.; Alexov, E. DelPhi: A comprehensive suite for DelPhi software and associated resources. BMC Biophys. 2012, 5, 9. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef]
- Radtke, S.; Haan, S.; Jörissen, A.; Hermanns, H.M.; Diefenbach, S.; Smyczek, T.; Schmitz-Vandeleur, H.; Heinrich, P.C.; Behrmann, I.; Haan, C. The Jak1 SH2 domain does not fulfill a classical SH2 function in Jak/STAT signaling but plays a structural role for receptor interaction and up-regulation of receptor surface expression. J. Biol. Chem. 2005, 280, 25760–25768. [Google Scholar] [CrossRef]
- Yoshikawa, S.; Maeda, C.; Iizuka, A.; Ikeya, T.; Yamashita, K.; Ashizawa, T.; Kanematsu, A.; Miyata, H.; Kikuchi, Y.; Urakami, K. Characterization of the Neoantigen Profile in a Tumor Mutation Burden-high Melanoma Patient With Multiple Metastases. Cancer Genom. Proteom. 2025, 22, 496–509. [Google Scholar] [CrossRef]
- Wei, Q.; Xu, J. Chronic eosinophilic leukemia with a novel JAK1 mutation responds well to the JAK1/2 inhibitor ruxolitinib. Blood 2024, 144, 238. [Google Scholar] [CrossRef]
- Gordon, G.M.; Lambert, Q.T.; Daniel, K.G.; Reuther, G.W. Transforming JAK1 mutations exhibit differential signalling, FERM domain requirements and growth responses to interferon-γ. Biochem. J. 2010, 432, 255–265. [Google Scholar] [CrossRef]
- Hernandez, L.M.; Montersino, A.; Niu, J.; Guo, S.; Faezov, B.; Sanders, S.S.; Dunbrack, R.L.; Thomas, G.M. Palmitoylation-dependent control of JAK1 kinase signaling governs responses to neuropoietic cytokines and survival in DRG neurons. J. Biol. Chem. 2023, 299. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, X.; Liu, K.; Chen, G.; Sun, S. Molecular Dynamics Simulation Analysis of JAK1 Initial Activation: Phosphorylation-Induced Conformational Dynamics and Domain Interactions. Life 2025, 15, 1316. https://doi.org/10.3390/life15081316
Peng X, Liu K, Chen G, Sun S. Molecular Dynamics Simulation Analysis of JAK1 Initial Activation: Phosphorylation-Induced Conformational Dynamics and Domain Interactions. Life. 2025; 15(8):1316. https://doi.org/10.3390/life15081316
Chicago/Turabian StylePeng, Xinyu, Kefu Liu, Guodong Chen, and Shengjie Sun. 2025. "Molecular Dynamics Simulation Analysis of JAK1 Initial Activation: Phosphorylation-Induced Conformational Dynamics and Domain Interactions" Life 15, no. 8: 1316. https://doi.org/10.3390/life15081316
APA StylePeng, X., Liu, K., Chen, G., & Sun, S. (2025). Molecular Dynamics Simulation Analysis of JAK1 Initial Activation: Phosphorylation-Induced Conformational Dynamics and Domain Interactions. Life, 15(8), 1316. https://doi.org/10.3390/life15081316





