Genome-Wide Identification and Comprehensive Analysis of AP2/ERF Gene Family in Adiantum nelumboides Under Abiotic Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Search and Identification of AP2/ERF Gene Sequence in A. nelumboides
2.2. Phylogenetic Analysis of AnAP2/ERF Gene Family
2.3. Conserved Domain and Gene Structure Analysis, Cis-Element Analysis of AnAP2/ERF Gene in Adiantum nelumboides
2.4. Differential Expression Analysis of AP2/ERF Gene Family in A. nelumboides
2.5. Plant Material and Treatments
- (1)
- Drought Stress Treatment in A. nelumboides: Uniform seedlings with optimal growth were selected and divided into experimental and control groups. The control group received regular watering, while the experimental group was irrigated with 250 mL of 20% PEG6000 solution. Both groups were maintained under controlled conditions: 12 h light (140 lx; ~20 μmol·m−2·s−1)/12 h dark photoperiod, 70% relative humidity, and 25 °C for 48 h. Leaf samples were collected at 0, 6, 12, 24, and 48 h for total RNA extraction [37].
- (2)
- Flooding Stress Treatment in A. nelumboides: Uniform seedlings were divided into experimental and control groups. The control group received regular watering, whereas the experimental group was fully submerged in water. Both groups were incubated under identical conditions: 12 h light (140 lx)/12 h dark, 70% humidity, 25 °C for 48 h. Leaf samples were harvested at 0, 6, 12, 24, and 48 h for total RNA isolation [35].
- (3)
- Phytohormone Treatment in A. nelumboides: Uniform seedlings were assigned to experimental and control groups. The control group was watered normally, while experimental groups were foliar-sprayed with 100 nM IAA; 100 μM gibberellic acid (GA3); 100 μM ABA. All groups were kept under 12 h light (140 lx)/12 h dark, 70% humidity, 25 °C for 48 h. Leaf sampling occurred at 0, 6, 12, 24, and 48 h for total RNA extraction [38].
2.6. Total RNA Extraction and Quantitative PCR Analysis
2.7. Data Analysis
3. Results
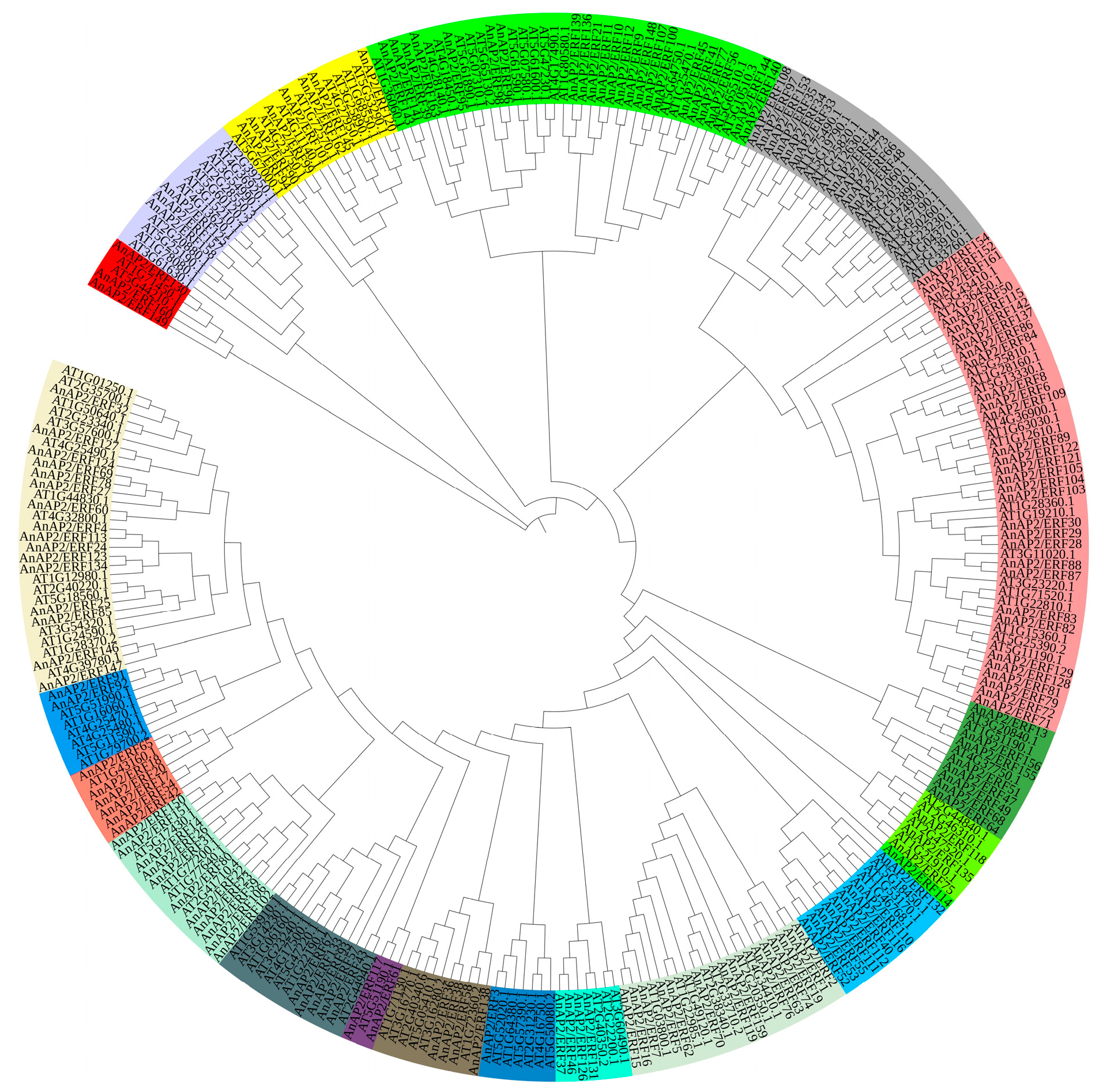
3.1. Identification of AnAP2/ERF Gene Family, Phylogenetic Analysis
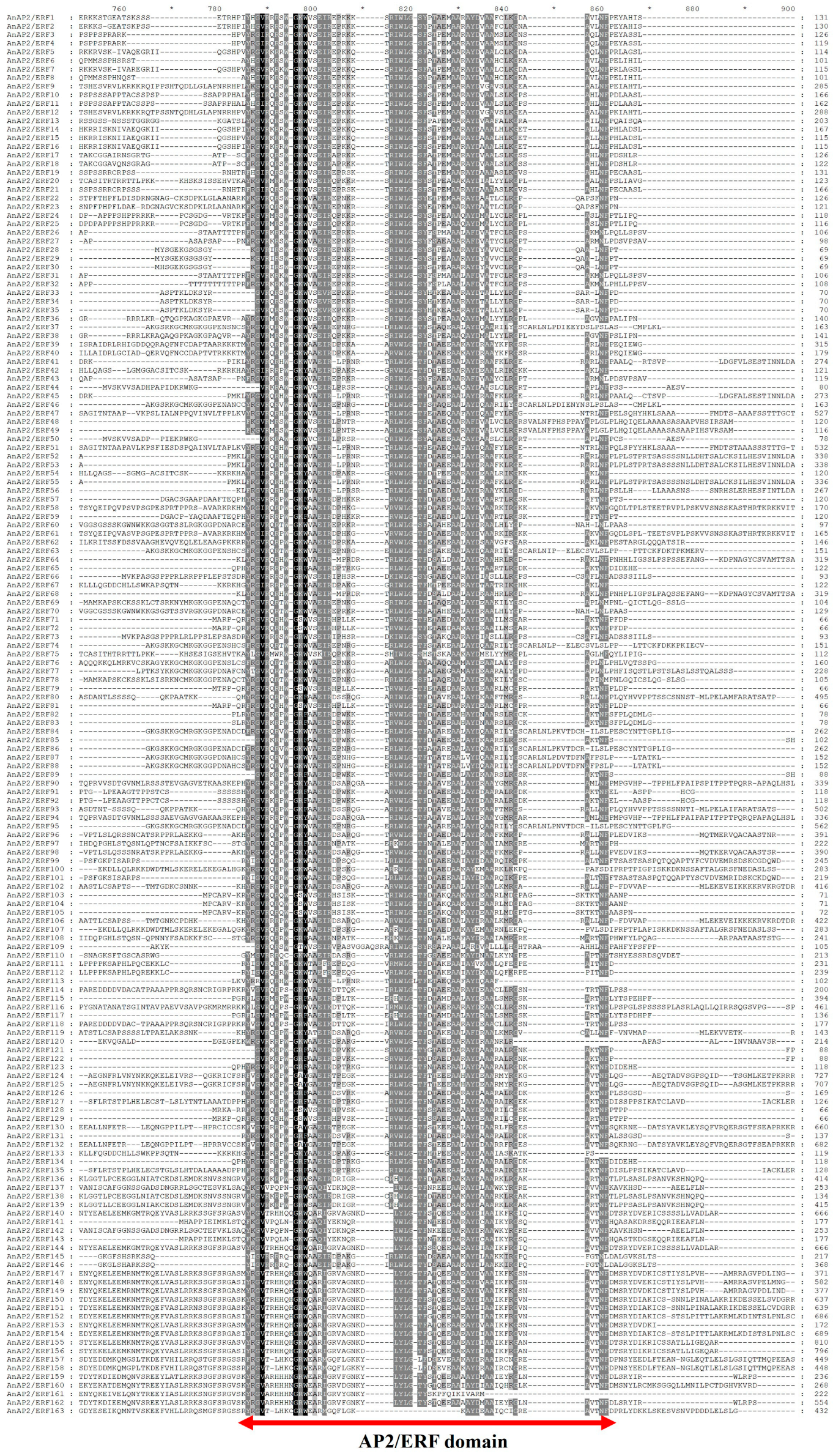
3.2. Conserved Domain and Gene Structure Analysis of AnAP2/ERFs
3.3. Analysis of Cis-Acting Elements of AnAP2/ERFs Family Gene Promoter
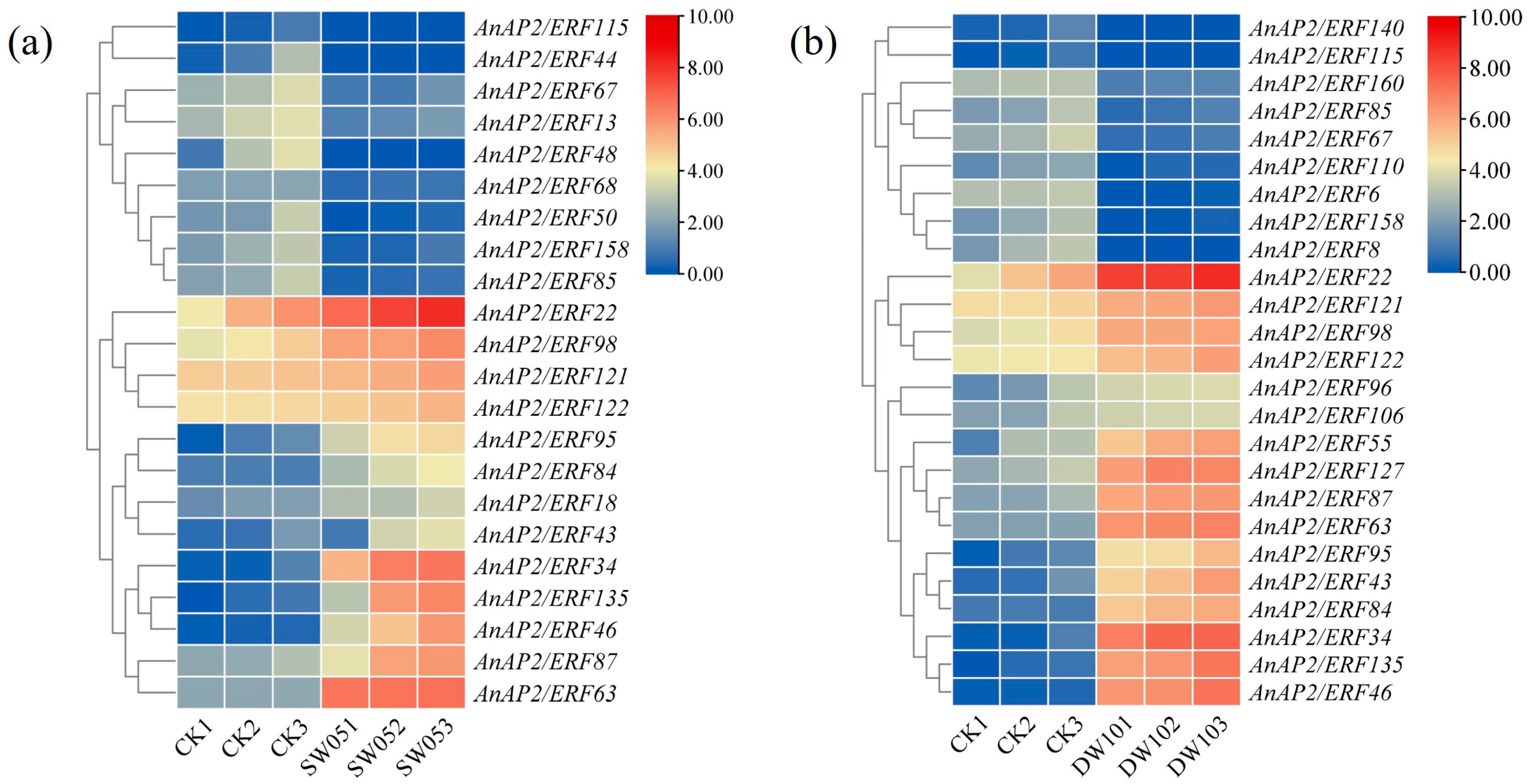
3.4. The Expression Profile of AnAP2/ERF Gene Under Drought and Flooding Stress
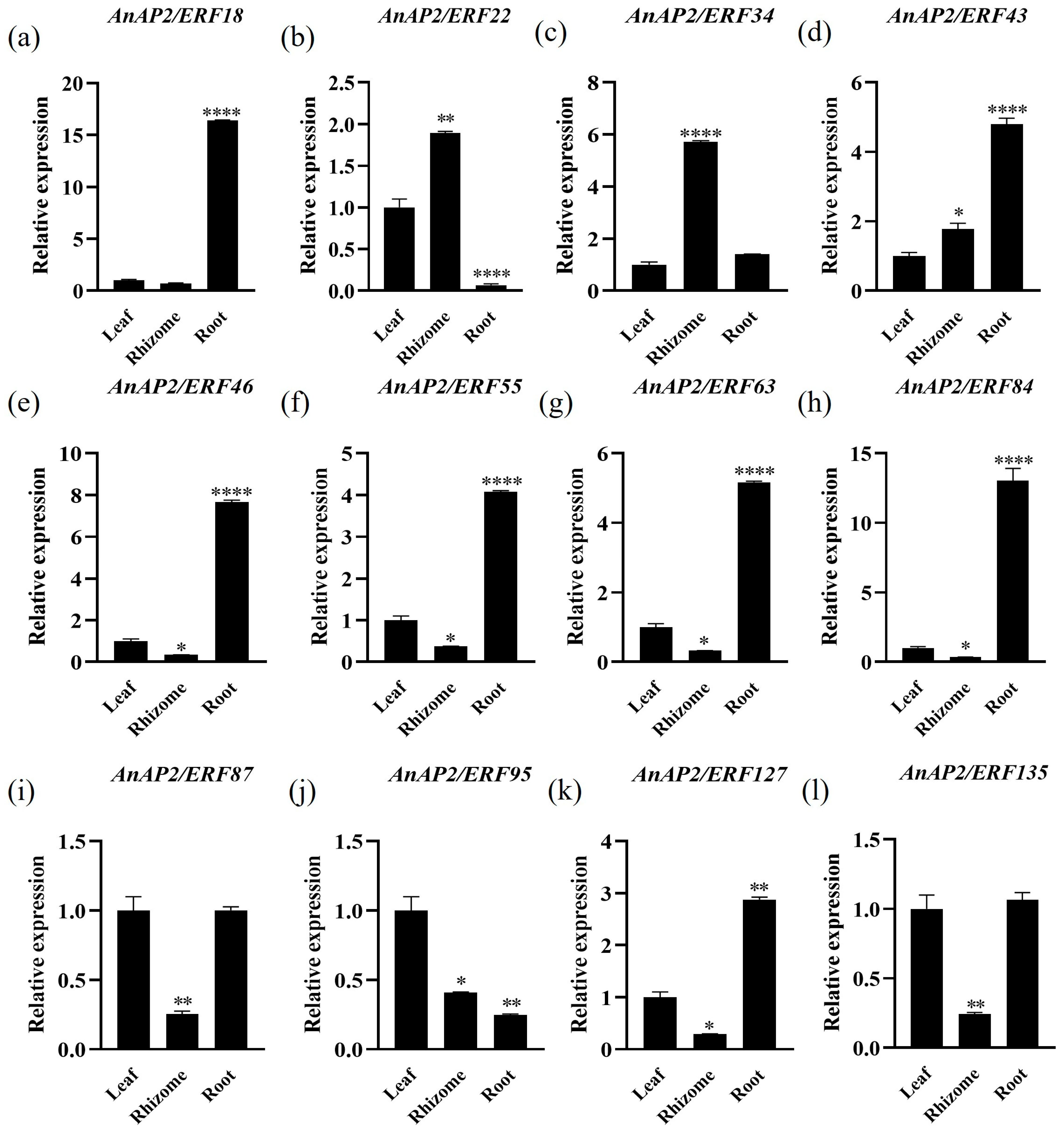
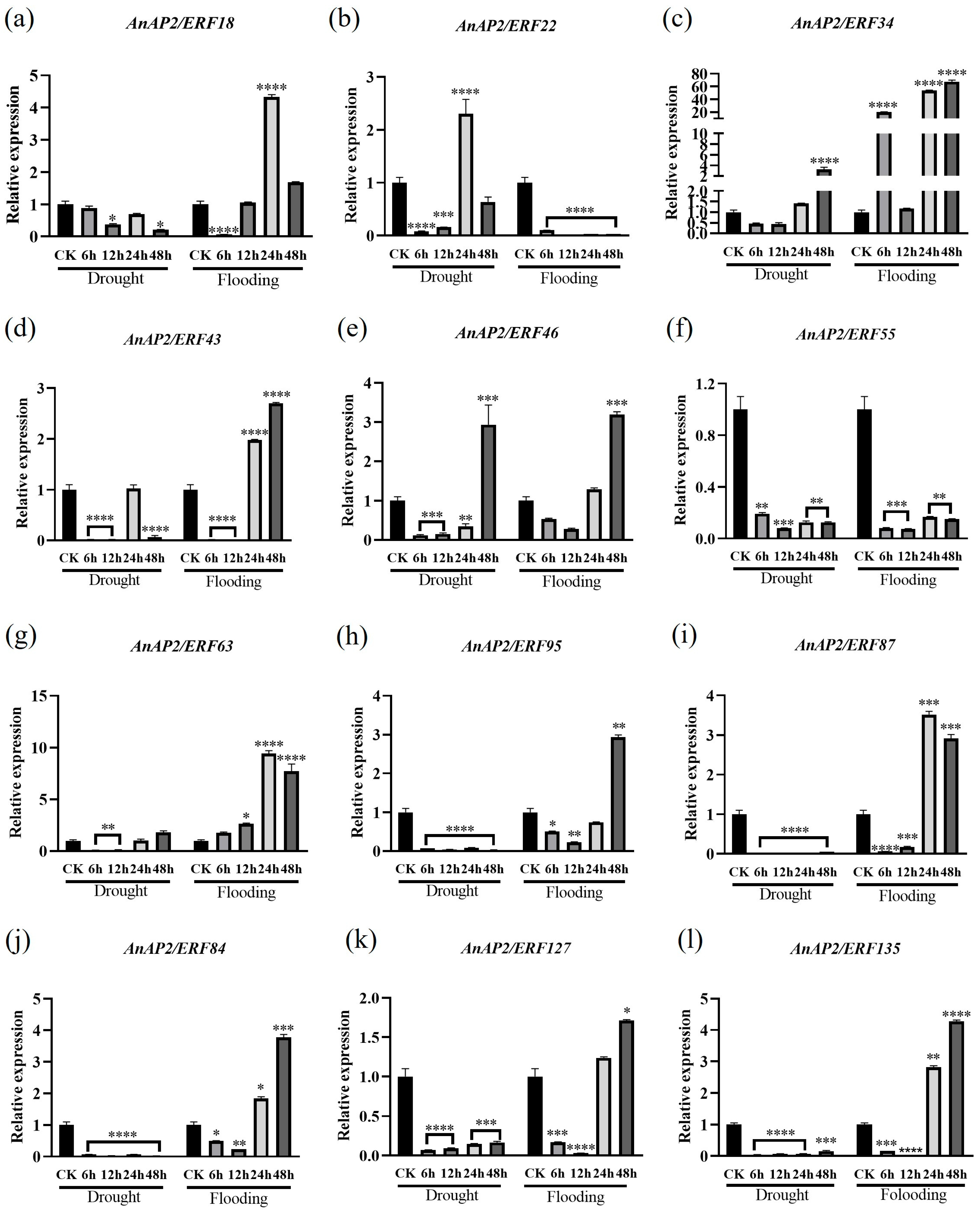
3.5. Expression Analysis of Selected AnAP2/ERFs After Drought and Flooding Stress
3.6. Expression Analysis of Screened Genes in Response to Hormones
3.7. Functional Diversification of AP2/ERF Transcription Factors: DNA-Binding Conservation and Stress-Adaptive Specialization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and Comparative Analysis of MYB and bHLH Plant Transcription Factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The Role of WRKY Transcription Factors in Plant Abiotic Stresses. Biochim. Biophys. Acta 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC Transcription Factors in Plant Abiotic Stress Responses. Biochim. Biophys. Acta 2012, 1819, 97–103. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.-L.; Xing, G.-M.; Liu, J.-X.; Duan, A.-Q.; Xu, Z.-S.; Li, M.-Y.; Zhuang, J.; Xiong, A.-S. Advances in AP2/ERF Super-Family Transcription Factors in Plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Tao, Q.; Niu, H.; Wang, Z.; Zhang, W.; Wang, H.; Wang, S.; Zhang, X.; Li, Z. Ethylene Responsive Factor ERF110 Mediates Ethylene-Regulated Transcription of a Sex Determination-Related Orthologous Gene in Two Cucumis Species. J. Exp. Bot. 2018, 69, 2953–2965. [Google Scholar] [CrossRef]
- Okamuro, J.K.; Caster, B.; Villarroel, R.; Van Montagu, M.; Jofuku, K.D. The AP2 Domain of APETALA2 Defines a Large New Family of DNA Binding Proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 1997, 94, 7076–7081. [Google Scholar] [CrossRef]
- Magnani, E.; Sjölander, K.; Hake, S. From Endonucleases to Transcription Factors: Evolution of the AP2 DNA Binding Domain in Plants. Plant Cell 2004, 16, 2265–2277. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Sharoni, A.M.; Nuruzzaman, M.; Satoh, K.; Shimizu, T.; Kondoh, H.; Sasaya, T.; Choi, I.-R.; Omura, T.; Kikuchi, S. Gene Structures, Classification and Expression Models of the AP2/EREBP Transcription Factor Family in Rice. Plant Cell Physiol. 2011, 52, 344–360. [Google Scholar] [CrossRef]
- Rashid, M.; Guangyuan, H.; Guangxiao, Y.; Hussain, J.; Xu, Y. AP2/ERF Transcription Factor in Rice: Genome-Wide Canvas and Syntenic Relationships between Monocots and Eudicots. Evol. Bioinform. Online 2012, 8, 321–355. [Google Scholar] [CrossRef]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) Transcription Factors: Mediators of Stress Responses and Developmental Programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K.; Agarwal, P.; Reddy, M.K.; Sopory, S.K. Role of DREB Transcription Factors in Abiotic and Biotic Stress Tolerance in Plants. Plant Cell Rep. 2006, 25, 1263–1274. [Google Scholar] [CrossRef]
- Sharma, M.K.; Kumar, R.; Solanke, A.U.; Sharma, R.; Tyagi, A.K.; Sharma, A.K. Identification, Phylogeny, and Transcript Profiling of ERF Family Genes during Development and Abiotic Stress Treatments in Tomato. Mol. Genet. Genom. 2010, 284, 455–475. [Google Scholar] [CrossRef]
- Hwang, J.E.; Lim, C.J.; Chen, H.; Je, J.; Song, C.; Lim, C.O. Overexpression of Arabidopsis Dehydration- Responsive Element-Binding Protein 2C Confers Tolerance to Oxidative Stress. Mol. Cells 2012, 33, 135–140. [Google Scholar] [CrossRef]
- Jin, Y.; Pan, W.; Zheng, X.; Cheng, X.; Liu, M.; Ma, H.; Ge, X. OsERF101, an ERF Family Transcription Factor, Regulates Drought Stress Response in Reproductive Tissues. Plant Mol. Biol. 2018, 98, 51–65. [Google Scholar] [CrossRef]
- Yu, Y.; Yu, M.; Zhang, S.; Song, T.; Zhang, M.; Zhou, H.; Wang, Y.; Xiang, J.; Zhang, X. Transcriptomic Identification of Wheat AP2/ERF Transcription Factors and Functional Characterization of TaERF-6-3A in Response to Drought and Salinity Stresses. Int. J. Mol. Sci. 2022, 23, 3272. [Google Scholar] [CrossRef]
- Kumar, V.; Kumar, A.; Tewari, K.; Garg, N.K.; Changan, S.S.; Tyagi, A. Isolation and Characterization of Drought and ABA Responsive Promoter of a Transcription Factor Encoding Gene from Rice. Physiol. Mol. Biol. Plants 2022, 28, 1813–1831. [Google Scholar] [CrossRef]
- Kong, L.; Song, Q.; Wei, H.; Wang, Y.; Lin, M.; Sun, K.; Zhang, Y.; Yang, J.; Li, C.; Luo, K. The AP2/ERF Transcription Factor PtoERF15 Confers Drought Tolerance via JA-Mediated Signaling in Populus. New Phytol. 2023, 240, 1848–1867. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nolan, T.; Jiang, H.; Tang, B.; Zhang, M.; Li, Z.; Yin, Y. The AP2/ERF Transcription Factor TINY Modulates Brassinosteroid-Regulated Plant Growth and Drought Responses in Arabidopsis. Plant Cell 2019, 31, 1788–1806. [Google Scholar] [CrossRef]
- Fukao, T.; Xu, K.; Ronald, P.C.; Bailey-Serres, J. A Variable Cluster of Ethylene Response Factor-like Genes Regulates Metabolic and Developmental Acclimation Responses to Submergence in Rice. Plant Cell 2006, 18, 2021–2034. [Google Scholar] [CrossRef] [PubMed]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.-J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The Ethylene Response Factors SNORKEL1 and SNORKEL2 Allow Rice to Adapt to Deep Water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef]
- Li, J.; Guo, X.; Zhang, M.; Wang, X.; Zhao, Y.; Yin, Z.; Zhang, Z.; Wang, Y.; Xiong, H.; Zhang, H.; et al. OsERF71 Confers Drought Tolerance via Modulating ABA Signaling and Proline Biosynthesis. Plant Sci. 2018, 270, 131–139. [Google Scholar] [CrossRef]
- Fu, J.; Zhu, C.; Wang, C.; Liu, L.; Shen, Q.; Xu, D.; Wang, Q. Maize Transcription Factor ZmEREB20 Enhanced Salt Tolerance in Transgenic Arabidopsis. Plant Physiol. Biochem. 2021, 159, 257–267. [Google Scholar] [CrossRef]
- Ma, Z.; Jin, Y.-M.; Wu, T.; Hu, L.; Zhang, Y.; Jiang, W.; Du, X. OsDREB2B, an AP2/ERF Transcription Factor, Negatively Regulates Plant Height by Conferring GA Metabolism in Rice. Front. Plant Sci. 2022, 13, 1007811. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, L.; Hu, H.; Tang, N.; Shi, L.; Xu, F.; Wang, S. Arabidopsis ERF012 Is a Versatile Regulator of Plant Growth, Development and Abiotic Stress Responses. Int. J. Mol. Sci. 2022, 23, 6841. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, O.; Piqueras, R.; Sánchez-Serrano, J.J.; Solano, R. ETHYLENE RESPONSE FACTOR1 Integrates Signals from Ethylene and Jasmonate Pathways in Plant Defense. Plant Cell 2003, 15, 165–178. [Google Scholar] [CrossRef]
- Tan, X.-L.; Fan, Z.-Q.; Shan, W.; Yin, X.-R.; Kuang, J.-F.; Lu, W.-J.; Chen, J.-Y. Association of BrERF72 with Methyl Jasmonate-Induced Leaf Senescence of Chinese Flowering Cabbage through Activating JA Biosynthesis-Related Genes. Hortic. Res. 2018, 5, 22. [Google Scholar] [CrossRef] [PubMed]
- PPG, I. A Community-Derived Classification for Extant Lycophytes and Ferns. J. Syst. Evol. 2016, 54, 563–603. [Google Scholar] [CrossRef]
- Liu, X.-Q.; Gituru, R.W.; Chen, L.-Q. Genetic Variation in the Endangered Fern Adiantum reniforme Var. Sinense (Adiantaceae) in China. Ann. Bot. Fenn. 2007, 44, 25–32. [Google Scholar]
- Liao, J.X.; Jiang, M.X.; Huang, H.D. Effects of Soil Moisture on Ecophysiological Characteristics of Adiantum Reniforme Var. Sinensis, an Endangered Fern Endemic to the Three Gorges Region in China. Am. Fern J. 2008, 98, 26–32. [Google Scholar] [CrossRef]
- Sun, W.; Shu, J.; Gu, Y.; Morigengaowa; Du, X.; Liu, B.; Yan, Y. (PDF) Conservation Genomics Analysis Revealed the Endangered Mechanism of Adiantum nelumboides. Biodiv. Sci. 2022, 30, 21508. [Google Scholar] [CrossRef]
- Yu, S.; Wang, B.; Sheng, L.; Li, Q.; Zuo, Y.; Wang, S.; Huang, X.; Zhao, B.; Deng, H.; Gu, F.; et al. Habitat Characteristics and Factors Threatening the Rare and Endangered Fern Adiantum nelumboides. Nord. J. Bot. 2022, 2022, e03492. [Google Scholar] [CrossRef]
- Wang, A.-H.; Sun, Y.; Schneider, H.; Zhai, J.-W.; Liu, D.-M.; Zhou, J.-S.; Xing, F.-W.; Chen, H.-F.; Wang, F.-G. Identification of the Relationship between Chinese Adiantum Reniforme Var. Sinense and Canary Adiantum Reniforme. BMC Plant Biol. 2015, 15, 36. [Google Scholar] [CrossRef][Green Version]
- Wu, D.; Li, L.; Ma, X.; Huang, G.; Yang, C. Morphological and Anatomical Adaptations to Dry, Shady Environments in Adiantum reniforme Var. Sinense (Pteridaceae). PeerJ 2020, 8, e9937. [Google Scholar][Green Version]
- Liang, Q.; Dun, B.; Li, L.; Ma, X.; Zhang, H.; Su, Y.; Wu, D. Metabolomic and Transcriptomic Responses of Adiantum (Adiantum nelumboides) Leaves under Drought, Half-Waterlogging, and Rewater Conditions. Front. Genet. 2023, 14, 1113470. [Google Scholar] [CrossRef]
- Zhong, Y.; Liu, Y.; Wu, W.; Chen, J.; Sun, C.; Liu, H.; Shu, J.; Ebihara, A.; Yan, Y.; Zhou, R.; et al. Genomic Insights into Genetic Diploidization in the Homosporous Fern Adiantum nelumboides. Genome Biol. Evol. 2022, 14, evac127. [Google Scholar] [CrossRef] [PubMed]
- Jian, H.; Sun, H.; Liu, R.; Zhang, W.; Shang, L.; Wang, J.; Khassanov, V.; Lyu, D. Construction of Drought Stress Regulation Networks in Potato Based on SMRT and RNA Sequencing Data. BMC Plant Biol. 2022, 22, 381. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, J.; Ji, F.; Cao, X.; Zhao, Q.; Cheng, C.; Ma, N.; Zhou, X.; Zhang, Z. Transcriptomic Profiling of Rose Flower under Treatment of Various Phytohormones and Plant Growth Regulators. Sci. Data 2022, 9, 669. [Google Scholar] [CrossRef]
- Yang, S.; Tang, X.-F.; Ma, N.-N.; Wang, L.-Y.; Meng, Q.-W. Heterology Expression of the Sweet Pepper CBF3 Gene Confers Elevated Tolerance to Chilling Stress in Transgenic Tobacco. J. Plant Physiol. 2011, 168, 1804–1812. [Google Scholar] [CrossRef]
- Lee, D.-K.; Jung, H.; Jang, G.; Jeong, J.S.; Kim, Y.S.; Ha, S.-H.; Do Choi, Y.; Kim, J.-K. Overexpression of the OsERF71 Transcription Factor Alters Rice Root Structure and Drought Resistance. Plant Physiol. 2016, 172, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-Binding Specificity of the ERF/AP2 Domain of Arabidopsis DREBs, Transcription Factors Involved in Dehydration- and Cold-Inducible Gene Expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Lee, M.; Jung, J.-H.; Han, D.-Y.; Seo, P.J.; Park, W.J.; Park, C.-M. Activation of a Flavin Monooxygenase Gene YUCCA7 Enhances Drought Resistance in Arabidopsis. Planta 2012, 235, 923–938. [Google Scholar] [CrossRef]
- Kim, J.I.; Baek, D.; Park, H.C.; Chun, H.J.; Oh, D.-H.; Lee, M.K.; Cha, J.-Y.; Kim, W.-Y.; Kim, M.C.; Chung, W.S.; et al. Overexpression of Arabidopsis YUCCA6 in Potato Results in High-Auxin Developmental Phenotypes and Enhanced Resistance to Water Deficit. Mol. Plant 2013, 6, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Kim, H.J.; Cho, H.S.; Jung, H.W.; Cha, J.-Y.; Yun, D.-J.; Oh, S.-W.; Chung, Y.-S. Overexpression of AtYUCCA6 in Soybean Crop Results in Reduced ROS Production and Increased Drought Tolerance. Plant Biotechnol. Rep. 2019, 13, 161–168. [Google Scholar] [CrossRef]
- Shi, H.; Chen, L.; Ye, T.; Liu, X.; Ding, K.; Chan, Z. Modulation of Auxin Content in Arabidopsis Confers Improved Drought Stress Resistance. Plant Physiol. Biochem. 2014, 82, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Yuan, S.; Xie, B.; Li, Q.; Wang, Q.; Shao, M. IAA Plays an Important Role in Alkaline Stress Tolerance by Modulating Root Development and ROS Detoxifying Systems in Rice Plants. Int. J. Mol. Sci. 2022, 23, 14817. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Gao, X.; Liu, Y.; Fu, B. Genome-Wide Identification and Expression Pattern Analysis of the Aux/IAA (Auxin/Indole-3-Acetic Acid) Gene Family in Alfalfa (Medicago sativa) and the Potential Functions under Drought Stress. BMC Genom. 2024, 25, 382. [Google Scholar] [CrossRef]
- Gao, Y.; Yang, J.; Duan, W.; Ma, X.; Qu, L.; Xu, Z.; Yang, Y.; Xu, J. NtRAV4 Negatively Regulates Drought Tolerance in Nicotiana Tabacum by Enhancing Antioxidant Capacity and Defence System. Plant Cell Rep. 2022, 41, 1775–1788. [Google Scholar] [CrossRef] [PubMed]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The Role of Gibberellin Signalling in Plant Responses to Abiotic Stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef]
- Wu, W.; Zhu, L.; Wang, P.; Liao, Y.; Duan, L.; Lin, K.; Chen, X.; Li, L.; Xu, J.; Hu, H.; et al. Transcriptome-Based Construction of the Gibberellin Metabolism and Signaling Pathways in Eucalyptus grandis × E. urophylla, and Functional Characterization of GA20ox and GA2ox in Regulating Plant Development and Abiotic Stress Adaptations. Int. J. Mol. Sci. 2023, 24, 7051. [Google Scholar] [CrossRef]
- Yang, F.; Lv, G. Combined Analysis of Transcriptome and Metabolome Reveals the Molecular Mechanism and Candidate Genes of Haloxylon Drought Tolerance. Front. Plant Sci. 2022, 13, 1020367. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, X.; Cai, K.; Zhang, Q.; Jiang, L.; Li, H.; Lv, Y.; Qu, G.; Zhao, X. Comparative Transcriptomic and Metabolic Analyses Reveal the Coordinated Mechanisms in Pinus Koraiensis under Different Light Stress Conditions. Int. J. Mol. Sci. 2022, 23, 9556. [Google Scholar] [CrossRef]
- Cui, F.; Brosché, M.; Lehtonen, M.T.; Amiryousefi, A.; Xu, E.; Punkkinen, M.; Valkonen, J.P.T.; Fujii, H.; Overmyer, K. Dissecting Abscisic Acid Signaling Pathways Involved in Cuticle Formation. Mol. Plant 2016, 9, 926–938. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Zhang, J.; Sun, J.; Zhang, Y.; Lu, M.; Xin, X.; Hu, J. Genome-Wide Characterization of Protein Phosphatase 2C Genes in Populus Euphratica and Their Expression Profiling under Multiple Abiotic Stresses. Tree Genet. Genomes 2018, 14, 80. [Google Scholar] [CrossRef]
- Yoo, M.-J.; Ma, T.; Zhu, N.; Liu, L.; Harmon, A.C.; Wang, Q.; Chen, S. Genome-Wide Identification and Homeolog-Specific Expression Analysis of the SnRK2 Genes in Brassica Napus Guard Cells. Plant Mol. Biol. 2016, 91, 211–227. [Google Scholar] [CrossRef]
- Vanjildorj, E.; Bae, T.-W.; Riu, K.-Z.; Yun, P.-Y.; Park, S.-Y.; Lee, C.-H.; Kim, S.-Y.; Lee, H.-Y. Transgenic Agrostis mongolica Roshev. with Enhanced Tolerance to Drought and Heat Stresses Obtained from Agrobacterium-Mediated Transformation. Plant Cell Tissue Organ Cult. 2006, 87, 109–120. [Google Scholar] [CrossRef]
- Zhu, Q.; Zhang, J.; Gao, X.; Tong, J.; Xiao, L.; Li, W.; Zhang, H. The Arabidopsis AP2/ERF Transcription Factor RAP2.6 Participates in ABA, Salt and Osmotic Stress Responses. Gene 2010, 457, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Ma, J.; Guo, F.; Qi, D.; Zhao, M.; Zhang, C.; Wang, L.; Song, B.; Liu, S.; He, S.; et al. The AP2/ERF GmERF113 Positively Regulates the Drought Response by Activating GmPR10-1 in Soybean. Int. J. Mol. Sci. 2022, 23, 8159. [Google Scholar] [CrossRef] [PubMed]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF Family Transcription Factors in Plant Abiotic Stress Responses. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Halkier, B.A.; Gershenzon, J. Biology and Biochemistry of Glucosinolates. Annu. Rev. Plant Biol. 2006, 57, 303–333. [Google Scholar] [CrossRef]
- Todaka, D.; Takahashi, F.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Chapter Three-ABA-Responsive Gene Expression in Response to Drought Stress: Cellular Regulation and Long-Distance Signaling. In Advances in Botanical Research; Seo, M., Marion-Poll, A., Eds.; Abscisic Acid in Plants; Academic Press: Cambridge, MA, USA, 2019; Volume 92, pp. 83–113. [Google Scholar]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-Dependent and ABA-Independent Signaling in Response to Osmotic Stress in Plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Ulate, B.; Vincenti, E.; Powell, A.L.T.; Cantu, D. Tomato Transcriptome and Mutant Analyses Suggest a Role for Plant Stress Hormones in the Interaction between Fruit and Botrytis Cinerea. Front. Plant Sci. 2013, 4, 142. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Gasch, P.; Fundinger, M.; Müller, J.T.; Lee, T.; Bailey-Serres, J.; Mustroph, A. Redundant ERF-VII Transcription Factors Bind to an Evolutionarily Conserved Cis-Motif to Regulate Hypoxia-Responsive Gene Expression in Arabidopsis. Plant Cell 2016, 28, 160–180. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, D.; Zhang, T.; Li, L.; Liang, Q.; Wang, J.; Xiao, Z.; Xiang, G.; Zhang, H.; Liu, J.; Huang, G. Genome-Wide Identification and Comprehensive Analysis of AP2/ERF Gene Family in Adiantum nelumboides Under Abiotic Stress. Life 2025, 15, 1269. https://doi.org/10.3390/life15081269
Wu D, Zhang T, Li L, Liang Q, Wang J, Xiao Z, Xiang G, Zhang H, Liu J, Huang G. Genome-Wide Identification and Comprehensive Analysis of AP2/ERF Gene Family in Adiantum nelumboides Under Abiotic Stress. Life. 2025; 15(8):1269. https://doi.org/10.3390/life15081269
Chicago/Turabian StyleWu, Di, Tonghua Zhang, Linbao Li, Qianyan Liang, Junchen Wang, Zhiqiang Xiao, Ganju Xiang, Haibo Zhang, Jihong Liu, and Guiyun Huang. 2025. "Genome-Wide Identification and Comprehensive Analysis of AP2/ERF Gene Family in Adiantum nelumboides Under Abiotic Stress" Life 15, no. 8: 1269. https://doi.org/10.3390/life15081269
APA StyleWu, D., Zhang, T., Li, L., Liang, Q., Wang, J., Xiao, Z., Xiang, G., Zhang, H., Liu, J., & Huang, G. (2025). Genome-Wide Identification and Comprehensive Analysis of AP2/ERF Gene Family in Adiantum nelumboides Under Abiotic Stress. Life, 15(8), 1269. https://doi.org/10.3390/life15081269




