The Complete Mitochondrial Genome of a Natural Triploid Crucian Carp Mutant, Carassius auratus var. suogu, and Its Phylogenetic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling, DNA Extraction, and Polymerase Chain Reaction (PCR)
2.2. De Novo Assembly and Annotation of the Mitochondrial Genome
2.3. Phylogenetic Analysis
2.4. Data Availability
3. Results
3.1. Genome Content and Organization
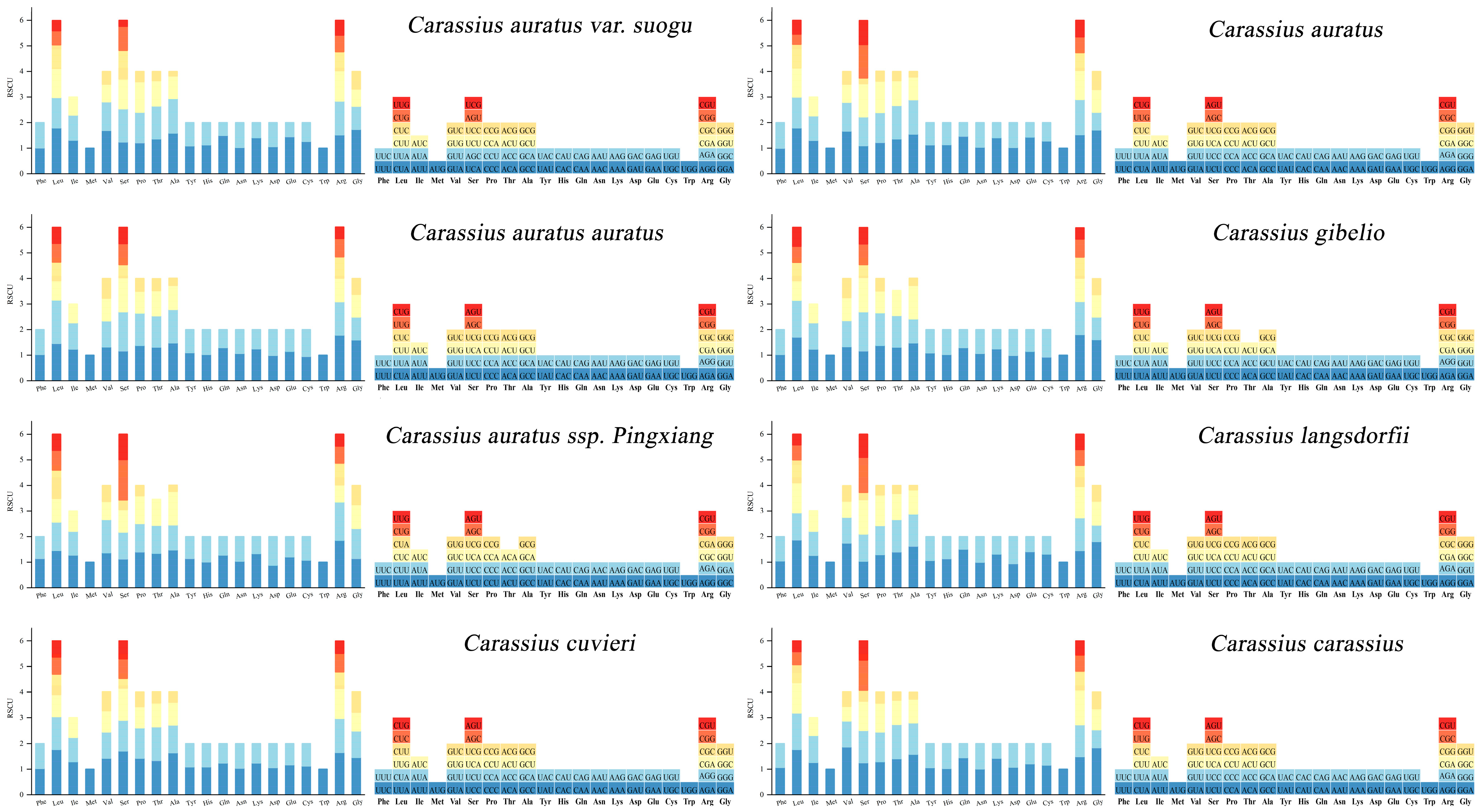
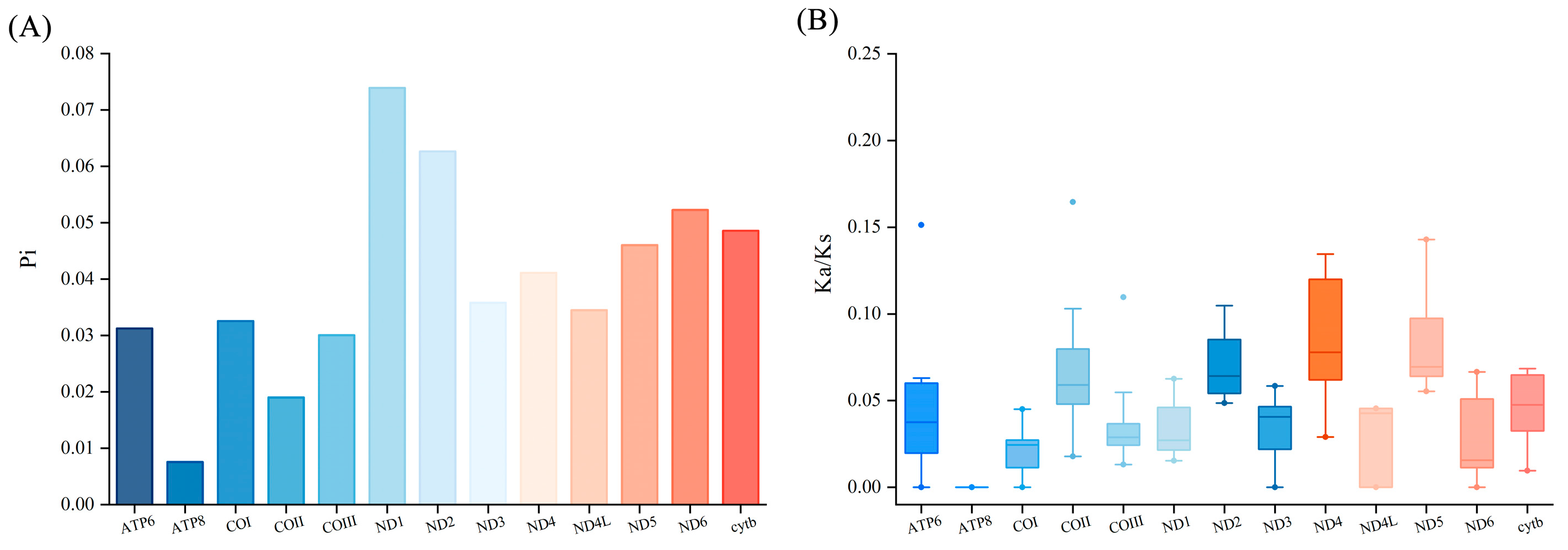
3.2. Protein-Coding Gene (PCGs) and Relative Synonymous Codon Usage (RSCU)
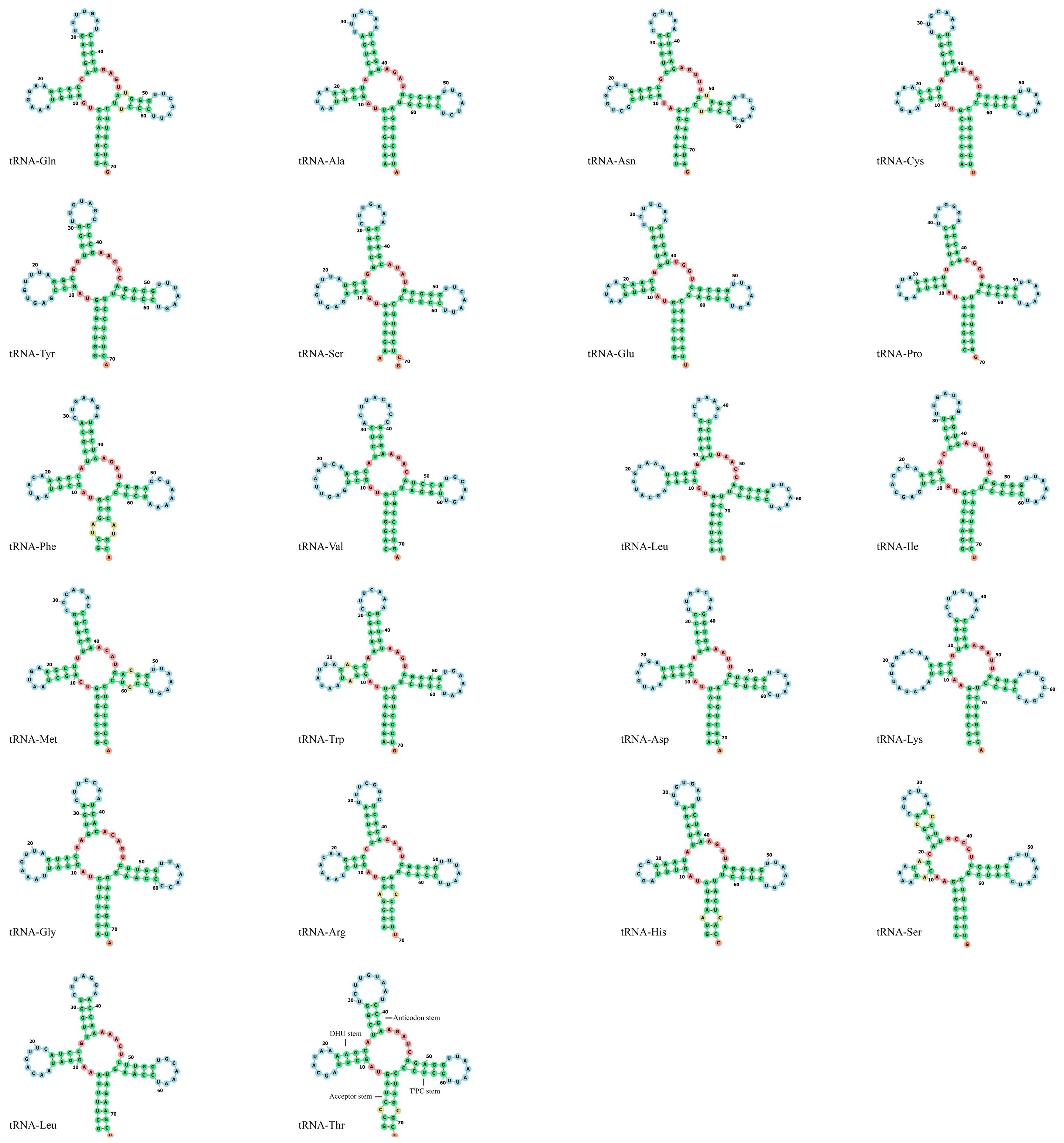
3.3. Transfer and Ribosomal RNA Genes, Non-Coding Region
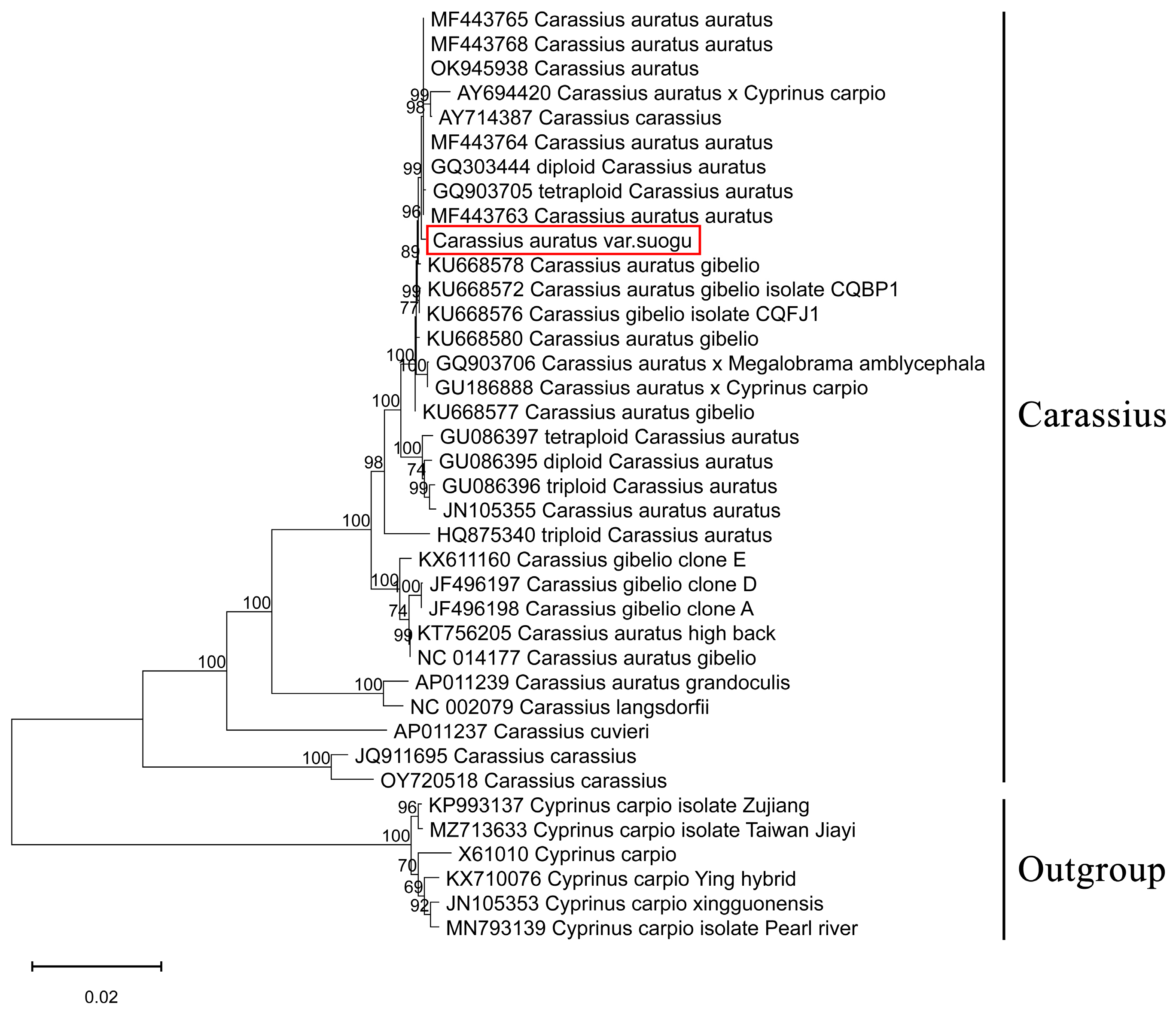
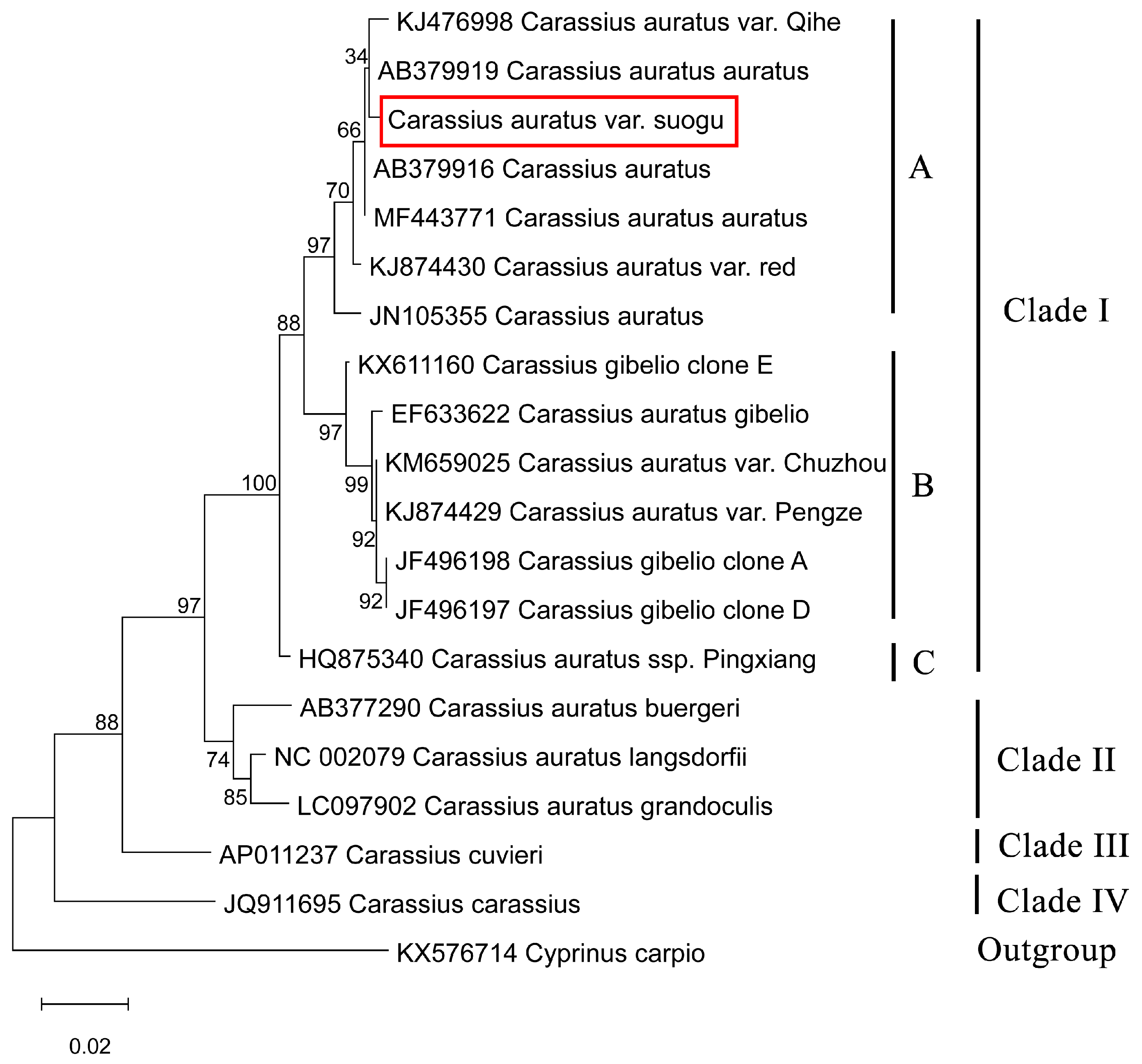
3.4. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; De Bruijn, M.H.L.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [PubMed]
- Smith, J.J.; Voss, S.R. Gene order data from a model amphibian (Ambystoma): New perspectives on vertebrate genome structure and evolution. BMC Genom. 2006, 7, 219. [Google Scholar] [CrossRef] [PubMed]
- Duchene, S.; Frey, A.; Alfaro-Núñez, A.; Dutton, P.H.; Gilbert, M.T.P.; Morin, P.A. Marine turtle mitogenome phylogenetics and evolution. Mol. Phylogenetics Evol. 2012, 65, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Wang, J.; He, S.; Zeng, L.; Peng, K.; Hong, Y. Complete mitochondrial genome of a natural triploid crucian carp mutant, Carassius auratus var. pingxiangnensis, and phylogenetic analysis of different ploidies in crucian carp. Genet. Mol. Res. 2014, 13, 5849–5864. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.Q.; Zeng, L.G.; Hong, Y.J.; Sheng, J.Q.; Wang, J.H.; Guan, J.F. Sequence Analysis of Mitochondrial Cytochrome b Gene from Natural Triploid Mutant, Carassius auratus var. pingxiangnensis. J. Zool. 2009, 44, 97–101, (In Chinese with English Abstract). [Google Scholar]
- Chen, S.; Wang, J.; Liu, S.; Qin, Q.; Xiao, J.; Duan, W.; Luo, K.; Liu, J.; Liu, Y. Study on biological characteristics of improved triploid crucian carp. Chin. Sci. 2009, 39, 479–484, (In Chinese with English Abstract). [Google Scholar]
- Liu, S.J.; Zhou, G.J.; Luo, K.K.; Qin, Q.B.; Duan, W.; Tao, M.; Zhang, C.; Yao, Z.Z.; Feng, H.; Liu, J. Research and Application of New and Improved Polyploid Fish; Scientific and Technological Achievement; Hunan Province, Hunan Normal University: Changsha, China, 2016; (In Chinese with English Abstract). [Google Scholar]
- Yu, H.X.; Zhang, H.M.; Lin, L.Y. A preliminary study on the biology and culture experiment of the gynogenetic crucian carp (Carassius auratus) of Guangdong. Acta Hydrobiol. Sin. 1987, 11, 287–288, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Yu, H.X.; Zhang, H.M. Preliminary report on the biology and culture experiment of Carassius auratus var. suogu. Fish. Sci. Technol. Inf. 1986, 3, 21, (In Chinese with English Abstract). [Google Scholar]
- Yang, Y.; Zhou, H.Q.; Shu, H.; Lan, Z.J.; Li, Q. Clone, expression, and screening of growth-associated SNPs of Carassius auratus var. suogu. Oceanol. Et Limnol. Sin. 2017, 48, 830–837, (In Chinese with English Abstract). [Google Scholar]
- Jiang, Y.; Yu, H.; Chen, B.; Liang, S.; Yang, D.; Lin, S. Artificial and natural gynogenesis in crucian carp. Acta Hydrobiol. Sin. 1982, 7, 471–477, 479–480, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Liu, L.G.; Zhao, J.; Chen, X.L. Research progress on fish gynogenesis. J. Aquac. 2002, 6, 39–43, (In Chinese with English Abstract). [Google Scholar]
- Zhang, Z.; Li, S.; Zhang, J.; Song, W.; Yang, J.; Mu, J. The complete mitochondrial genome of an endangered minnow Aphyocypris lini (Cypriniformes: Xenocyprididae): Genome characterization and phylogenetic consideration. Biologia 2021, 76, 3311–3321. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Iwasaki, W.; Fukunaga, T.; Isagozawa, R.; Yamada, K.; Maeda, Y.; Satoh, T.P.; Sado, T.; Mabuchi, K.; Takeshima, H.; Miya, M.; et al. MitoFish and MitoAnnotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol. Biol. Evol. 2013, 30, 2531–2540. [Google Scholar] [CrossRef] [PubMed]
- Peden, J.F. Analysis of Codon Usage; University of Nottingham: Nottingham, UK, 2000. [Google Scholar]
- Zhong, Z.; Norvienyeku, J.; Chen, M.; Bao, J.; Lin, L.; Chen, L.; Lin, Y.; Wu, X.; Cai, Z.; Zhang, Q.; et al. Directional selection from host plants is a major force driving host specificity in Magnaporthe species. Sci. Rep. 2016, 6, 25591. [Google Scholar] [CrossRef] [PubMed]
- Vivas-Toro, I.; Ortega, J.; Baeza, J.A. The complete mitochondrial genome of the Honduran white bat Ectophylla alba (Allen 1982) (Chiroptera: Phyllostomidae). Gene 2021, 802, 145868. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Darty, K.; Denise, A.; Ponty, Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 2009, 25, 1974. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K.; Battistuzzi, F.U. MEGA12: Molecular Evolutionary Genetic Analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef] [PubMed]
- Baek, I.G.; Kim, Y.H.; Han, H.S.; Huynh, D.T.; Bang, I.C. Complete mitochondrial genome sequence and phylogenetic analysis of the hybrid flat fish Platichthys stellatus (♀)× Platichthys bicoloratus (♂). Mitochondrial DNA Part B 2024, 9, 1317–1321. [Google Scholar] [CrossRef] [PubMed]
- Fritzsch, G.; Schlegel, M.; Stadler, P.F. Alignments of mitochondrial genome arrangements: Applications to metazoan phylogeny. J. Theor. Biol. 2006, 240, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.L. Mitochondrial genome organization and vertebrate phylogenetics. Genet. Mol. Biol. 2000, 23, 745–752. [Google Scholar] [CrossRef]
- Asakawa, S.; Kumazawa, Y.; Araki, T.; Himeno, H.; Miura, K.I.; Watanabe, K. Strand-specific nucleotide composition bias in echinoderm and vertebrate mitochondrial genomes. J. Mol. Evol. 1991, 32, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.P.; Miya, M.; Mabuchi, K.; Nishida, M. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 2016, 17, 719. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, Q.; Lu, G.; Xu, J.; Yang, Q.; Li, S. Complete mitochondrial genome of the grass carp (Ctenopharyngodon idella, Teleostei): Insight into its phylogenic position within Cyprinidae. Gene 2008, 424, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Huang, F.; Lo, T. The complete nucleotide sequence and gene organization of carp (Cyprinus carpio) mitochondrial genome. J. Mol. Evol. 1994, 38, 138–155. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Liang, L.Q.; Sun, X.W. The complete mitochondrial genome of the crucian carp, Carassius carassius (Cypriniformes, Cyprinidae). Mitochondrial DNA 2012, 23, 350–351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yue, B.; Jiang, W.; Song, Z. The complete mitochondrial genome of rock carp Procypris rabaudi (Cypriniformes: Cyprinidae) and phylogenetic implications. Mol. Biol. Rep. 2009, 36, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Liu, T.-J.; Hao, G.; Ge, X.-J.; Yan, H.-F. Comparative analyzes of three complete Primula mitogenomes with insights into mitogenome size variation in Ericales. BMC Genom. 2022, 23, 770. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Li, B.; Ma, L.; Zhao, Y.; Kong, X. The complete mitogenome of natural triploid Carassius auratus in Qihe River. Mitochondrial DNA Part A 2016, 27, 605–606. [Google Scholar] [CrossRef] [PubMed]
- Miya, M.; Takeshima, H.; Endo, H.; Ishiguro, N.B.; Inoue, J.G.; Mukai, T.; Satoh, T.P.; Yamaguchi, M.; Kawaguchi, A.; Mabuchi, K.; et al. Major patterns of higher teleostean phylogenies: A new perspective based on 100 complete mitochondrial DNA sequences. Mol. Phylogenetics Evol. 2003, 26, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Inoue, J.G.; Miya, M.; Tsukamoto, K.; Nishida, M. Evolution of the deep-sea gulper eel mitochondrial genomes: Large-scale gene rearrangements originated within the eels. Mol. Biol. Evol. 2003, 20, 1917–1924. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Xu, W.; Zhang, G.; Liu, Z.; Liu, H. Characterization of the complete mitochondrial genomes of four tarantulas (Arachnida: Theraphosidae) with phylogenetic analysis. Gene 2025, 933, 148954. [Google Scholar] [CrossRef] [PubMed]
- Jühling, F.; Pütz, J.; Bernt, M.; Donath, A.; Middendorf, M.; Florentz, C.; Stadler, P.F. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 2012, 40, 2833–2845. [Google Scholar] [CrossRef] [PubMed]
- Parvathy, S.T.; Udayasuriyan, V.; Bhadana, V. Codon usage bias. Mol. Biol. Rep. 2022, 49, 539–565. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Zou, T.; Chen, Y.; Chen, L.; Liu, S.; Tao, M.; Zhang, C.; Zhao, R.; Zhou, Y.; Long, Y.; et al. Coexistence of diploid, triploid and tetraploid crucian carp (Carassius auratus) in natural waters. BMC Genet. 2011, 12, 20, (In Chinese with English Abstract). [Google Scholar] [CrossRef] [PubMed]
- Takada, M.; Tachihara, K.; Kon, T.; Yamamoto, G.; Iguchi, K.; Miya, M.; Nishida, M. Biogeography and evolution of the Carassius auratus-complex in East Asia. BMC Evol. Biol. 2010, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.W. Fauna of Cyprinidae in China; Shanghai Scientific & Technical Publishers: Shanghai, China, 1977; pp. 395–438, (In Chinese with English Abstract). [Google Scholar]
- Yamamoto, G.; Takada, M.; Iguchi, K.; Nishida, M. Genetic constitution and phylogenetic relationships of Japanese crucian carps (Carassius). Ichthyol. Res. 2010, 57, 215–222. [Google Scholar] [CrossRef]
- Lu, C.Y.; Yang, Y.H.; Dong, G.X.; Hao, J.; Sun, X.W.; Liang, L.Q.; Lei, Q.Q. Comparative studies of genetic diversity between C. auratus gibelio (Bloch) and C. Auratus Auratus (L.) in the same reservoir. Chin. J. Fish. 2006, 19, 42–50, (In Chinese with English Abstract). [Google Scholar]
- Li, Q.; Zheng, P.; Huang, W.W.; Peng, S.H.; Zhou, J.W. Sequence cloning and phylogenetic analysis of Cyt b gene and control region in mtdna of carassius auratus var. Suogu. Hunan Agric. Sci. 2023, 10, 7–10+2, (In Chinese with English Abstract). [Google Scholar]
- Hu, Y.T.; Jiang, H.; Duan, G.Q.; Lin, J.; Pan, T.S.; Zhou, X.H.; Wang, H. Sequence and phylogenetic analysis of the complete mitochondrial genome of Chuzhou Crucian Carp (Carassius auratus). Prog. Fish. Sci. 2015, 36, 63–70, (In Chinese with English Abstract). [Google Scholar]


| Gene/Element | Strand | Position | Size (bp) | Start Codon | Stop Codon a | Anticodon | Intergenic Nucleotide b |
|---|---|---|---|---|---|---|---|
| tRNA-Phe | H | 1–69 | 69 | GAA | 0 | ||
| 12S rRNA | H | 70–1023 | 954 | 0 | |||
| tRNA-Val | H | 1024–1095 | 72 | TAC | 0 | ||
| 16S rRNA | H | 1096–2777 | 1682 | 0 | |||
| tRNA-Leu(UUR) | H | 2778–2853 | 76 | TAA | +1 | ||
| ND1 | H | 2855–3829 | 975 | ATG | TAA | +4 | |
| tRNA-IIe | H | 3834–3905 | 72 | GAT | −2 | ||
| tRNA-Gln | L | 3904–3974 | 71 | TTG | +1 | ||
| tRNA-Met | H | 3976–4044 | 69 | CAT | +10 | ||
| ND2 | H | 4055–5090 | 1036 | ACG | T-- | 0 | |
| tRNA-Trp | H | 5091–5161 | 71 | TCA | +2 | ||
| tRNA-Ala | L | 5164–5232 | 69 | TGC | +1 | ||
| tRNA-Asn | L | 5234–5306 | 73 | GTT | +33 | ||
| tRNA-Cys | L | 5340–5408 | 69 | GCA | 1 | ||
| tRNA-Tyr | L | 5408–5478 | 71 | GTA | +1 | ||
| COI | H | 5480–7030 | 1551 | GTG | TAA | 0 | |
| tRNA-Ser(UCN) | L | 7031–7101 | 71 | TGA | +3 | ||
| tRNA-Asp | H | 7105–7176 | 72 | GTC | +2 | ||
| COII | H | 7189–7879 | 691 | ATG | T-- | 0 | |
| tRNA-Lys | H | 7880–7955 | 76 | TTT | +1 | ||
| ATPase 8 | H | 7957–8121 | 165 | ATG | TAG | −7 | |
| ATPase 6 | H | 8115–8486 | 372 | ATG | TAG | +310 | |
| COIII | H | 8797–9581 | 785 | ATG | TA- | 0 | |
| tRNA-Gly | H | 9582–9653 | 72 | TCC | 0 | ||
| ND3 | H | 9654–10,002 | 349 | ATG | T-- | 0 | |
| tRNA-Arg | H | 10,003–10,072 | 70 | TCG | 0 | ||
| ND4L | H | 10,073–10,369 | 297 | ATG | TAA | −7 | |
| ND4 | H | 10,363–11,743 | 1381 | ATG | T-- | 0 | |
| tRNA-His | H | 11,744–11,812 | 69 | GTG | 0 | ||
| tRNA-Ser(AGY) | H | 11,813–11,881 | 69 | GCT | +1 | ||
| tRNA-Leu(CUN) | H | 11,883–11,955 | 73 | TAG | +3 | ||
| ND5 | H | 11,959–13,782 | 1824 | ATG | TAA | −4 | |
| ND6 | L | 13,779–14,300 | 522 | ATG | TAG | 0 | |
| tRNA-Glu | L | 14,301–14,369 | 69 | TTC | +5 | ||
| Cyt-b | H | 14,375–15,515 | 1141 | ATG | T-- | 0 | |
| tRNA-Thr | H | 15,516–15,587 | 72 | TGT | −1 | ||
| tRNA-Pro | L | 15,587–15,656 | 70 | TGG | 0 | ||
| D-loop | H | 15,657–16,580 | 924 | 0 |
| Gene | AT Content (%)/AT Skew of CDS | |||||||
|---|---|---|---|---|---|---|---|---|
| C. auratus var. suogu | C. auratus | C. auratus auratus | C. gibelio | |||||
| ND1 | 58.46 | 0.039 | 58.55 | 0.038 | 58.48 | 0.039 | 58.48 | 0.039 |
| ND2 | 57.80 | 0.047 | 57.89 | 0.046 | 57.82 | 0.047 | 57.82 | 0.047 |
| COI | 56.80 | 0.023 | 56.76 | 0.024 | 56.81 | 0.023 | 56.81 | 0.023 |
| COII | 57.31 | 0.032 | 57.42 | 0.031 | 57.35 | 0.032 | 57.35 | 0.032 |
| ATP8 | 58.18 | 0.048 | 58.18 | 0.047 | 58.18 | 0.048 | 58.18 | 0.048 |
| ATP6 | 57.46 | 0.035 | 57.53 | 0.034 | 57.49 | 0.035 | 57.49 | 0.035 |
| COIII | 56.94 | 0.028 | 56.89 | 0.027 | 56.91 | 0.028 | 56.91 | 0.028 |
| ND3 | 58.17 | 0.041 | 58.22 | 0.040 | 58.20 | 0.041 | 58.20 | 0.041 |
| ND4L | 57.89 | 0.033 | 57.97 | 0.032 | 57.93 | 0.033 | 57.93 | 0.033 |
| ND4 | 57.06 | 0.029 | 57.12 | 0.028 | 57.09 | 0.029 | 57.09 | 0.029 |
| ND5 | 57.24 | 0.031 | 57.31 | 0.030 | 57.27 | 0.031 | 57.27 | 0.031 |
| ND6 | 58.24 | 0.043 | 58.31 | 0.042 | 58.28 | 0.043 | 58.28 | 0.043 |
| cytb | 56.62 | 0.025 | 56.68 | 0.026 | 56.65 | 0.025 | 56.65 | 0.025 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Deng, B.; Lin, S.; Ye, S.; Zheng, P.; Cai, G.; Liang, W.; Han, C.; Li, Q. The Complete Mitochondrial Genome of a Natural Triploid Crucian Carp Mutant, Carassius auratus var. suogu, and Its Phylogenetic Analysis. Life 2025, 15, 1156. https://doi.org/10.3390/life15081156
Zhou Y, Deng B, Lin S, Ye S, Zheng P, Cai G, Liang W, Han C, Li Q. The Complete Mitochondrial Genome of a Natural Triploid Crucian Carp Mutant, Carassius auratus var. suogu, and Its Phylogenetic Analysis. Life. 2025; 15(8):1156. https://doi.org/10.3390/life15081156
Chicago/Turabian StyleZhou, Yicheng, Binhua Deng, Shengyue Lin, Shuzheng Ye, Peng Zheng, Guojun Cai, Weiqian Liang, Chong Han, and Qiang Li. 2025. "The Complete Mitochondrial Genome of a Natural Triploid Crucian Carp Mutant, Carassius auratus var. suogu, and Its Phylogenetic Analysis" Life 15, no. 8: 1156. https://doi.org/10.3390/life15081156
APA StyleZhou, Y., Deng, B., Lin, S., Ye, S., Zheng, P., Cai, G., Liang, W., Han, C., & Li, Q. (2025). The Complete Mitochondrial Genome of a Natural Triploid Crucian Carp Mutant, Carassius auratus var. suogu, and Its Phylogenetic Analysis. Life, 15(8), 1156. https://doi.org/10.3390/life15081156




