The Effect of cdk1 Gene Knockout on Heat Shock-Induced Polyploidization in Loach (Misgurnus anguillicaudatus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish
2.2. Artificial Induction of Ovulation and Fertilization
2.3. Induction of Tetraploid Loaches
2.4. Polyploid Detection and Survival Rate Calculation
2.5. Preparation of Single Embryo Chromosome Specimens
- Using a pipette, a single embryo is taken and placed in a Petri dish. Using a syringe, the egg membrane and yolk of the fertilized egg at the blastocyst stage are removed, leaving the remaining embryonic tissue.
- Transfer the remaining part of the embryo using a pipette to a small beaker containing 0.0025% colchicine solution and treat for 45 min. Then, add 0.8% sodium citrate for hypotonic treatment for 20 min, with the hypotonic solution being replaced once during this period.
- Fix the sample three times with pre-cooled Carnoy’s fixative (methanol–acetic acid = 3:1), for 15 min each time. After fixation, store it at −20 °C overnight.
- The next day, remove the samples, treat with 50% acetic acid for 5 min for dissociation, and then mechanically shake it to prepare a single-cell suspension. Add fresh Carnoy’s fixative and mix evenly before dropping it onto slides, which should then be quickly dried by flame and left to stand at room temperature.
- Use the Wright–Jenner staining solution to stain the chromosome specimens.
- Randomly select 50 mid-stage division cells to count the chromosome numbers and calculate the induction success rate.
2.6. Preparation of Juvenile Fish Chromosome Preparation
- Transfer the 3-day-old experimental fish larvae to a solution containing 100 mg/L colchicine and 0.7% sodium citrate at room temperature.
- Fix with pre-cooled Carnoy’s fixative (methanol: acetic acid = 3:1) for three times, 15 min each time, and then place it in a −20 °C refrigerator for freezing overnight.
- The next day, take out the samples, add 50% acetic acid for dissociation for 5 min, and then mechanically shake the cells to prepare a single-cell suspension. Add fresh Carnoy’s fixative and mix evenly before dropping it onto slides, which should be then quickly dried by flame and left to stand at room temperature.
- After staining with the Wright–Jenner method, rinse the samples with distilled water, air-dry at room temperature, and image them using a microscopic image analysis system.
2.7. qPCR
2.8. Data Analysis
3. Results
3.1. The Optimal Conditions for Inducing Tetraploidy in Heat Shock
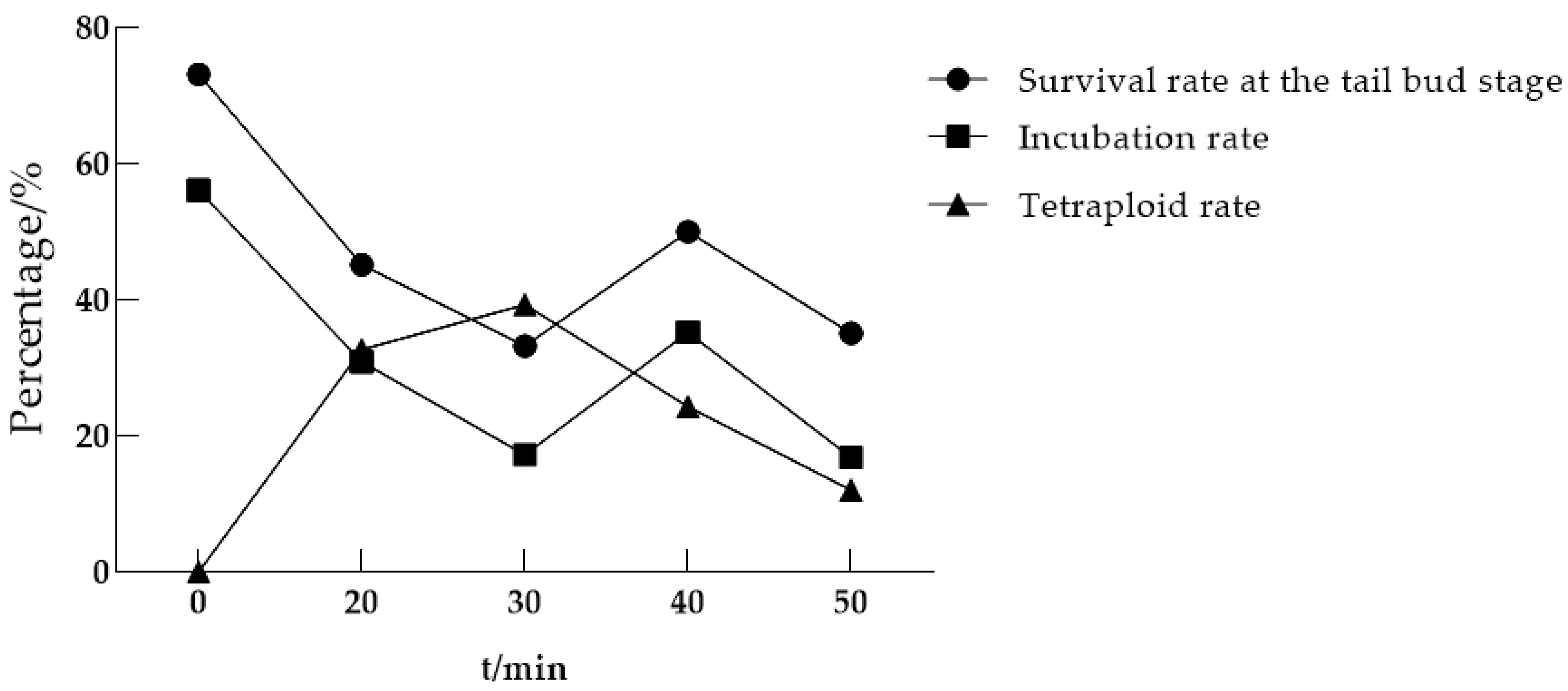
3.1.1. The Optimal Time for Initiating Heat Shock Treatment After Fertilization
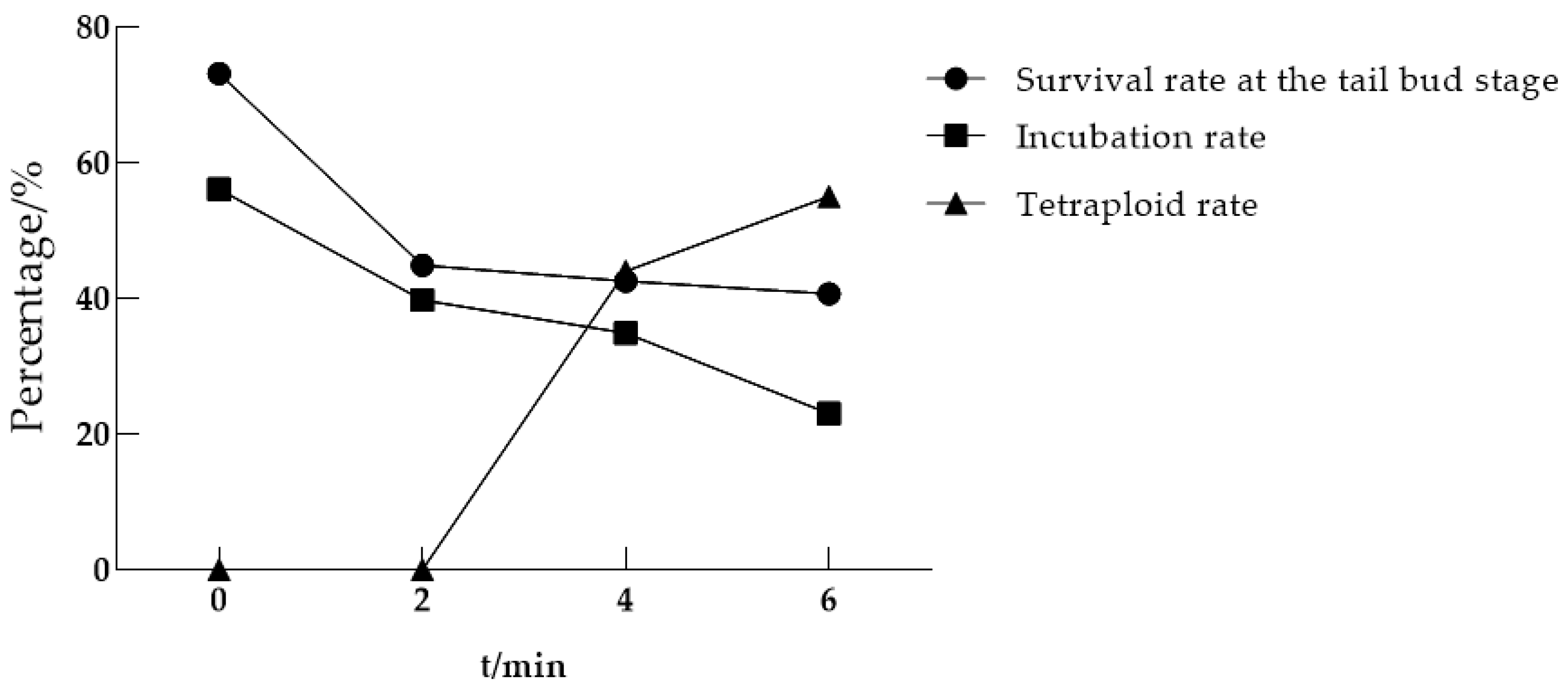
3.1.2. Optimal Continuous Processing Time
3.2. Knocking out the cdk1 Gene Is Conducive to Inducing Tetraploidy
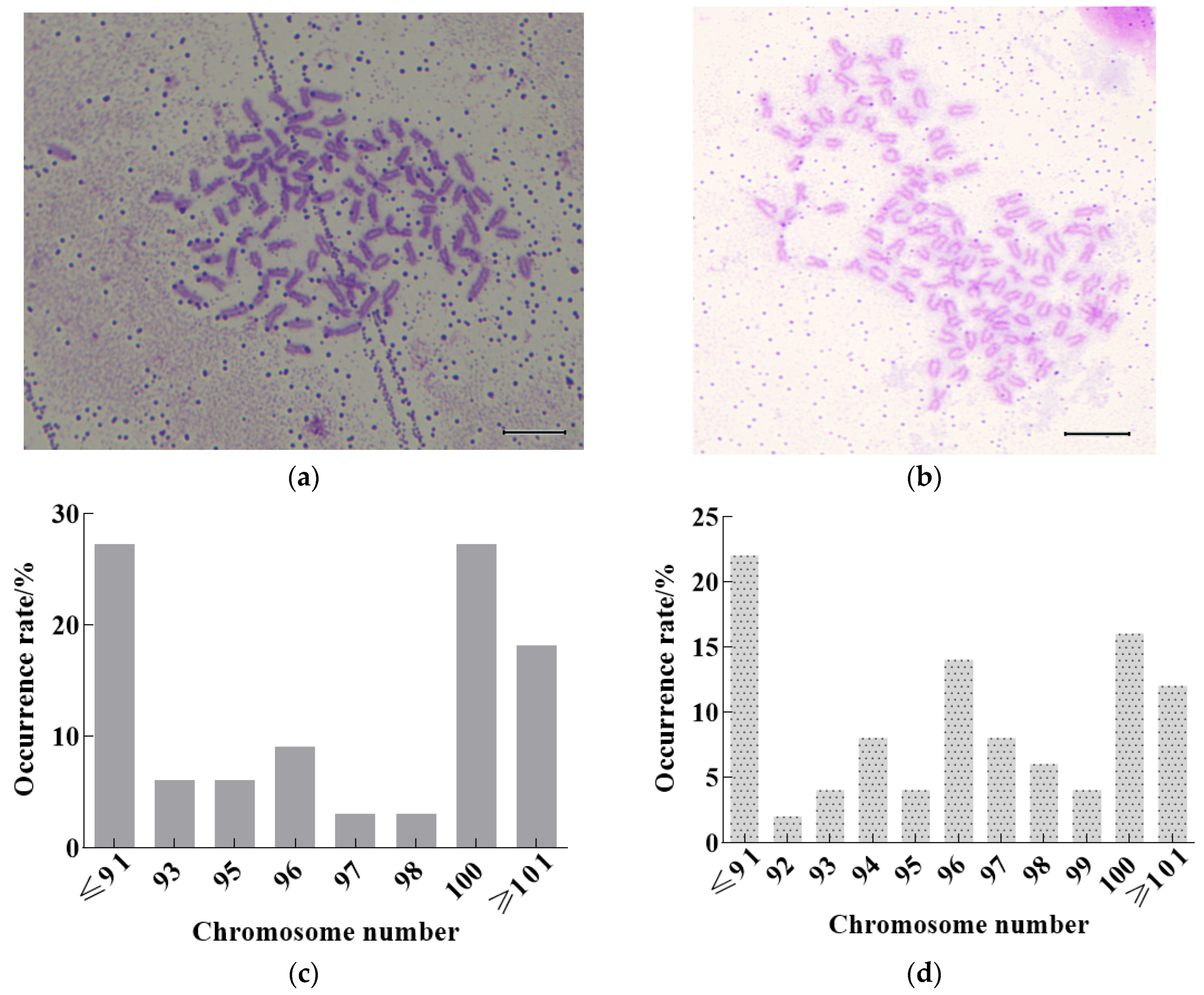
3.3. Observation and Analysis of Chromosomes in Heat Shock of 2n cdk1+/+ and 2n cdk1−/−
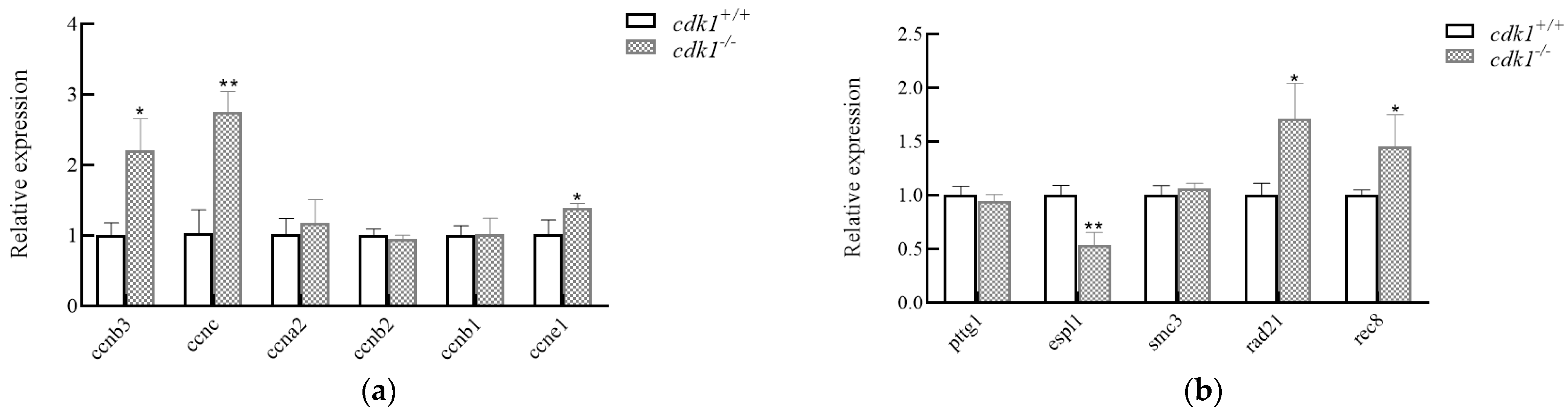
3.4. Expression of Related Genes During the Process of Heat Shock Chromosome Doubling
4. Discussion
4.1. The Effect of Heat Shock on Artificially Induced Tetraploid Loaches
4.2. The Effect of cdk1 on Chromosome Doubling in Diploid Loaches
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Piferrer, F.; Beaumont, A.; Falguière, J.C.; Flajšhans, M.; Haffray, P.; Colombo, L. Polyploid fish and shellfish: Production, biology and applications to aquaculture for performance improvement and genetic containment. Aquaculture 2009, 293, 125–156. [Google Scholar] [CrossRef]
- Zhou, L.; Gui, J. Natural and artificial polyploids in aquaculture. Aquac. Fish. 2017, 2, 103–111. [Google Scholar] [CrossRef]
- Arm, K. Genetics of the loach, Misgurnus anguillicaudatus: Recent progress and perspective. Folia Biol. 2003, 51, 107–117. [Google Scholar]
- Abbas, K.; Li, M.Y.; Wang, W.M.; Zhou, X.Y. First record of the natural occurrence of hexaploid Dojo loach Misgurnus anguillicaudatus in Hubei Province. J. Fish Biol. 2009, 75, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Arai, K.; Fujimoto, T. Genomic Constitution and Atypical Reproduction in Polyploid and Unisexual Lineages of the Misgurnus Dojo loach, a Teleost Fish. Cytogenet. Genome Res. 2013, 140, 226–240. [Google Scholar] [CrossRef]
- Brown, N.R.; Korolchuk, S.; Martin, M.P.; Stanley, W.A.; Moukhametzianov, R.; Noble, M.E.; Endicott, J.A. CDK1 structures reveal conserved and unique features of the essential cell cycle CDK. Nat. Commun. 2015, 6, 6769. [Google Scholar] [CrossRef]
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Lu, L.X.; Ohi, M.D.; Creamer, K.M.; English, C.; Partridge, J.F.; Ohi, R.; Gould, K.L. Cdk1 phosphorylation of the kinetochore protein Nsk1 prevents error-prone chromosome segregation. J. Cell Biol. 2011, 195, 583–593. [Google Scholar] [CrossRef]
- Vázquez-Novelle, M.D.; Sansregret, L.; Dick, A.E.; Smith, C.A.; McAinsh, A.D.; Gerlich, D.W.; Petronczki, M. Cdk1 Inactivation Terminates Mitotic Checkpoint Surveillance and Stabilizes Kinetochore Attachments in Anaphase. Curr. Biol. 2014, 24, 638–645. [Google Scholar] [CrossRef]
- Mirchenko, L.; Uhlmann, F. Sli15(INCENP) dephosphorylation prevents mitotic checkpoint reengagement due to loss of tension at anaphase onset. Curr. Biol. 2010, 20, 1396–1401. [Google Scholar] [CrossRef]
- Oliveira, R.A.; Nasmyth, K. Getting through anaphase: Splitting the sisters and beyond. Biochem. Soc. Trans. 2010, 38, 1639–1644. [Google Scholar] [CrossRef]
- Qi, Q.R.; Zhao, X.Y.; Zuo, R.J. Involvement of atypical transcription factor E2F8 in the polyploidization during mouse and human decidualization. Cell Cycle 2015, 14, 1842–1858. [Google Scholar] [CrossRef]
- Zhang, Y.; Lv, M.; Jiang, H. Mitotic defects lead to unreduced sperm formation in cdk1−/− mutants. Int. J. Biol. Macromol. 2023, 242, 125171. [Google Scholar] [CrossRef]
- Ma, R.; Liu, X.H.; Meng, Y.Q. Protein nutrition on sub-adult triploid rainbow trout (1): Dietary requirement and effect on anti-oxidative capacity, protein digestion and absorption. Aquaculture 2019, 507, 428–434. [Google Scholar] [CrossRef]
- Nan, P.; Du, Q.; Yan, S. Induction of Tetraploidy Mud Loach (Misgurnus anguillicaudatus). J. Henan Norm. Univ. (Nat. Sci.) 2008, 36, 103–106. [Google Scholar]
- Chang, Z.; Du, Q. Studies on the Induction of Tetraploidy Loach (Paramisgurnus dabryanus). J. Henan Norm. Univ. (Nat. Sci.) 2002, 30, 70–73. [Google Scholar]
- Sakao, S.; Fujimoto, T.; Kimura, S.; Yamaha, E.; Arai, K. Drastic mortality in tetraploid induction results from the elevation of ploidy in masu salmon Oncorhynchus masou. Aquaculture 2006, 252, 147–160. [Google Scholar] [CrossRef]
- de Alvarenga, É.R.; Fernandes, A.F.A.; Lopes, L.R.; de Alvarenga, E.R.; Fernandes, A.F.A.; Lopes, L.R.; Soares, T.E.; de Oliveira Alves, G.F.; da Costa, F.F.B.; de Sales, S.C.M.; et al. Attempt to produce a Nile tilapia tetraploid line by heat shock induction. Aquaculture 2020, 529, 735647. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, M.; Jiang, M.; Peng, L.; Wan, C.; Liu, J.; Liu, W.; Zhao, R.; Zhao, X.; Hu, W.; et al. Autotetraploid cell Line induced by SP600125 from crucian carp and its developmental potentiality. Sci. Rep. 2016, 6, 21814. [Google Scholar] [CrossRef] [PubMed]
- Hayward, D.; Alfonso-Pérez, T.; Gruneberg, U. Orchestration of the spindle assembly checkpoint by CDK1-cyclin B1. FEBS Lett. 2019, 593, 2889–2907. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Fan, M.; Candas, D.; Zhang, T.Q.; Qin, L.; Eldridge, A.; Wachsmann-Hogiu, S.; Ahmed, K.M.; Chromy, B.A.; Nantajit, D.; et al. Cyclin B1/Cdk1 Coordinates Mitochondrial Respiration for Cell-Cycle G2/M Progression. Dev. Cell 2014, 29, 217–232. [Google Scholar] [CrossRef]
- Jones, K.T. Cohesin and Cdk1: An anaphase barricade. Nat. Cell Biol. 2010, 12, 106–108. [Google Scholar] [CrossRef]
- Brito, D.A.; Rieder, C.L. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr. Biol. 2006, 16, 1194–1200. [Google Scholar] [CrossRef]
- Wheatley, S.P. CDK1 Inactivation Regulates Anaphase Spindle Dynamics and Cytokinesis In Vivo. J. Cell Biol. 1997, 138, 385–393. [Google Scholar] [CrossRef]
- Schmidt, A.K. Aberrant CDK1 Activity in S Phase Induces Chromosomal Instability by Increasing Mitotic Microtubule Polymerisation Rates in Colorectal Cancer Cells. Ph.D. Thesis, Georg-August Universität, Göttingen, Germany, 2021. [Google Scholar]
- Vázquez-Novelle, M.D.; Mirchenko, L.; Uhlmann, F.; Petronczki, M. The ‘anaphase problem’: How to disable the mitotic checkpoint when sisters split. Biochem. Soc. Trans. 2010, 38, 1660–1666. [Google Scholar] [CrossRef]
- Hervé, S. The Role of Centromere Dysfunction in Genome Instability. Ph.D. Thesis, Université Paris Cité, Paris, France, 2020. [Google Scholar]
- Chotiner, J.Y.; Wolgemuth, D.J.; Wang, P.J. Functions of cyclins and CDKs in mammalian gametogenesis. Biol. Reprod. 2019, 101, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Huang, Q.; Guo, X.; Liu, J.; Feng, D.; Cao, X. Cloning and expression patterns of espl1gene in loach (Misgurnus anguillicaudatus) during gonad development. Isr. J. Aquac.-Bamidgeh 2024, 76, 13–21. [Google Scholar]
- Michaelis, C.; Ciosk, R.; Nasmyth, K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 1997, 91, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Kumada, K.; Yao, R.; Kawaguchi, T.; Karasawa, M.; Hoshikawa, Y.; Ichikawa, K.; Sugitani, Y.; Imoto, I.; Inazawa, J.; Sugawara, M.; et al. The selective continued linkage of centromeres from mitosis to interphase in the absence of mammalian separase. J. Cell Biol. 2006, 172, 835–846. [Google Scholar] [CrossRef]
- Shepard, J.L.; Amatruda, J.F.; Finkelstein, D.; Ziai, J.; Finley, K.R.; Stern, H.M.; Chiang, K.; Hersey, C.; Barut, B.; Freeman, J.L.; et al. A mutation in separase causes genome instability and increased susceptibility to epithelial cancer. Genes Dev. 2007, 21, 55–59. [Google Scholar] [CrossRef]
- Panigrahi, A.K.; Pati, D. Road to the crossroads of life and death: Linking sister chromatid cohesion and separation to aneuploidy, apoptosis and cancer. Crit. Rev. Oncol./Hematol. 2009, 72, 181–193. [Google Scholar] [CrossRef] [PubMed]
| Name | Sequence (5′-3′) | Notes |
|---|---|---|
| β-actin-F | GACCATGCTGTGCAGAGTCGGATA | * |
| β-actin-R | GGGCTGAAGGGACACTTGGGTAATA | * |
| gapdh-F | CCGGCCCATCCATCGTCCAC | * |
| gapdh-R | CTGCTGCATGGCCAGGTATGGT | * |
| ccnb3-F | CCATGCTCATCGCTGCCAAGTT | % |
| ccnb3-R | CTTTGCGTAGCGTCGGAGGAAC | % |
| ccnc-F | CACTGGCGTGGAGGATTGTCAA | % |
| ccnc-R | CAACGGACAGCTCTGCAAACCA | % |
| ccna2-F | TGAACGCACGGTCAAGTGAACA | % |
| ccna2-R | CACGCAGGTATCGGTGGATGTC | % |
| ccnb2-F | ATGAAGGTGATGCCGACATGCC | % |
| ccnb2-R | TCAGCCAGTCAACCAGGAGAGC | % |
| ccnb1-F | AGCCAGTCGCACCTCACTTTCT | % |
| ccnb1-R | CCCGGAGCCAAGGATTCTCAGA | % |
| ccne1-F | CCGCAGGCTACGTTCGTACAAA | % |
| ccne1-R | TCCACTTCACGCACTCCTCCAA | % |
| pttg1-F | GAGAATGGCAGACTGACAA | # |
| pttg1-R | AGACTGAGATGGCACACT | # |
| espl1-F | GGAACAGTCTCCAGGTTAGCG | # |
| espl1-R | ACCACCTCCTCCACAAAC | # |
| smc3-F | ATGCGACCCTGCTCCGTTCT | # |
| smc3-R | ATGTGATTACGGCAGAGCAGGC | # |
| rad21-F | CTGGCAGATTGTAACGAGGC | # |
| rad21-R | GCCCCGTGAATTATGTCCAC | # |
| rec8-F | AACTCCAGCCTCTTCGGTGGTT | # |
| rec8-R | GGGCAGCAGAGAGTGGAAGGTA | # |
| The Time to Start Processing After Fertilization/Min | Eggs Processed/Eggs | Survival Rate at the Tail Bud Stage/% | Incubation Rate/% | Tetraploid Rate/% |
|---|---|---|---|---|
| 0 (Control group) | 410 | 73.17 | 56.10 | 0.00 |
| 20 | 686 | 45.19 | 30.90 | 32.69 |
| 30 | 707 | 33.24 | 17.26 | 39.33 |
| 40 | 690 | 50.00 | 35.22 | 24.33 |
| 50 | 570 | 35.09 | 16.84 | 12.00 |
| Processing Time/Min | Eggs Processed/Eggs | Survival Rate at the Tail Bud Stage/% | Incubation Rate/% | Tetraploid Rate/% |
|---|---|---|---|---|
| 0 (Control group) | 410 | 73.17 | 56.10 | 0 |
| 2 | 156 | 44.87 | 39.74 | 0 |
| 4 | 235 | 42.55 | 34.89 | 44 |
| 6 | 295 | 40.68 | 23.05 | 55 |
| Hybrid Type | Test the Sample | Sample Quantity | Diploid | Tetraploid | Induction Rate/% |
|---|---|---|---|---|---|
| cdk1+/+ ♂ × cdk1+/+ ♀ | Embryo | 14 | 8 | 6 | 42.9% b |
| Juvenile fish | 12 | 9 | 3 | 25.0% a | |
| cdk1+/− ♂ × cdk1+/− ♀ | Embryo | 20 | 11 | 9 | 45.0% b |
| Juvenile fish | 10 | 5 | 5 | 50.0% b | |
| cdk1−/− ♂ × cdk1−/− ♀ | Embryo | 10 | 4 | 6 | 60.0% c |
| cdk1−/− ♂ × cdk1−/− ♀ (Control group) | Embryo | 10 | 10 | 0 | 0.0% a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, H.; Lei, Q.; Ma, W.; Wang, J.; Gong, J.; Guo, X.; Cao, X. The Effect of cdk1 Gene Knockout on Heat Shock-Induced Polyploidization in Loach (Misgurnus anguillicaudatus). Life 2025, 15, 1223. https://doi.org/10.3390/life15081223
Jiang H, Lei Q, Ma W, Wang J, Gong J, Guo X, Cao X. The Effect of cdk1 Gene Knockout on Heat Shock-Induced Polyploidization in Loach (Misgurnus anguillicaudatus). Life. 2025; 15(8):1223. https://doi.org/10.3390/life15081223
Chicago/Turabian StyleJiang, Hanjun, Qi Lei, Wenhao Ma, Junru Wang, Jing Gong, Xusheng Guo, and Xiaojuan Cao. 2025. "The Effect of cdk1 Gene Knockout on Heat Shock-Induced Polyploidization in Loach (Misgurnus anguillicaudatus)" Life 15, no. 8: 1223. https://doi.org/10.3390/life15081223
APA StyleJiang, H., Lei, Q., Ma, W., Wang, J., Gong, J., Guo, X., & Cao, X. (2025). The Effect of cdk1 Gene Knockout on Heat Shock-Induced Polyploidization in Loach (Misgurnus anguillicaudatus). Life, 15(8), 1223. https://doi.org/10.3390/life15081223





