DNA Fingerprint Profile of Zizania spp. Plant, Monitoring Its Leaves with Screening of Their Biological Activity: Antimicrobial, Antioxidant and Cytotoxicity
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material Collection and Identification
2.2. Genomic DNA Extraction and Quality Assessment
2.2.1. PCR Amplification and Genetic Fingerprinting
- 2.5 µL 10× DreamTaq™ buffer (Thermo Scientific, USA)
- 2.0 mM MgCl2
- 0.2 mM dNTP mix (Thermo Scientific)
- 0.4 µM primer
- 1 U DreamTaq™ DNA polymerase
- 40 ng of template DNA
- Nuclease-free water to volume
- Initial denaturation: 94 °C for 4 min
- 35 cycles of: Denaturation: 94 °C for 30 s, Annealing: 36–48 °C (primer-dependent) for 45 s, Extension: 72 °C for 90 s, Final extension: 72 °C for 7 min
2.2.2. Gel Electrophoresis and Band Scoring
2.3. Phytochemical Extraction and Analysis
2.3.1. Leaf Extraction
2.3.2. Phytochemical Screening Procedures
- Flavonoids–detected via alkaline reagent test (yellow coloration turns colorless upon acid addition).
- Tannins–tested with ferric chloride (appearance of greenish-black precipitate).
- Terpenoids–Salkowski test using chloroform and concentrated sulfuric acid (reddish-brown interface);
- Saponins–frothing test (stable foam greater than 1 cm after 10 min shaking).
- Alkaloids––tested using Mayer’s reagent (cream-colored precipitate).
- Glycosides–Keller–Killiani test (reddish-brown ring at interface).
2.4. GC-MS and HPLC Analysis
Sample Preparation
2.5. HPLC Conditions
2.6. Biological Activity Assays
Antioxidant Activity (DPPH Method)
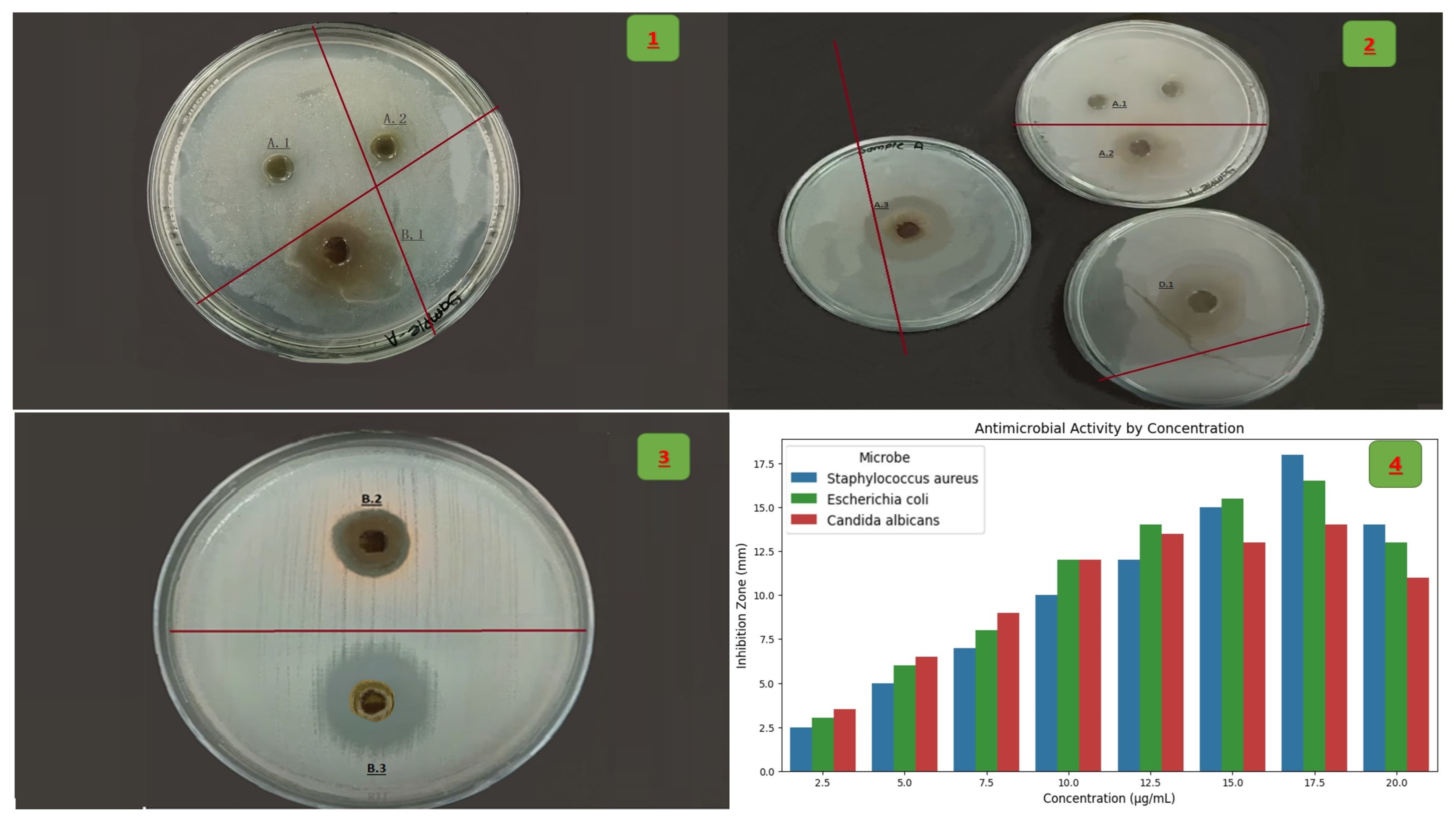
2.7. Antimicrobial Assay
- Staphylococcus aureus (S. aureus) (ATCC 25923), Escherichia coli (E. coli) (ATCC 25922), C. albicans (ATCC 10231)
Minimum Inhibitory Concentration (MIC)
2.8. Cell Line and Culture Conditions
2.8.1. MTT Assay Procedure
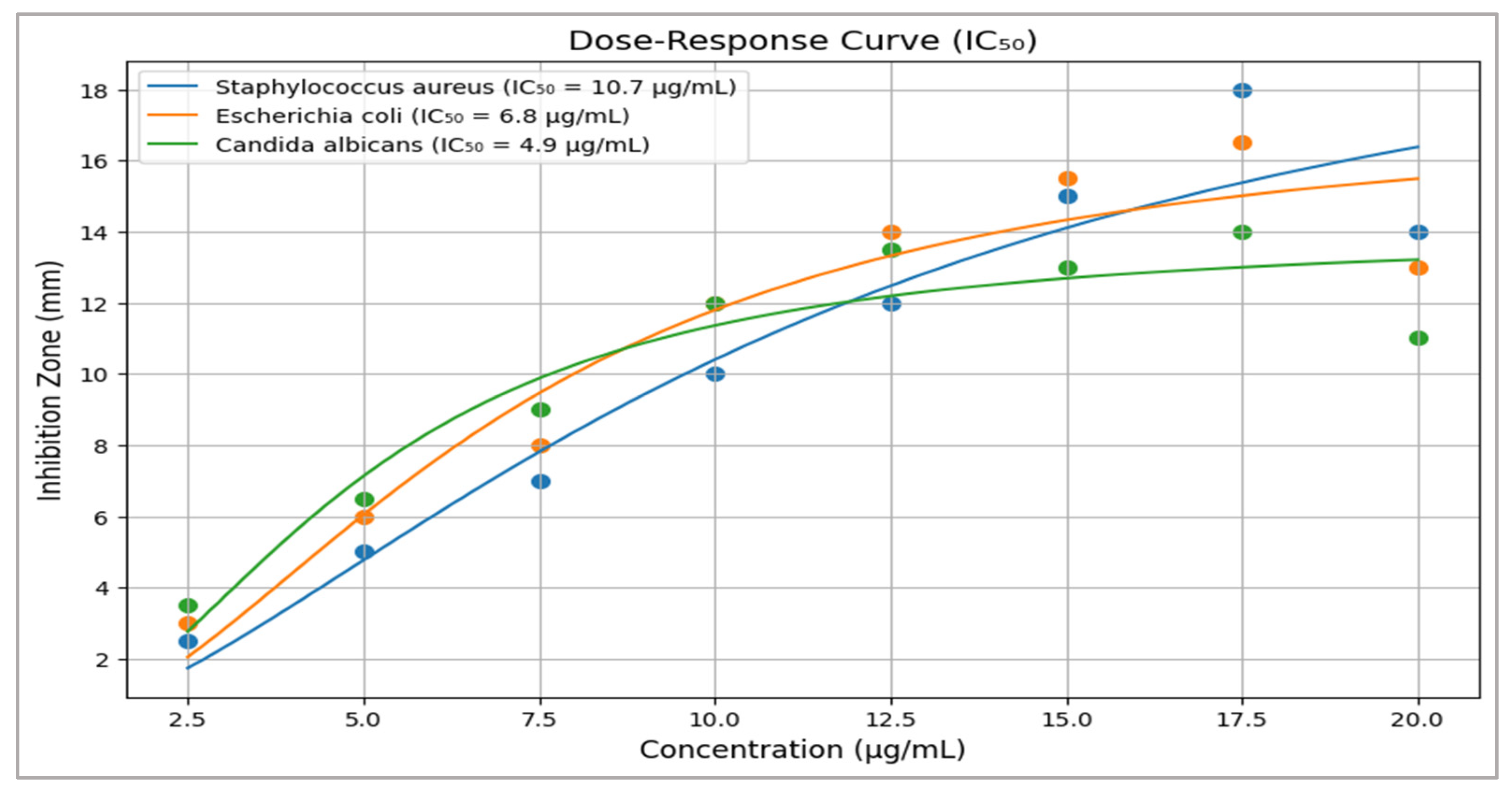
2.8.2. IC50 Calculation
2.9. Statistical Analysis
2.9.1. Genetic Data Analysis
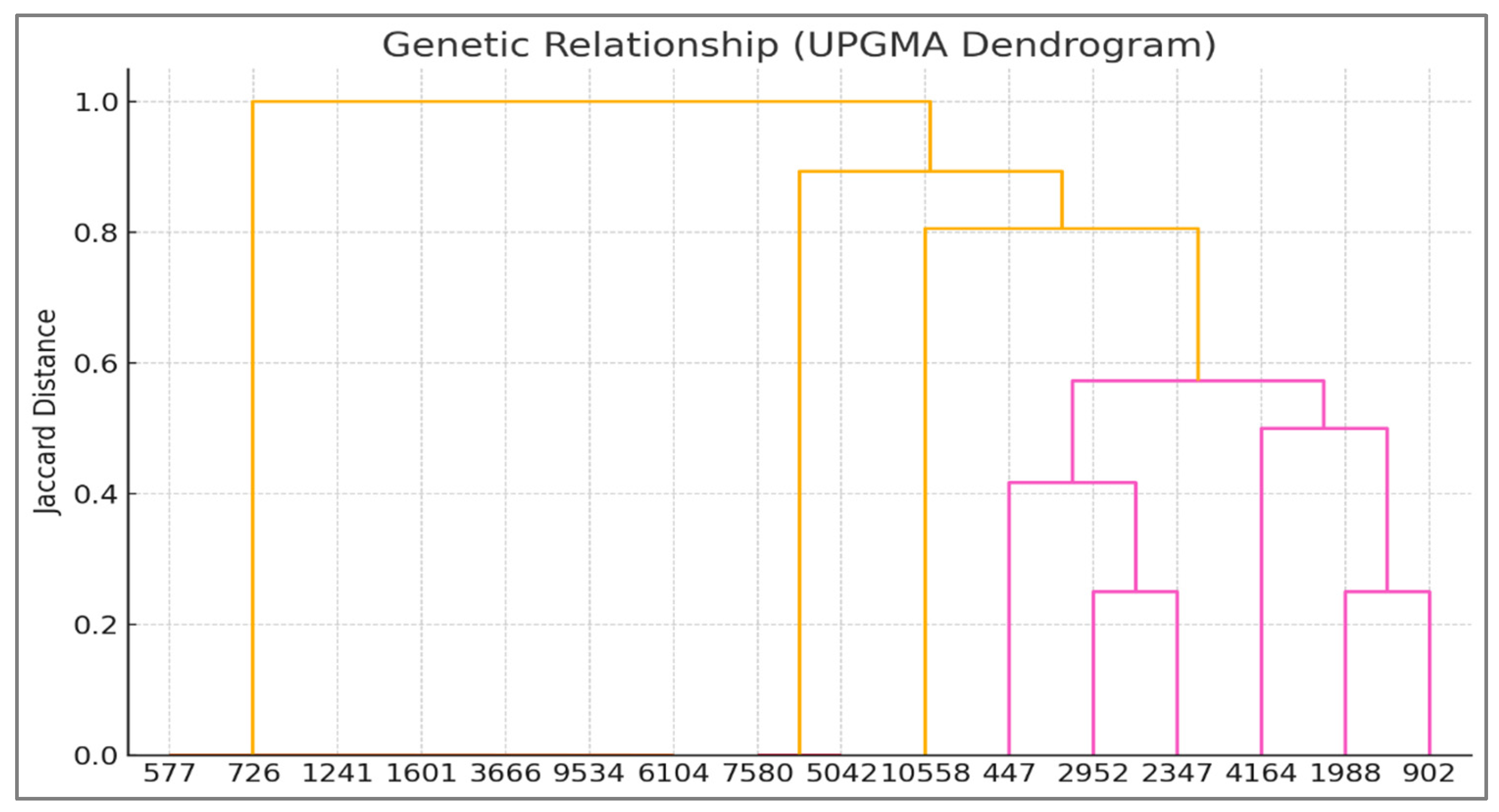
- UPGMA Dendrogram: Hierarchical clustering was performed using the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) based on Jaccard genetic distances derived from ISSR/RAPD band presence/absence data. Bootstrap values (577–10,558 replicates) assessed node robustness. This analysis grouped Z. texana (L8–L9) and unknowns (L10–L11) into a cohesive clade (Jaccard distance = 0.2), distinct from Z. latifolia (distance = 0.4) [24].
- Principal Component Analysis (PCA): Genetic divergence was visualized along two principal axes (PC1: 48% variance; PC2: 32%) using covariance matrices of the banding patterns. Unknowns clustered with Z. texana along PC1, confirming conspecificity [25].
- Jaccard Similarity Coefficient: Pairwise genetic similarity (85%) between Z. texana and unknowns was calculated from shared ISSR/RAPD markers (e.g., 750 bp and 1500 bp) [26].
- K-means Clustering: Multivariate analysis grouped specimens into clusters based on banding profiles, aligning unknowns with Z. texana along PC1 [27].
- Binary Presence/Absence Matrix: ISSR/RAPD bands were coded as binary traits (1 = present, 0 = absent) to identify diagnostic markers. Analyses were conducted using Arlequin v3.5 [28].
2.9.2. Phytochemical and Bioactivity Analysis
- ANOVA with Tukey’s Post Hoc Test: Antimicrobial inhibition zones (agar disc diffusion) were compared across concentrations (5–20 mg/mL) and microbial species (C. albicans, E. coli, and S. aureus). Significance was set at p < 0.05 [29].
- Four-Parameter Logistic Model: Dose–response curves for DPPH radical scavenging (IC50 = 38.6 µg/mL) and cytotoxicity (e.g., MCF-7 IC50 = 28.3 µg/mL) were modeled using GraphPad Prism v 9.0. Parameters included slope, bottom/top asymptotes, and EC50 [30].
- GC-MS and HPLC: Compound identification (α-pineneand and quercetin) utilized retention time matching and spectral libraries (NIST 2020). Concentrations were quantified via peak area integration.
2.9.3. Software and Validation
3. Results
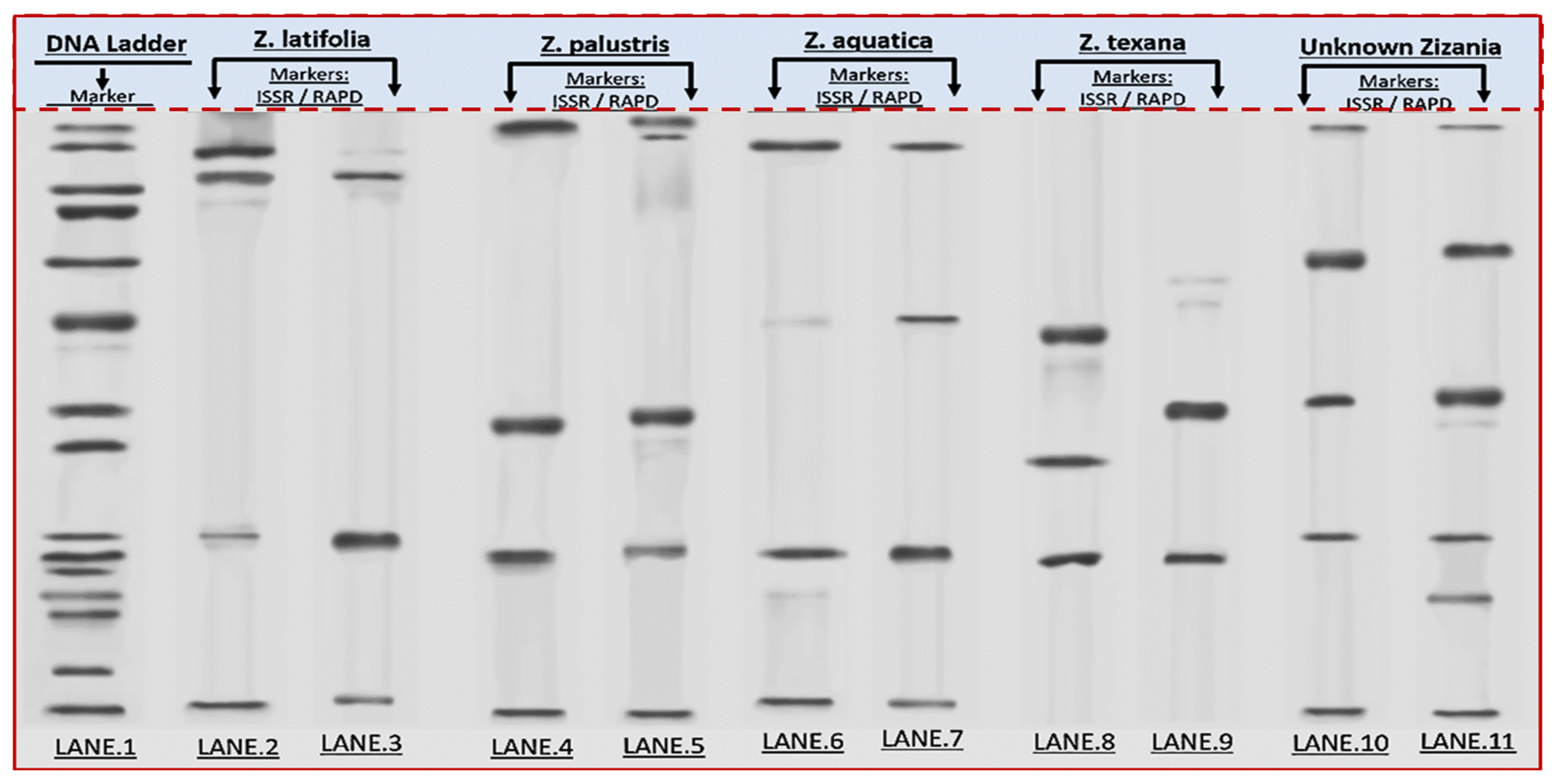
3.1. Gel Electrophoresis and Genetic Relationships
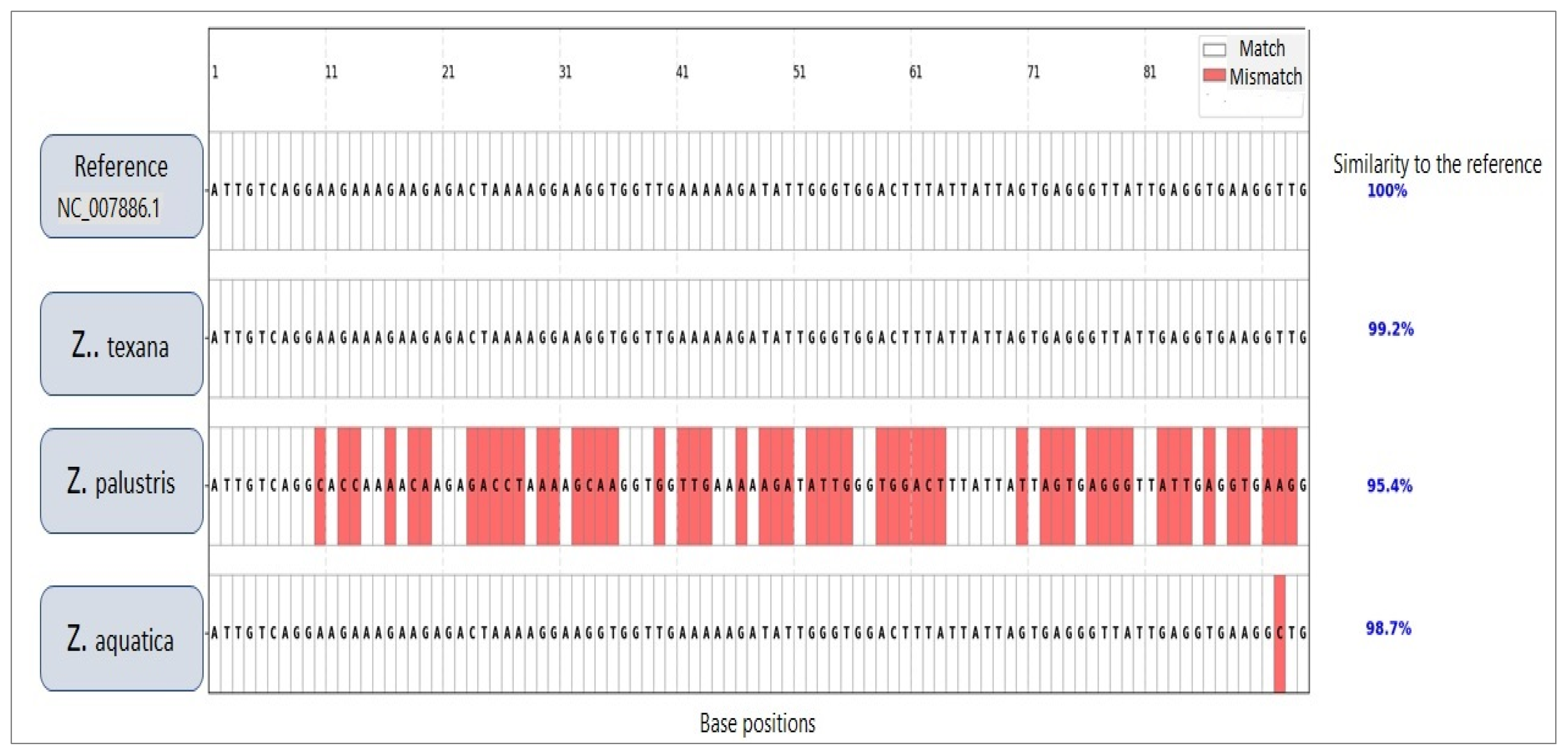
3.2. Integrated Analysis of Unknown Specimens and Taxonomic Classification
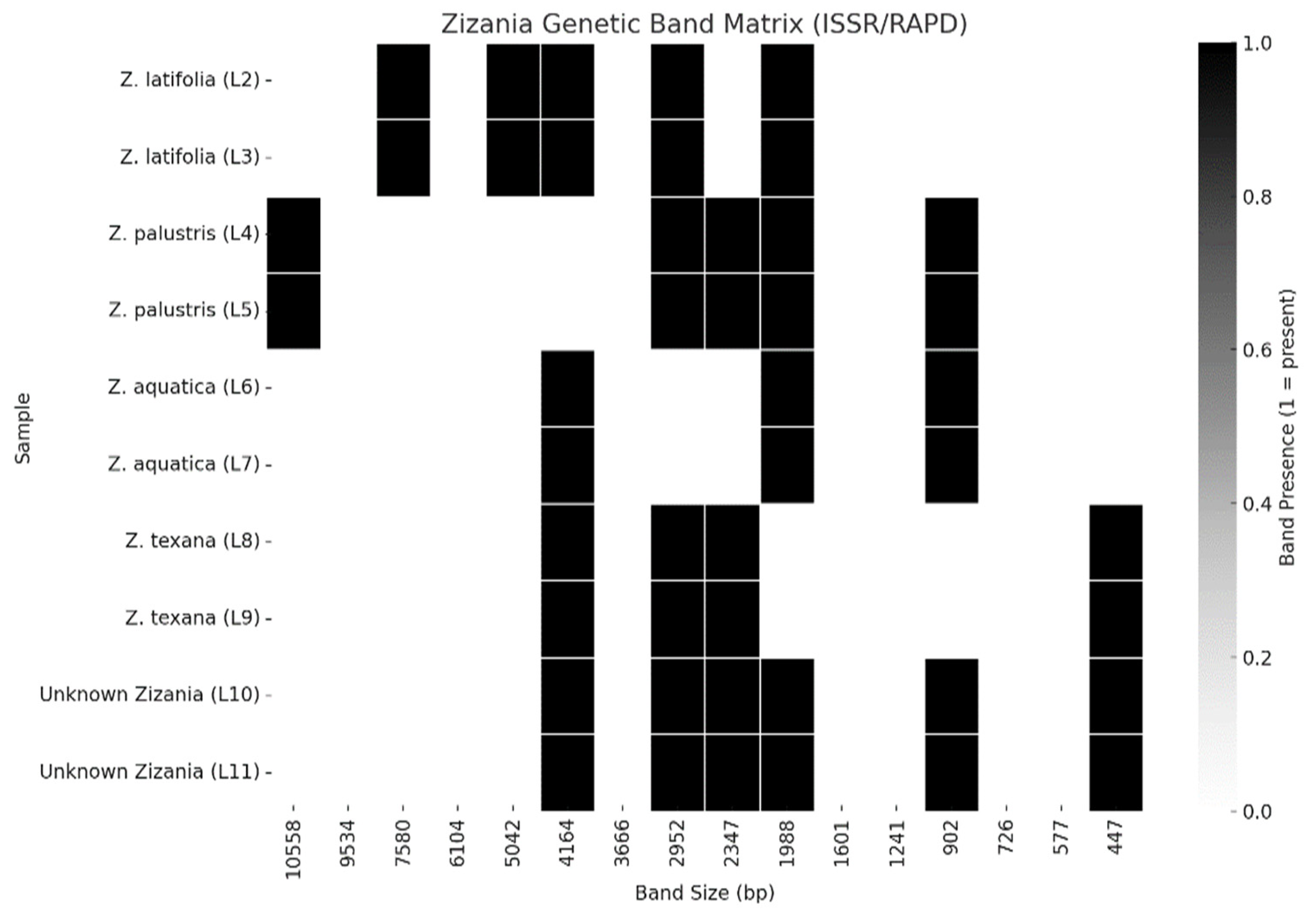
3.3. Genetic Band Matrix (ISSR/RAPD)
3.4. UPGMA Dendrogram (Jaccard Distance)
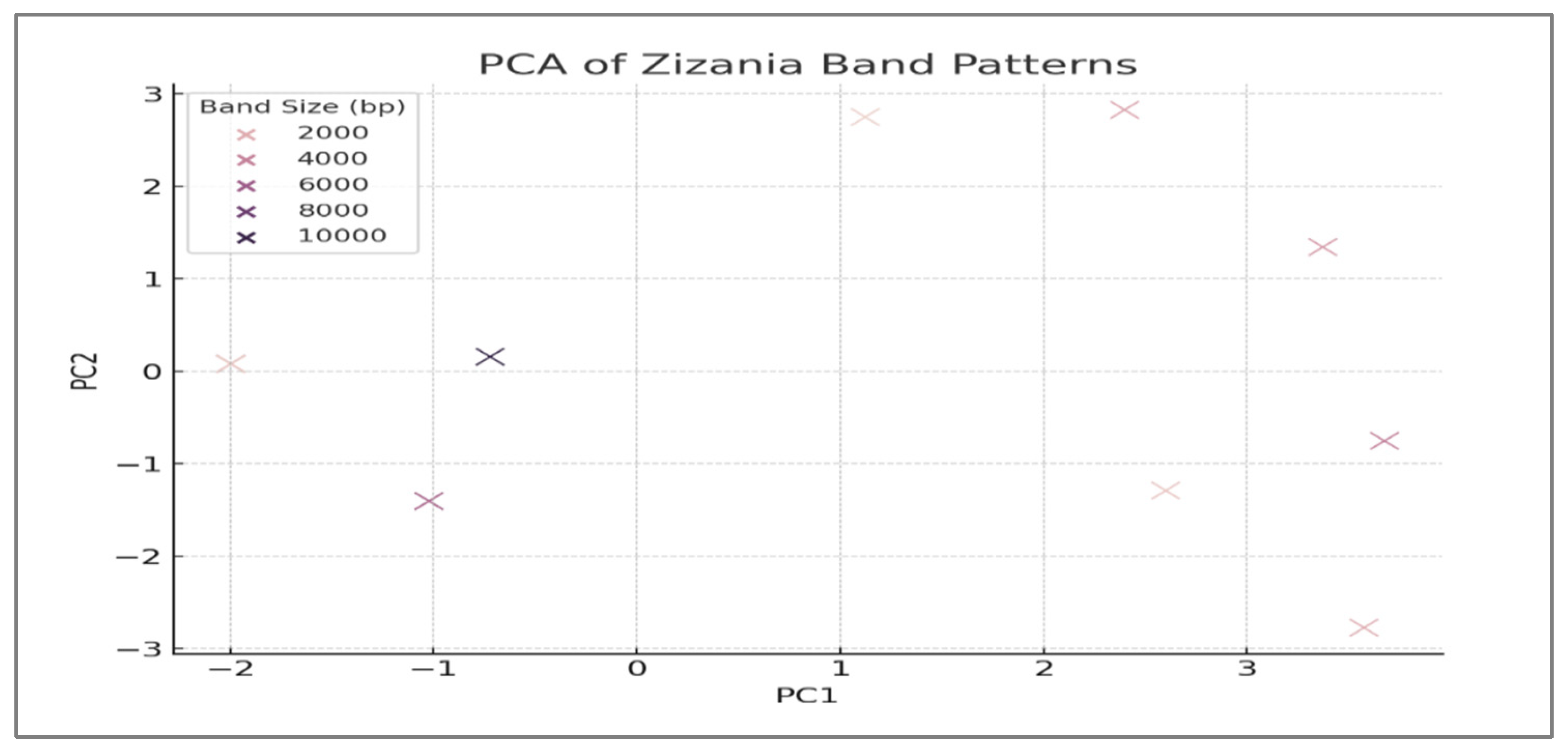
3.5. Principal Component Analysis (PCA)
- Z. texana and unknown samples cluster together in the positive PC1 region;
- Z. latifolia isolates along negative PC2, driven by species-specific bands (e.g., 1200 bp band);
- Z. palustris and Z. aquatica occupy intermediate positions. Band that positions significantly contribute to variance are annotated with arrows.
3.6. Morphological Characterization of the Plant Through Distinctive Structures
3.7. Qualitative Phytochemical Analysis
3.7.1. Phytochemical Screening
3.7.2. Dominant Bioactive Compounds
3.7.3. Synergistic Secondary Metabolites
3.7.4. Structural and Defensive Components
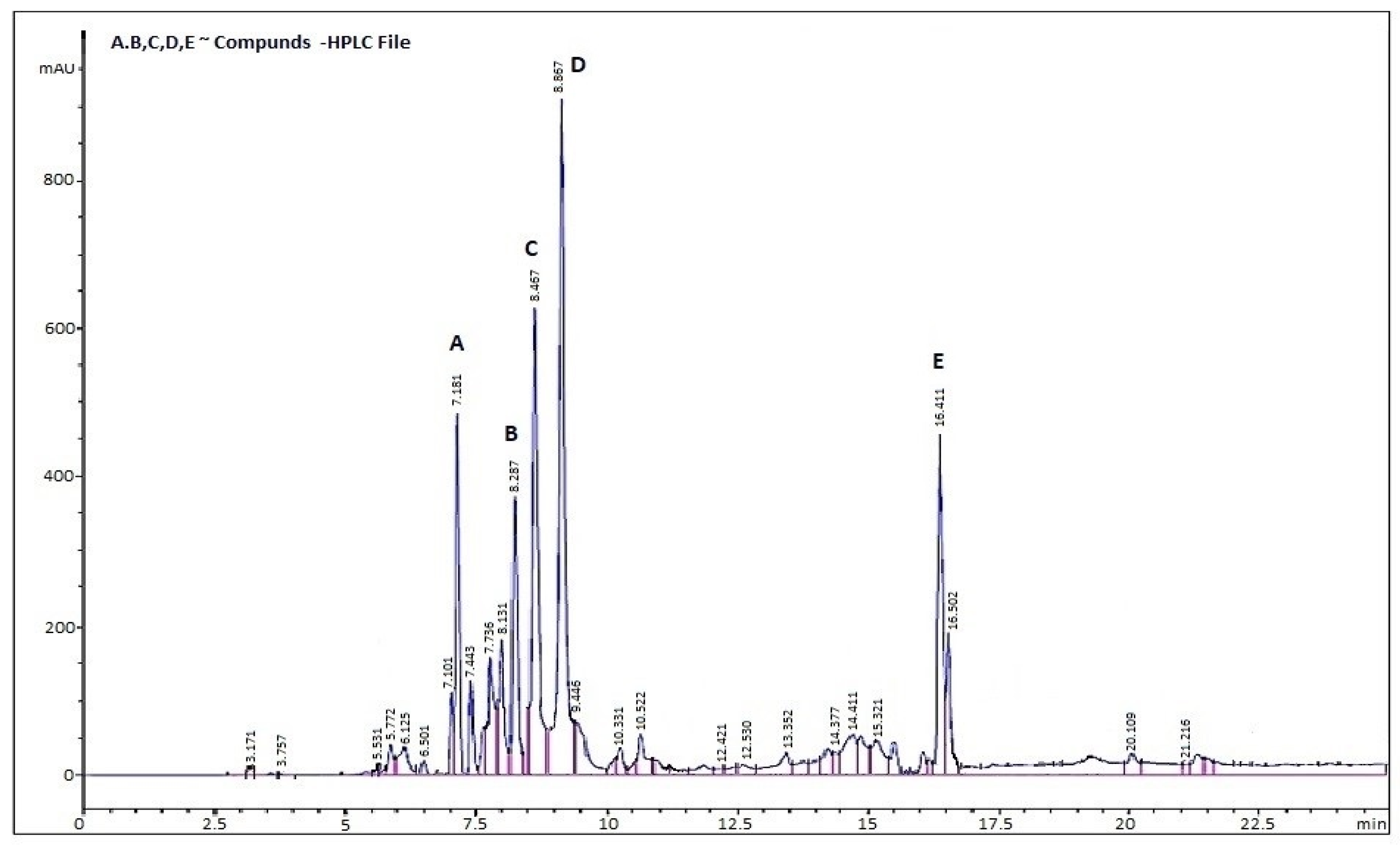
3.7.5. High-Performance Liquid (HPLC)
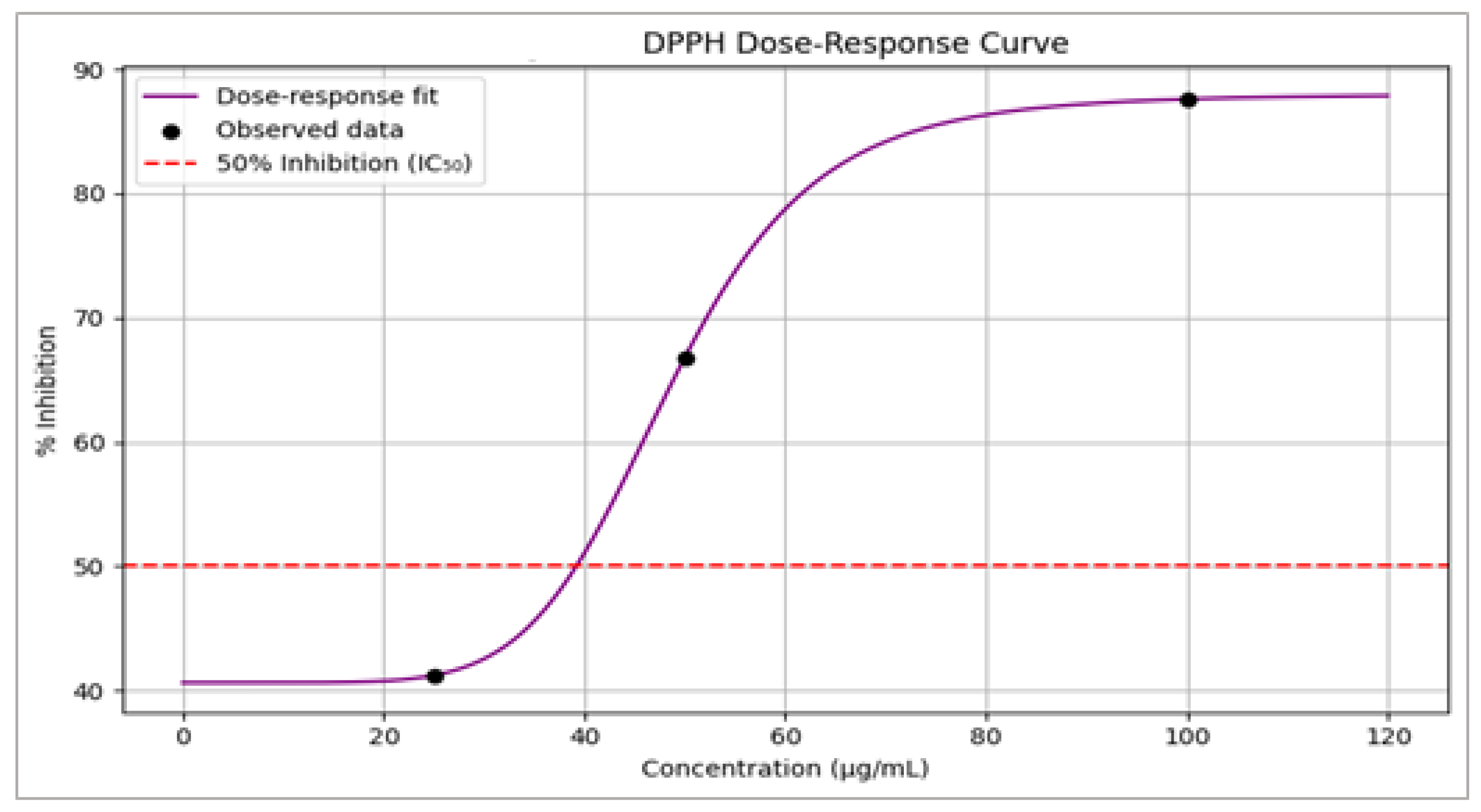
3.8. DPPH Radical Scavenging Activity
3.9. Cytotoxic Activity
4. Discussion
4.1. Genetic Characterization and Taxonomic Classification
4.2. Phytochemical Complexity and Bioactive Synergy
4.3. Antimicrobial and Cytotoxic Mechanisms
4.4. Antioxidant Capacity and Phytochemical Synergy
4.5. Mechanistic Insights and Ecological Relevance
4.6. Bioactivity and Functional Implications
4.7. Comparative Analysis with Related Species
4.8. Cytotoxic Effects
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Z. texana | Zizania texana |
| Z. latifolia | Zizania latifolia |
| Z. palustris | Zizania palustris |
| Z. aquatica | Zizania aquatica |
| C. albicans | Candida albicans |
| S. aureus | Staphylococcus aureus |
| E. coli | Escherichia coli |
| DMEM | Dulbecco’s Modified Eagle Medium |
| FBS | Fetal Bovine Serum |
| UPGMAM | Unweighted Pair Group Method with Arithmetic Mean |
| PCA | Principal Component Analysis |
References
- Ahmed, N.; Thompson, S.; Hardy, B.; Turchini, G.M. An ecosystem approach to wild rice-fish cultivation. Rev. Fish. Sci. Aquac. 2021, 29, 549–565. [Google Scholar] [CrossRef]
- Quayyum, H.A. Interference of Aquatic Plants Associated with Wild Rice (Zizania palustril L.). Ph.D. Thesis, Lakehead University, Thunder Bay, ON, Canada, 1995. [Google Scholar]
- Shaltout, K. Status of the Egyptian biodiversity: A bibliography (2000–2018). In Contribution to the Sixth National Report on Biological Diversity in Egypt; UNDP: New York, NY, USA, 2018. [Google Scholar]
- Salgotra, R.K.; Chauhan, B.S. Genetic diversity, conservation, and utilization of plant genetic resources. Genes 2023, 14, 174. [Google Scholar] [CrossRef]
- Babu, K.N.; Sheeja, T.E.; Minoo, D.; Rajesh, M.K.; Samsudeen, K.; Suraby, E.J.; Kumar, I.P.V. Random amplified polymorphic DNA (RAPD) and derived techniques. In Molecular Plant Taxonomy: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2021; pp. 219–247. [Google Scholar]
- Omar, H.S.; Elsayed, T.R.; Reyad, N.E.H.A.; Shamkh, I.M.; Sedeek, M.S. Gene-targeted molecular phylogeny, phytochemical analysis, antibacterial and antifungal activities of some medicinal plant species cultivated in Egypt. Phytochem. Anal. 2021, 32, 724–739. [Google Scholar] [CrossRef]
- Dar, R.A.; Shahnawaz, M.; Ahanger, M.A.; Majid, I.U. Exploring the diverse bioactive compounds from medicinal plants: A review. J. Phytopharm 2023, 12, 189–195. [Google Scholar] [CrossRef]
- Ezzougari, R.; Saouab, F.E.; Laasli, S.E.; Benamar, K.; Khadiri, M.; Farhaoui, A.; Echchgadda, G.; Lahlali, R. Breeding Strategies for Stress-Tolerant and Climate-Smart Medicinal Plants. In Biotechnology, Multiple Omics, and Precision Breeding in Medicinal Plants; CRC Press: Boca Raton, FL, USA, 2025; pp. 1–19. [Google Scholar]
- Botet, R.; Keurentjes, J.J. The role of transcriptional regulation in hybrid vigor. Front. Plant Sci. 2020, 11, 410. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.N.; Qi, Q.Q.; Li, W.H.; Li, Y.L.; Zhang, Y.; Wang, H.M.; Zhang, Y.F.; Ye, Z.H.; Guo, D.P.; Qian, Q.; et al. Domestication, breeding, omics research, and important genes of Zizania latifolia and Zizania palustris. Front. Plant Sci. 2023, 14, 1183739. [Google Scholar] [CrossRef] [PubMed]
- Negri, S.; Commisso, M.; Bisson, L.; Zorzi, G.; Pietrolucci, F.; Dusi, V.; Ramos, C.; Pinzauti, F.; Nigris, S.; Baldan, B.; et al. Phylogenetic-guided bioprospection of the Italian flora: From the exploitation of bioactive phytochemicals to the study of chemo-evolutionary dynamics. In Book of Abstracts; The University of Verona: Verona, Italy, 2024; p. 109. [Google Scholar]
- Čertner, M.; Lučanová, M.; Sliwinska, E.; Kolář, F.; Loureiro, J. Plant material selection, collection, preservation, and storage for nuclear DNA content estimation. Cytom. Part A 2022, 101, 737–748. [Google Scholar] [CrossRef]
- Aboul-Maaty, N.A.F.; Oraby, H.A.S. Extraction of high-quality genomic DNA from different plant orders applying a modified CTAB-based method. Bull. Natl. Res. Cent. 2019, 43, 25. [Google Scholar] [CrossRef]
- Somasundaram, S.M.; Subbaraya, U.; Durairajan, S.G.; Rajendran, S.; Gopalakrishnan, J.; Hameed, B.S.; Palani, D.; Suthanthiram, B. Comparison of two different electrophoretic methods in studying the genetic diversity among plantains (Musa spp.) using ISSR markers. Electrophoresis 2019, 40, 1265–1272. [Google Scholar] [CrossRef]
- Harborne, J.B. Phytochemical Methods: A Guide to Modern Techniques of Plant Analysis; Chapman and Hall; Springer: Berlin/Heidelberg, Germany, 1998. [Google Scholar]
- Sofowora, A. Medicinal Plants and Traditional Medicine in Africa; Spectrum Books Ltd.: Ibadan, Nigeria, 2006. [Google Scholar]
- Pandey, A.; Tripathi, S. Concept of standardization, extraction and pre phytochemical screening strategies for herbal drug. J. Pharmacogn. Phytochem. 2014, 2, 115–119. [Google Scholar]
- Uraku, A.J. Leaves by gas chromatography-mass spectrometry (GC-MS) method. Res. J. Phytochem. 2015, 9, 175–187. [Google Scholar]
- Zhang, X.; Zhou, X.; Liu, X.; Li, X.; Whang, W. Development and application of an HPLC–UV procedure to determine multiple flavonoids and phenolics in Acanthopanax leaf extracts. J. Chromatogr. Sci. 2016, 54, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Gawron-Gzella, A.; Dudek-Makuch, M.; Matlawska, I. DPPH radical scavenging activity and phenolic compound content in different leaf extracts from selected blackberry species. Acta Biol. Cracoviensia Ser. Bot. 2012, 54, 32–38. [Google Scholar] [CrossRef]
- Othman, M.; San Loh, H.; Wiart, C.; Khoo, T.J.; Lim, K.H.; Ting, K.N. Optimal methods for evaluating antimicrobial activities from plant extracts. J. Microbiol. Methods 2011, 84, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Plumb, J.A. Cell sensitivity assays: The MTT assay. In Cancer Cell Culture: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2004; pp. 165–169. [Google Scholar]
- Carmichael, J.; Mitchell, J.B.; DeGraff, W.G.; Gamson, J.; Gazdar, A.F.; Johnson, B.E.; Glatstein, E.; Minna, J.D. Chemosensitivity testing of human lung cancer cell lines using the MTT assay. Br. J. Cancer 1988, 57, 540–547. [Google Scholar] [CrossRef]
- Sokal, R.R.; Michener, C.D. A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 1958, 38, 1409–1438. [Google Scholar]
- Jolliffe, I.T. Principal Component Analysis, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- Chierichetti, F.; Kumar, R.; Pandey, S.; Vassilvitskii, S. Finding the jaccard median. In Proceedings of the Twenty-First Annual ACM-SIAM Symposium on Discrete Algorithms, Austin, TX, USA, 17–19 January 2010; pp. 293–311. [Google Scholar]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A k-means clustering algorithm. J. R. Stat. Society Ser. C (Appl. Stat.) 1979, 28, 100–108. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Zar, J.H. Biostatistical Analysis, 5th ed.; Pearson: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-response analysis using R. PLoS ONE 2015, 10, e0146021. [Google Scholar] [CrossRef]
- Zaitchik, B.F.; LeRoux, L.G.; Kellogg, E.A. Development of male flowers in Zizania aquatica (North American wild-rice; Gramineae). Int. J. Plant Sci. 2000, 161, 345–351. [Google Scholar] [CrossRef]
- Walters, C.; Richards, C.M.; Volk, G.M. Genebank conservation of germplasm collected from wild species. In North American Crop Wild Relatives, Volume 1: Conservation Strategies; Springer: Berlin/Heidelberg, Germany, 2018; pp. 245–280. [Google Scholar]
- Lu, R.; Chen, M.; Feng, Y.; Yuan, N.; Zhang, Y.; Cao, M.; Liu, J.; Wang, Y.; Hang, Y.; Sun, X. Comparative plastome analyses and genomic resource development in wild rice (Zizania spp., Poaceae) using genome skimming data. Ind. Crops Prod. 2022, 186, 115244. [Google Scholar] [CrossRef]
- Ye, Z.; Liu, J.; Jin, Y.; Cui, H.; An, X.; Fu, X.; Yu, X. Physiological and proteomic changes in Zizania latifolia under chilling stress. Biologia 2017, 72, 1291–1299. [Google Scholar] [CrossRef]
- Kahler, A.L. Genome Organization and Genetic Diversity of Wildrice (Zizania palustris L.); University of Minnesota: Minneapolis, MN, USA, 2010. [Google Scholar]
- Yan, N.; Yang, T.; Yu, X.T.; Shang, L.G.; Guo, D.P.; Zhang, Y.; Meng, L.; Qi, Q.Q.; Li, Y.L.; Du, Y.M.; et al. Chromosome-level genome assembly of Zizania latifolia provides insights into its seed shattering and phytocassane biosynthesis. Commun. Biol. 2022, 5, 36. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Priya, P.; Singh, N.; Kumar, M.; Choudhary, B.K.; Kumar, L.; Singh, I.S.; Kumar, N. RAPD and ISSR marker-based comparative evaluation of genetic diversity among Indian germplasms of Euryale ferox: An aquatic food plant. Appl. Biochem. Biotechnol. 2016, 180, 1345–1360. [Google Scholar] [CrossRef] [PubMed]
- Ngangkham, U.; Dash, S.; Parida, M.; Samantaray, S.; Nongthombam, D.; Yadav, M.K.; Kumar, A.; Chidambaranathan, P.; Katara, J.L.; Patra, B.C.; et al. The potentiality of rice microsatellite markers in assessment of cross-species transferability and genetic diversity of rice and its wild relatives. 3 Biotech 2019, 9, 217. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Walters, C.; Antolin, M.F.; Alexander, M.L.; Lutz, S.; Ge, S.; Wen, J. Phylogeny and biogeography of the eastern Asian–North American disjunct wild-rice genus (Zizania L., Poaceae). Mol. Phylogenetics Evol. 2010, 55, 1008–1017. [Google Scholar] [CrossRef]
- Zhang, D.; Li, K.; Gao, J.; Liu, Y.; Gao, L.Z. The complete plastid genome sequence of the wild rice Zizania latifolia and comparative chloroplast genomics of the rice tribe Oryzeae, Poaceae. Front. Ecol. Evol. 2016, 4, 88. [Google Scholar] [CrossRef]
- Luo, X.; Gu, C.; Gao, S.; Li, M.; Zhang, H.; Zhu, S. Complete mitochondrial genome assembly of Zizania latifolia and comparative genome analysis. Front. Plant Sci. 2024, 15, 1381089. [Google Scholar] [CrossRef]
- Lacoul, P.; Freedman, B. Environmental influences on aquatic plants in freshwater ecosystems. Environ. Rev. 2006, 14, 89–136. [Google Scholar] [CrossRef]
- Castellani, M.B.; Dalla Vecchia, A.; Bolpagni, R.; Natale, R.; Piaser, E.; Lastrucci, L.; Coppi, A.; Villa, P. Genetic drift versus natural selection affecting the evolution of spectral and functional traits of two key macrophytes: Phragmites australis and Nuphar lutea. Freshw. Biol. 2023, 68, 1739–1750. [Google Scholar] [CrossRef]
- Jamdade, R.; Upadhyay, M.; Al Shaer, K.; Al Harthi, E.; Al Sallani, M.; Al Jasmi, M.; Al Ketbi, A. Evaluation of Arabian vascular plant barcodes (rbcL and matK): Precision of unsupervised and supervised learning methods towards accurate identification. Plants 2021, 10, 2741. [Google Scholar] [CrossRef]
- Hu, J.; Qi, Q.; Zhu, Y.; Wen, C.; Olatunji, O.J.; Jayeoye, T.J.; Eze, F.N. Unveiling the anticancer, antimicrobial, antioxidative properties, and UPLC-ESI-QTOF-MS/GC–MS metabolite profile of the lipophilic extract of siam weed (Chromolaena odorata). Arab. J. Chem. 2023, 16, 104834. [Google Scholar] [CrossRef]
- Zhang, J.; Zhong, H.; Li, F.; Yu, K.; Chen, J. A Potential Natural Enemy of Rust Fungus Uromyces coronatus of an Aquatic Vegetable Water-Oat Zizania latifolia. Entomol. News 2025, 132, 174–183. [Google Scholar] [CrossRef]
- Lombrea, A.; Antal, D.; Ardelean, F.; Avram, S.; Pavel, I.Z.; Vlaia, L.; Mut, A.M.; Diaconeasa, Z.; Dehelean, C.A.; Soica, C.; et al. A recent insight regarding the phytochemistry and bioactivity of Origanum vulgare L. essential oil. Int. J. Mol. Sci. 2020, 21, 9653. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Simoneit, B.R.; Jara, B.; Jaffé, R. Gas chromatography mass spectrometry based profiling of alkyl coumarates and ferulates in two species of cattail (Typha domingensis P., and Typha latifolia L.). Phytochem. Lett. 2015, 13, 91–98. [Google Scholar] [CrossRef]
- Fonseca Do Carmo, P.H.; Pinheiro Lage, A.C.; Garcia, M.T.; Soares da Silva, N.; Santos, D.A.; Mylonakis, E.; Junqueira, J.C. Resveratrol-coated gold nanorods produced by green synthesis with activity against Candida albicans. Virulence 2024, 15, 2416550. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Martínez, F.J.; Barrajón-Catalán, E.; Encinar, J.A.; Rodríguez-Díaz, J.C.; Micol, V. Antimicrobial capacity of plant polyphenols against gram-positive bacteria: A comprehensive review. Curr. Med. Chem. 2020, 27, 2576–2606. [Google Scholar] [CrossRef]
- Lawal, R.; James, A.B.; Shaibu, R.; Okoye, O.; Odetunde, S.; Ajibare, A. Cytotoxic effects of Curcuma longa leaves on MCF-7 and HepG2 cells. Fountain J. Nat. Appl. Sci. 2022, 11, 31–35. [Google Scholar] [CrossRef]
- Chien, S.-Y.; Wu, Y.-C.; Chung, J.-G.; Yang, J.-S.; Lu, H.-F.; Tsou, M.-F.; Wood, W.; Kuo, S.-J.; Chen, D.-R. Quercetin-induced apoptosis acts through mitochondrial-and caspase-3-dependent pathways in human breast cancer MDA-MB-231 cells. Hum. Exp. Toxicol. 2009, 28, 493–503. [Google Scholar] [CrossRef]
- Zhong, C.; Li, H.-D.; Liu, D.-Y.; Xu, F.-B.; Wu, J.; Lin, X.-M.; Guo, R.-P. Clinical study of hepatectomy combined with Jianpi Huayu therapy for hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2014, 15, 5951–5957. [Google Scholar] [CrossRef]
- Yousaf, H. Evaluation And Comparison Of The Antioxidant And Free Radical Scavenging Properties Of Medicinal Plants By Using The Dpph Assay In-Vitro. bioRxiv 2023. [Google Scholar] [CrossRef]
- Sumczynski, D.; Kotásková, E.; Orsavová, J.; Valášek, P. Contribution of individual phenolics to antioxidant activity and in vitro digestibility of wild rices (Zizania aquatica L.). Food Chem. 2017, 218, 107–115. [Google Scholar] [CrossRef]
- Chu, M.J.; Du, Y.M.; Liu, X.M.; Yan, N.; Wang, F.Z.; Zhang, Z.F. Extraction of proanthocyanidins from Chinese wild rice (Zizania latifolia) and analyses of structural composition and potential bioactivities of different fractions. Molecules 2019, 24, 1681. [Google Scholar] [CrossRef] [PubMed]
- Shahidi, F.; Danielski, R.; Rhein, S.O.; Meisel, L.A.; Fuentes, J.; Speisky, H.; Schwember, A.R.; de Camargo, A.C. Wheat and rice beyond phenolic acids: Genetics, identification database, antioxidant properties, and potential health effects. Plants 2022, 11, 3283. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Cao, Y.; Bao, B.; Zhang, L.; Ding, A. Antioxidant capacity of Typha angustifolia extracts and two active flavonoids. Pharm. Biol. 2017, 55, 1283–1288. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, S.M.; Hassan, K.M.; Abdelhamid, A.N.; Yousef, E.E.; Abdellatif, Y.M.; Abu-Hussien, S.H.; Nasser, M.A.; Elshalakany, W.A.; Darwish, D.B.; Abdulmajeed, A.M.; et al. Exogenous paclobutrazol reinforces the antioxidant and antimicrobial properties of lavender (Lavandula officinalis L.) oil through modulating its composition of oxygenated terpenes. Plants 2022, 11, 1607. [Google Scholar] [CrossRef]
- Gebashe, F.; Aremu, A.O.; Gruz, J.; Finnie, J.F.; Van Staden, J. Phytochemical profiles and antioxidant activity of grasses used in South African traditional medicine. Plants 2020, 9, 371. [Google Scholar] [CrossRef]




| RAPD Primers (Operon Technologies, Alameda, CA, USA) | ISSR Primers (ISSR Primers (Genetic Engineering Research Institute, Agricultural Research Center, Egypt)) |
|---|---|
| OPA-03: 5′-AGTCAGCCAC-3′ | UBC-807: 5′-(AG)8T-3′ |
| OPA-11: 5′-CAATCGCCGT-3′ | UBC-812: 5′-(GA)8A-3′ |
| OPB-04: 5′-GGACTGGAGT-3′ | UBC-826: 5′-(AC)8C-3′ |
| OPB-14: 5′-TCCGCTCTGG-3′ | UBC-841: 5′-(GA)8YC-3′ |
| OPC-07: 5′-GTCCCGACGA-3′ | UBC-873: 5′-(GACA)4-3′ |
| Species | PC1 Mean ± SD | PC2 Mean ± SD | Diagnostic Bands (bp) | Cluster Association |
|---|---|---|---|---|
| Z. texana | 1.82 ± 0.31 | 0.29 ± 0.42 | 2400, 4800 | Positive PC1 |
| Z. palustris | 0.21 ± 0.35 | 1.48 ± 0.39 | 3600, 7200 | Neutral |
| Z. aquatica | −0.52 ± 0.38 | −1.01 ± 0.41 | 6000, 8400 | Neutral |
| Z. latifolia | −1.23 ± 0.42 | 0.81 ± 0.36 | 1200, 9600 | Negative PC2 |
| Unknowns | 1.75 ± 0.33 | 0.42 ± 0.38 | 2400, 4800 | Positive PC1 |
| Phytochemical Class | Qualitative Presence |
|---|---|
| Flavonoids | +++ (Strong) |
| Tannins | ++ (Moderate) |
| Terpenoids | ++ (Moderate) |
| Saponins | + (Slight) |
| Alkaloids | ++ (Moderate) |
| Glycosides | + (Slight) |
| No. | Retention Time (Min) | Concentration % | Compound Name | Molecular Formula | Biological Activity |
|---|---|---|---|---|---|
| 1 | 3.97 | 13.65 | α-Pinene | C10H16 | Antimicrobial, anti-inflammatory |
| 2 | 5.86 | 3.92 | Camphene | C10H16 | Fragrance, mild antibacterial |
| 3 | 6.67 | 10.1 | β-Pinene | C10H16 | Antioxidant, anti-inflammatory |
| 4 | 7.66 | 5.41 | Myrcene | C10H16 | Analgesic, antioxidant |
| 5 | 9.95 | 6.21 | Limonene | C10H16 | Anti-cancer, insecticidal |
| 6 | 10.49 | 9.52 | Linalool | C10H18O | Antibacterial, anxiolytic |
| 7 | 12.45 | 4.43 | Terpineol | C10H18O | Antiseptic, antioxidant |
| 8 | 14.51 | 6.88 | Palmitic acid | C16H32O2 | Antioxidant, antibacterial |
| 9 | 16.18 | 6.39 | Phytol | C20H40O | Anti-inflammatory, precursor to vitamins E/K |
| 10 | 17.48 | 11.21 | β-Caryophyllene | C15H24 | Cytotoxic, anti-inflammatory |
| 11 | 18.22 | 3.47 | Caryophyllene oxide | C15H24O | Antifungal, anti-cancer |
| 12 | 19.39 | 3.92 | Neophytadiene | C20H38 | Antioxidant, antimicrobial |
| 13 | 20.81 | 4.11 | Stearic acid | C18H36O2 | Emollient, lipid metabolism |
| 14 | 20.82 | 3.43 | Squalene | C30H50 | Antioxidant, chemopreventive |
| 15 | 22.94 | 4.41 | Nonacosane | C29H60 | Cuticular wax, defense compound |
| 16 | 24.65 | 2.94 | Dotriacontane | C32H66 | Hydrocarbon, structural leaf wax |
| Peak No. | Retention Time (min) | Compound Identified | Concentration (µg/mL) |
|---|---|---|---|
| A | 7.181 | Gallic acid | 23.4 ± 1.2 |
| B | 8.287 | Caffeic acid | 15.6 ± 0.9 |
| C | 8.467 | Rutin | 37.8 ± 1.8 |
| D | 8.867 | Quercetin | 42.1 ± 2.0 |
| E | 16.411 | Kaempferol | 17.9 ± 1.1 |
| Microbe | Mean IC50 ± SD |
|---|---|
| S. aureus | 10.7 ± 1.3 |
| E. coli | 6.8 ± 0.9 |
| C. albicans | 4.9 ± 0.6 |
| Concentration (µg/mL) | Inhibition (% ± SD) |
|---|---|
| 25 | 41.2 ± 1.5 |
| 50 | 66.8 ± 2.1 |
| 100 | 87.6 ± 1.2 |
| Concentration (µg/mL) | MCF-7 | HepG2 | A549 |
|---|---|---|---|
| 6.25 | 11.4 ± 0.8 | 10.2 ± 1.1 | 8.6 ± 0.9 |
| 12.5 | 25.7 ± 1.3 | 21.9 ± 1.2 | 20.2 ± 1.0 |
| 25 | 49.6 ± 1.6 | 45.3 ± 1.5 | 41.0 ± 1.3 |
| 50 | 70.3 ± 2.0 | 66.1 ± 1.8 | 60.4 ± 1.6 |
| 100 | 87.9 ± 2.2 | 84.5 ± 2.0 | 80.2 ± 1.9 |
| 200 | 94.6 ± 2.4 | 91.3 ± 2.2 | 88.7 ± 2.1 |
| Cancer Cells Type | IC50 Values ± SD |
|---|---|
| MCF-7 | 28.3 ± 1.5 µg/mL |
| HepG2 | 31.4 ± 1.8 µg/mL |
| A549 | 36.9 ± 2.0 µg/mL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Shammari, L.A. DNA Fingerprint Profile of Zizania spp. Plant, Monitoring Its Leaves with Screening of Their Biological Activity: Antimicrobial, Antioxidant and Cytotoxicity. Life 2025, 15, 1240. https://doi.org/10.3390/life15081240
Al Shammari LA. DNA Fingerprint Profile of Zizania spp. Plant, Monitoring Its Leaves with Screening of Their Biological Activity: Antimicrobial, Antioxidant and Cytotoxicity. Life. 2025; 15(8):1240. https://doi.org/10.3390/life15081240
Chicago/Turabian StyleAl Shammari, Latifah A. 2025. "DNA Fingerprint Profile of Zizania spp. Plant, Monitoring Its Leaves with Screening of Their Biological Activity: Antimicrobial, Antioxidant and Cytotoxicity" Life 15, no. 8: 1240. https://doi.org/10.3390/life15081240
APA StyleAl Shammari, L. A. (2025). DNA Fingerprint Profile of Zizania spp. Plant, Monitoring Its Leaves with Screening of Their Biological Activity: Antimicrobial, Antioxidant and Cytotoxicity. Life, 15(8), 1240. https://doi.org/10.3390/life15081240






