Tapping into Metabolomics for Understanding Host and Rotavirus Group A Interactome
Abstract
1. Introduction
2. Materials and Method
2.1. Ethical Approval
2.2. Sample Population and Collection
2.3. Metabolic Molecular Profiling
2.3.1. Reagents and Chemicals
2.3.2. Preparation of Samples
2.3.3. Two-Dimensional Gas Chromatography with Time-of-Flight Mass Spectrometer System
2.3.4. Data Processing for Peak Identification
2.4. Integration of Metabolomics with Lipidomic and Proteomics Metadata Analyses
2.5. Data Analysis
3. Results
3.1. Saliva Quantification
3.2. Univariate and Chemometrics Analysis
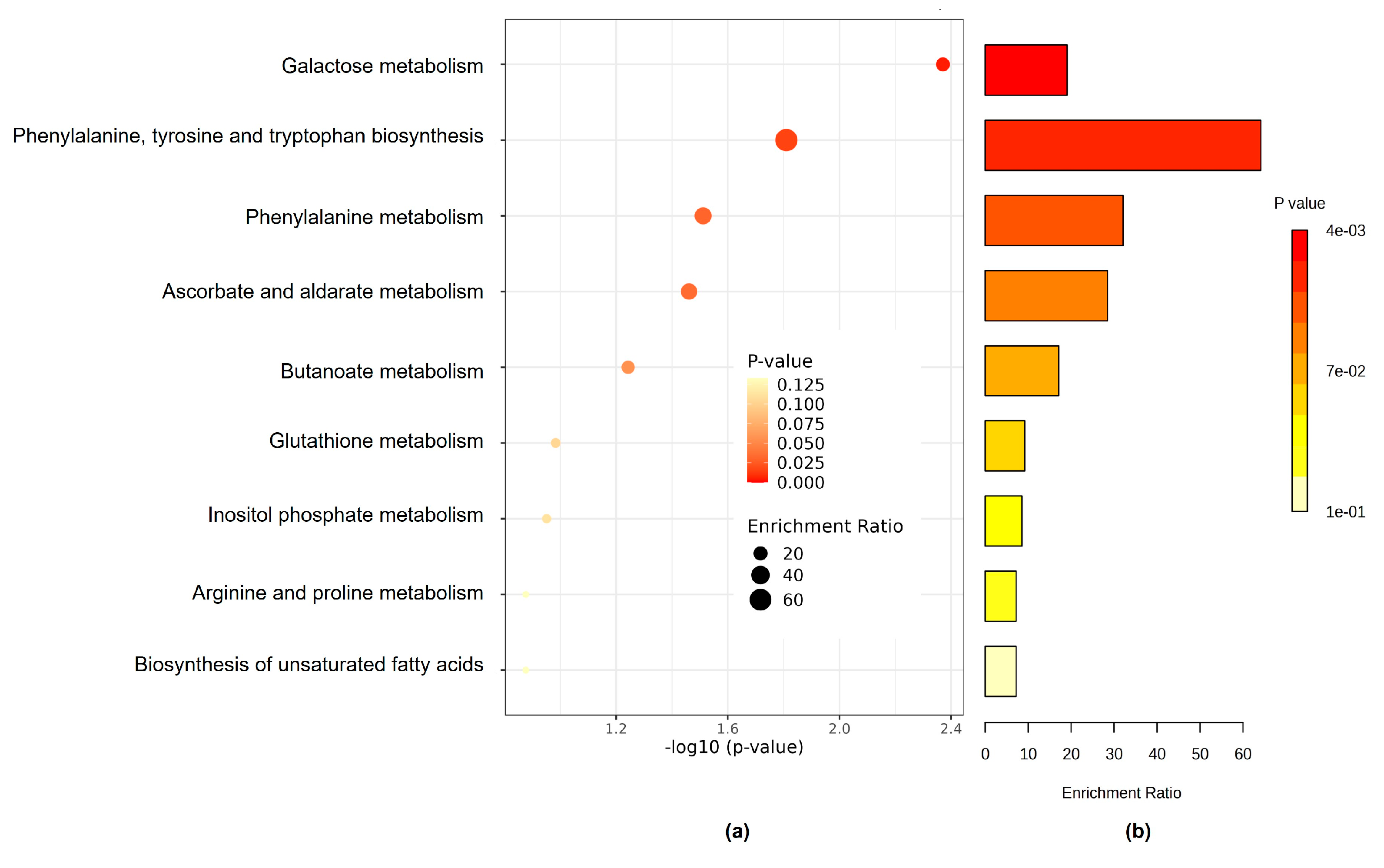
3.3. The Kyoto Encyclopedia of Genes and Genome Pathway
3.4. Integration of Metabolomics with Lipidomic and Proteomics Metadata Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crawford, S.E.; Ramani, S.; Tate, J.E.; Parashar, U.D.; Svensson, L.; Hagbom, M.; Franco, M.A.; Greenberg, H.B.; O’Ryan, M.; Kang, G.; et al. Rotavirus infection. Nat. Rev. Dis. Primers 2017, 9, 17083. [Google Scholar] [CrossRef] [PubMed]
- Bishop, R.F.; Davidson, G.P.; Holmes, I.H.; Ruck, B.J. Virus particles in epithelial cells of duodenal mucosa from children with acute non-bacterial gastroenteritis. Lancet 1973, 8, 1281–1283. [Google Scholar] [CrossRef] [PubMed]
- Desselberger, U. Genome rearrangements of rotaviruses. Adv. Virus Res. 1996, 46, 69–95. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Attoui, H.; Bányai, K.; Brussaard, C.; Danthi, P.; Del Vas, M.; Dermody, T.; Duncan, R.; Fang, Q.; Johne, R.; et al. ICTV Virus Taxonomy Profile: Sedoreoviridae 2022. J. Gen. Virol. 2022, 103, 001782. [Google Scholar] [CrossRef]
- Rodríguez, J.; Chichón, F.; Martín-Forero, E.; González-Camacho, F.; Carrascosa, J.; Castón, J.; Luque, D. New Insights into Rotavirus Entry Machinery: Stabilization of Rotavirus Spike Conformation Is Independent of Trypsin Cleavage. PLoS Pathog. 2014, 10, e1004157. [Google Scholar] [CrossRef]
- Greenberg, H.; Estes, M. Rotaviruses: From Pathogenesis to Vaccination. Gastroenterology 2009, 136, 1939–1951. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Heylen, E.; Zeller, M.; Rahman, M.; Lemey, P.; Van Ranst, M. Phylodynamic Analyses of Rotavirus Genotypes G9 and G12 Underscore Their Potential for Swift Global Spread. Mol. Biol. Evol. 2010, 27, 2431–2436. [Google Scholar] [CrossRef]
- Dóró, R.; László, B.; Martella, V.; Leshem, E.; Gentsch, J.; Parashar, U.; Bányai, K. Review of Global Rotavirus Strain Prevalence Data from Six Years Post Vaccine Licensure Surveillance: Is There Evidence of Strain Selection from Vaccine Pressure? Infect. Genet. Evol. 2014, 28, 446–461. [Google Scholar] [CrossRef] [PubMed]
- Santos, N.; Hoshino, Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 2005, 15, 29–56. [Google Scholar] [CrossRef]
- Unicomb, L.E.; Podder, G.; Gentsch, J.R.; Woods, P.A.; Hasan, K.Z.; Faruque, A.S.; Albert, M.J.; Glass, R.I. Evidence of high-frequency genomic reassortment of group A rotavirus strains in Bangladesh: Emergence of type G9 in 1995. J. Clin. Microbiol. 1999, 37, 1885–1891. [Google Scholar] [CrossRef]
- Iturriza-Gómara, M.; Isherwood, B.; Desselberger, U.; Gray, J. Reassortment in vivo: Driving force for diversity of human rotavirus strains isolated in the United Kingdom between 1995 and 1999. J. Virol. 2001, 75, 3696–3705. [Google Scholar] [CrossRef] [PubMed]
- Palombo, E.A. Genetic and antigenic diversity of human rotaviruses: Potential impact on the success of candidate vaccines. FEMS Microbiol. Lett. 1999, 181, 1–8. [Google Scholar] [CrossRef]
- Huang, P.; Xia, M.; Tan, M.; Zhong, W.; Wei, C.; Wang, L.; Morrow, A.; Jiang, X. Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J. Virol. 2012, 86, 4833–4843. [Google Scholar] [CrossRef]
- Ravn, V.; Dabelsteen, E. Tissue distribution of histo-blood group antigens. APMIS 2000, 108, 1–28. [Google Scholar] [CrossRef]
- Koda, Y.; Soejima, M.; Kimura, H. The polymorphisms of fucosyltransferases. Leg. Med. 2001, 3, 2–14. [Google Scholar] [CrossRef]
- Marionneau, S.; Cailleau-Thomas, A.; Rocher, J.; Le Moullac-Vaidye, B.; Ruvoën, N.; Clément, M.; Le Pendu, J. ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie 2001, 83, 565–573. [Google Scholar] [CrossRef] [PubMed]
- de Mattos, L.C. Structural diversity and biological importance of ABO, H, Lewis and secretor histo-blood group carbohydrates. Rev. Bras. Hematol. Hemoter. 2016, 38, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Troeger, C.; Khalil, I.A.; Rao, P.C.; Cao, S.; Blacker, B.F.; Ahmed, T.; Armah, G.; Bines, J.E.; Brewer, T.G.; Colombara, D.V.; et al. Rotavirus Vaccination and the Global Burden of Rotavirus Diarrhea Among Children Younger Than 5 Years. JAMA Pediatr. 2018, 172, 958–965. [Google Scholar] [CrossRef]
- Khan, M.M.; Ernst, O.; Manes, N.P.; Oyler, B.L.; Fraser, I.D.C.; Goodlett, D.R.; Nita-Lazar, A. Multi-Omics Strategies Uncover Host-Pathogen Interactions. ACS Infect. Dis. 2019, 5, 493–505. [Google Scholar] [CrossRef]
- Gomes, F.; Alfson, K.; Junqueira, M. Editorial: The application of OMICS technologies to interrogate host-virus interactions. Front. Cell. Infect. Microbiol. 2022, 12, 1050012. [Google Scholar] [CrossRef]
- Mwenda, J.M.; Ntoto, K.M.; Abebe, A.; Enweronu-Laryea, C.; Amina, I.; Mchomvu, J.; Kisakye, A.; Mpabalwani, E.M.; Pazvakavambwa, I.; Armah, G.E.; et al. Burden and epidemiology of rotavirus diarrhea in selected African countries: Preliminary results from the African Rotavirus Surveillance Network. J. Infect. Dis. 2010, 202, S5–S11. [Google Scholar] [CrossRef] [PubMed]
- Rakau, K.; Gededzha, M.; Peenze, I.; Huang, P.; Tan, M.; Steele, A.D.; Seheri, L.M. The Association between Symptomatic Rotavirus Infection and Histo-Blood Group Antigens in Young Children with Diarrhea in Pretoria, South Africa. Viruses 2022, 14, 2735. [Google Scholar] [CrossRef]
- Mueller, D.C.; Piller, M.; Niessner, R.; Scherer, M.; Scherer, G. Untargeted metabolomic profiling in saliva of smokers and nonsmokers by a validated GC-TOF-MS method. J. Proteome Res. 2014, 13, 1602–1613. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef] [PubMed]
- Thyrsted, J.; Holm, C.K. Virus-induced metabolic reprogramming and innate sensing hereof by the infected host. Curr. Opin. Biotechnol. 2021, 68, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, K.A.; Sanchez, E.L.; Camarda, R.; Lagunoff, M. Dengue virus induces and requires glycolysis for optimal replication. J. Virol. 2015, 89, 2358–2366. [Google Scholar] [CrossRef]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef]
- Gardner, A.; Carpenter, G.; So, P.W. Salivary metabolomics: From diagnostic biomarker discovery to investigating biological function. Metabolites 2020, 10, 47. [Google Scholar] [CrossRef]
- Kashyap, B.; Kullaa, A. Salivary metabolites produced by oral microbes in oral diseases and oral squamous cell carcinoma: A Review. Metabolites 2024, 14, 277. [Google Scholar] [CrossRef]
- Ciurli, A.; Mohammed, Y.; Ammon, C.; Derks, R.J.E.; Olivier-Jimenez, D.; Ducarmon, Q.R.; Slingerland, M.; Neefjes, J.; Giera, M. Spatially and temporally resolved metabolome of the human oral cavity. iScience 2024, 27, 108884. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Hoefsloot, H.C.; Smit, S.; Vis, D.J.; Smilde, A.K.; van Velzen, E.J.; van Duijnhoven, J.P.; van Dorsten, F.A. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef]
- Kjeldahl, K.; Bro, R. Some common misunderstandings in chemometrics. J. Chemom. 2010, 24, 558–564. [Google Scholar] [CrossRef]
- Mazzara, S.; Sinisi, A.; Cardaci, A.; Rossi, R.L.; Muratori, L.; Abrignani, S.; Bombaci, M. Two of them do it better: Novel serum biomarkers improve autoimmune Hepatitis diagnosis. PLoS ONE 2015, 10, e0137927. [Google Scholar] [CrossRef] [PubMed]
- Vinaixa, M.; Samino, S.; Saez, I.; Duran, J.; Guinovart, J.J.; Yanes, O. A Guideline to univariate statistical analysis for LC/MS-based untargeted metabolomics-derived data. Metabolites 2012, 2, 775–795. [Google Scholar] [CrossRef]
- Patti, G.J.; Tautenhahn, R.; Siuzdak, G. Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nat. Protoc. 2012, 7, 508–516. [Google Scholar] [CrossRef]
- Boye, T.L.; Hammerhøj, A.; Nielsen, O.H.; Wang, Y. Metabolomics for enhanced clinical understanding of inflammatory bowel disease. Life Sci. 2024, 359, 123238. [Google Scholar] [CrossRef]
- Canani, R.B.; Costanzo, M.D.; Leone, L.; Pedata, M.; Meli, R.; Calignano, A. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J. Gastroenterol. 2011, 17, 1519–1528. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef]
- Liu, H.; Wang, J.; He, T.; Becker, S.; Zhang, G.; Li, D.; Ma, X. Butyrate: A Double-Edged Sword for Health? Adv. Nutr. 2018, 9, 21–29. [Google Scholar] [CrossRef]
- Kasubuchi, M.; Hasegawa, S.; Hiramatsu, T.; Ichimura, A.; Kimura, I. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients 2015, 7, 2839–2849. [Google Scholar] [CrossRef]
- Bjerrum, J.T.; Wang, Y.; Hao, F.; Coskun, M.; Ludwig, C.; Günther, U.; Nielsen, O.H. Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn’s disease and healthy individuals. Metabolomics 2015, 11, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.R.; Holmes, E.; Khan, F.; Kochhar, S.; Scanlan, P.; Shanahan, F.; Wilson, I.D.; Wang, Y. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J. Proteome Res. 2007, 6, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hu, N.; Jiang, Q.; Zhu, L.; Zhang, M.; Jiang, J.; Xiong, M.; Yang, M.; Yang, J.; Shen, L.; et al. Protective effects of sodium butyrate on rotavirus inducing endoplasmic reticulum stress-mediated apoptosis via PERK-eIF2α signaling pathway in IPEC-J2 cells. J. Anim. Sci. Biotechnol. 2021, 12, 69. [Google Scholar] [CrossRef]
- Dong, X.; Wang, Y.; Zhu, X.; Shen, L.; Chen, L.; Niu, L.; Gan, M.; Zhang, S.; Zhang, M.; Jiang, J.; et al. Sodium butyrate protects against rotavirus-induced intestinal epithelial barrier damage by activating AMPK-Nrf2 signaling pathway in IPEC-J2 cells. Int. J. Biol. Macromol. 2023, 228, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Kim, S.Y.; Chun, Y.S.; Chun, Y.J.; Shin, S.Y.; Choi, C.H.; Choi, H.K. Characteristics of fecal metabolic profiles in patients with irritable bowel syndrome with predominant diarrhea investigated using 1H-NMR coupled with multivariate statistical analysis. Neurogastroenterol. Motil. 2020, 32, e13830. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, Z. Unraveling the causal link: Fatty acids and inflammatory bowel disease. Front. Immunol. 2024, 15, 1405790. [Google Scholar] [CrossRef]
- Weng, Y.J.; Gan, H.Y.; Li, X.; Huang, Y.; Li, Z.C.; Deng, H.M.; Chen, S.Z.; Zhou, Y.; Wang, L.S.; Han, Y.P.; et al. Correlation of diet, microbiota and metabolite networks in inflammatory bowel disease. J. Dig. Dis. 2019, 20, 447–459. [Google Scholar] [CrossRef]
- Li, H.; Wen, Z. Effects of ulcerative colitis and Crohn’s disease on neurodegenerative diseases: A Mendelian randomization study. Front. Genet. 2022, 13, 846005. [Google Scholar] [CrossRef]
- Haikonen, R.; Kärkkäinen, O.; Koistinen, V.; Hanhineva, K. Diet- and microbiota-related metabolite, 5-aminovaleric acid betaine (5-AVAB), in health and disease. Trends Endocrinol. Metab. 2022, 33, 463–480. [Google Scholar] [CrossRef]
- Liebich, H.M.; Pickert, A.; Stierle, U.; Wöll, J. Gas chromatography-mass spectrometry of saturated and unsaturated dicarboxylic acids in urine. J. Chromatogr. 1980, 199, 181–189. [Google Scholar] [CrossRef]
- Sonmez, G.; Mutlu, H.; Ozturk, E.; Sildiroglu, H.O.; Keskin, A.T.; Basekim, C.C.; Kizilkaya, E. Magnetic resonance imaging findings of adult-onset glutaric aciduria type I. Acta Radiol. 2007, 48, 557–559. [Google Scholar] [CrossRef]
- Kharouba, M.; Patel, D.D.; Jaber, R.H.; Mahmoud, S.H. Metabolomic Analysis in Neurocritical Care Patients. Metabolites 2023, 13, 745. [Google Scholar] [CrossRef]
- Lai, K.; Klapa, M.I. Alternative pathways of galactose assimilation: Could inverse metabolic engineering provide an alternative to galactosemic patients? Metab. Eng. 2004, 6, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Sheng, L.; Madika, A.; Lau, M.S.H.; Zhang, Y.; Minton, N.P. Metabolic engineering for the production of acetoin and 2,3-butanediol at elevated temperature in Parageobacillus thermoglucosidasius NCIMB 11955. Front. Bioeng. Biotechnol. 2023, 11, 1191079. [Google Scholar] [CrossRef]
- Mitra, S.; Datta Chaudhuri, R.; Sarkar, R.; Banerjee, S.; Mukherjee, A.; Sharma, R.; Gope, A.; Kitahara, K.; Miyoshi, S.I.; Chawla-Sarkar, M. Rotavirus rewires host cell metabolic pathways toward glutamine catabolism for effective virus infection. Gut Microbes 2024, 16, 2428425. [Google Scholar] [CrossRef] [PubMed]
- Ramani, S.; Hu, L.; Venkataram Prasad, B.V.; Estes, M.K. Diversity in Rotavirus-Host glycan interactions: A “Sweet” Spectrum. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Raev, S.A.; Amimo, J.O.; Saif, L.J.; Vlasova, A.N. Intestinal mucin-type O-glycans: The major players in the host-bacteria-rotavirus interactions. Gut Microbes 2023, 15, 2197833. [Google Scholar] [CrossRef]
- Amimo, J.O.; Raev, S.A.; Chepngeno, J.; Mainga, A.O.; Guo, Y.; Saif, L.; Vlasova, A.N. Rotavirus Interactions with host intestinal epithelial cells. Front. Immunol. 2021, 12, 793841. [Google Scholar] [CrossRef]
- Stojanovic, B.S.; Stojanovic, B.; Milovanovic, J.; Arsenijević, A.; Dimitrijevic Stojanovic, M.; Arsenijevic, N.; Milovanovic, M. The Pivotal Role of Galectin-3 in Viral Infection: A Multifaceted Player in Host–Pathogen Interactions. Int. J. Mol. Sci. 2023, 24, 9617. [Google Scholar] [CrossRef]
- Hakerdi, L.A.; Wallace, L.; Smyth, G.; Madden, N.; Clark, A.; Hendroff, U.; McGovern, M.; Connellan, S.; Gillman, B.; Treacy, E.P. Determination of the lactose and galactose content of common foods: Relevance to galactosemia. Food Sci. Nutr. 2022, 10, 3789–3800. [Google Scholar] [CrossRef]
- Jiang, L.; Tang, A.; Song, L.; Tong, Y.; Fan, H. Advances in the development of antivirals for rotavirus infection. Front. Immunol. 2023, 14, 1041149. [Google Scholar] [CrossRef] [PubMed]
- Dickman, K.G.; Hempson, S.J.; Anderson, J.; Lippe, S.; Zhao, L.; Burakoff, R.; Shaw, R.D. Rotavirus alters paracellular permeability and energy metabolism in Caco-2 cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2000, 279, G757–G766. [Google Scholar] [CrossRef] [PubMed]
- Gaunt, E.R.; Zhang, Q.; Cheung, W.; Wakelam, M.J.O.; Lever, A.M.L.; Desselberger, U. Lipidome analysis of rotavirus-infected cells confirms the close interaction of lipid droplets with viroplasms. J. Gen. Virol. 2013, 94, 1576–1586. [Google Scholar] [CrossRef] [PubMed]
- Liebisch, G.; Fahy, E.; Aoki, J.; Dennis, E.A.; Durand, T.; Ejsing, C.S.; Fedorova, M.; Feussner, I.; Griffiths, W.J.; Köfeler, H.; et al. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J. Lipid Res. 2020, 61, 1539–1555. [Google Scholar] [CrossRef]
- Cohen, L.J.; Esterhazy, D.; Kim, S.H.; Lemetre, C.; Aguilar, R.R.; Gordon, E.A.; Pickard, A.J.; Cross, J.R.; Emiliano, A.B.; Han, S.M.; et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 2017, 549, 48–53. [Google Scholar] [CrossRef]
- An, D.; Oh, S.F.; Olszak, T.; Neves, J.F.; Avci, F.Y.; Erturk-Hasdemir, D.; Lu, X.; Zeissig, S.; Blumberg, R.S.; Kasper, D.L. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 2014, 156, 123–133. [Google Scholar] [CrossRef]
- Raque, M.; Raev, S.A.; Guo, Y.; Kick, M.K.; Saif, L.J.; Vlasova, A.N. Host Cell Response to Rotavirus Infection with Emphasis on Virus-Glycan Interactions, Cholesterol Metabolism, and Innate Immunity. Viruses 2023, 15, 1406. [Google Scholar] [CrossRef]
- Sen, A.; Namsa, N.D.; Feng, N.; Greenberg, H.B. Rotavirus Reprograms Multiple Interferon Receptors and Restricts Their Intestinal Antiviral and Inflammatory Functions. J. Virol. 2020, 94, e01775-19. [Google Scholar] [CrossRef]
- Mishra, S.P.; Karunakar, P.; Taraphder, S.; Yadav, H. Free Fatty Acid Receptors 2 and 3 as microbial metabolite sensors to shape host health: Pharmacophysiological View. Biomedicines 2020, 6, 154. [Google Scholar] [CrossRef]
- Chun, E.; Lavoie, S.; Fonseca-Pereira, D.; Bae, S.; Michaud, M.; Hoveyda, H.R.; Fraser, G.L.; Gallini Comeau, C.A.; Glickman, J.N.; Fuller, M.H.; et al. Metabolite-sensing receptor FFAR2 regulates colonic group 3 innate lymphoid cells and gut immunity. Immunity 2019, 51, 871–884.e6. [Google Scholar] [CrossRef]
- Kaemmerer, E.; Plum, P.; Klaus, C.; Weiskirchen, R.; Liedtke, C.; Adolf, M.; Schippers, A.; Wagner, N.; Reinartz, A.; Gassler, N. Fatty acid binding receptors in intestinal physiology and pathophysiology. World J. Gastrointest. Pathophysiol. 2010, 1, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Carretta, M.D.; Quiroga, J.; López, R.; Hidalgo, M.A.; Burgos, R.A. Participation of short-chain fatty acids and their receptors in gut inflammation and colon cancer. Front. Physiol. 2021, 12, 662739. [Google Scholar] [CrossRef] [PubMed]
- Hajjar, G.; Santos, M.C.B.; Bertrand-Michel, J.; Canlet, C.; Castelli, F.; Creusot, N.; Dechaumet, S.; Diémé, B.; Giacomoni, F.; Giraudeau, P.; et al. Scaling-up metabolomics: Current state and perspectives. Trends Anal. Chem. 2023, 167, 117225. [Google Scholar] [CrossRef]
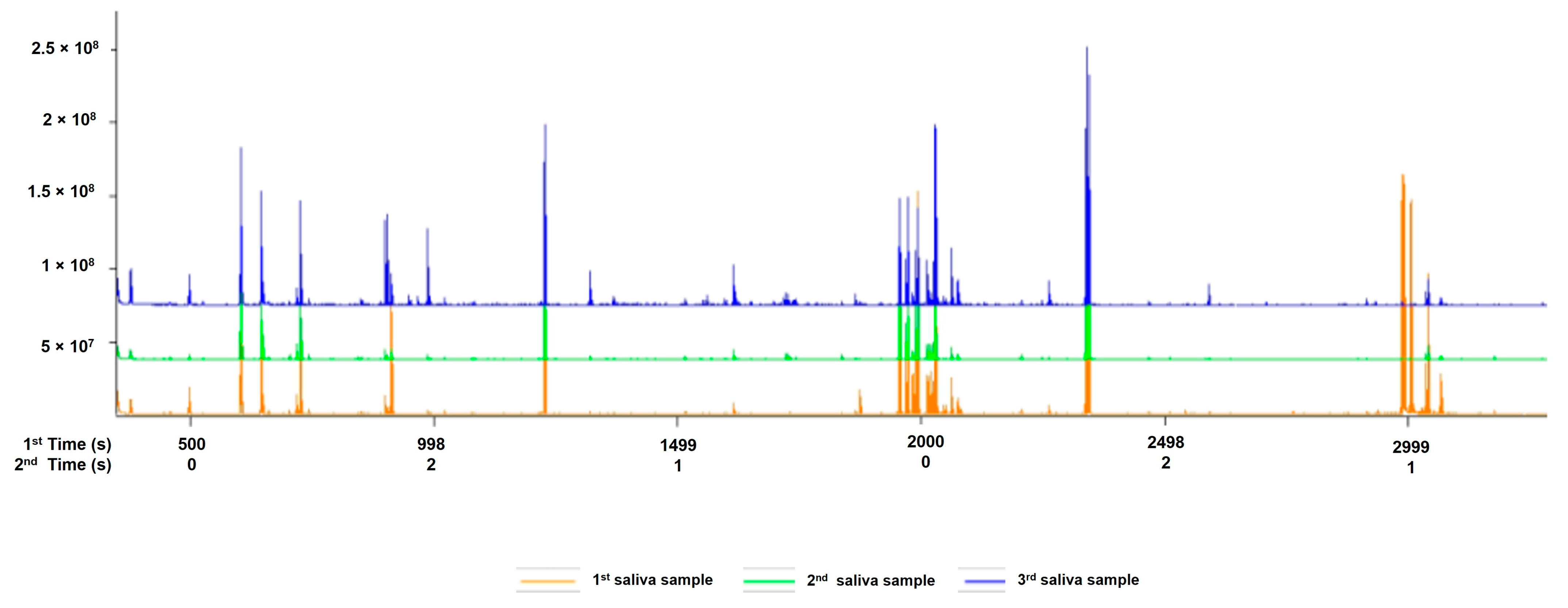


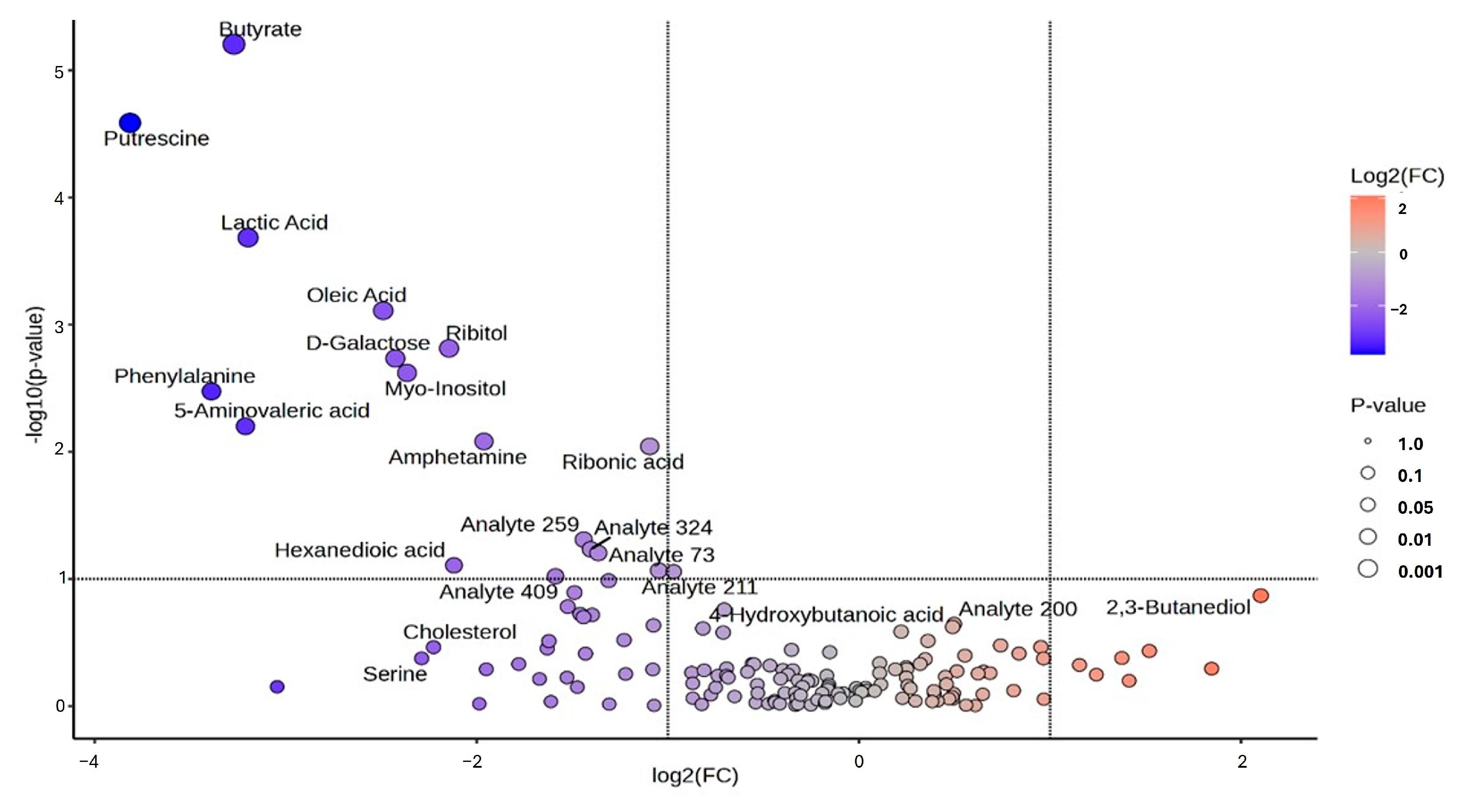
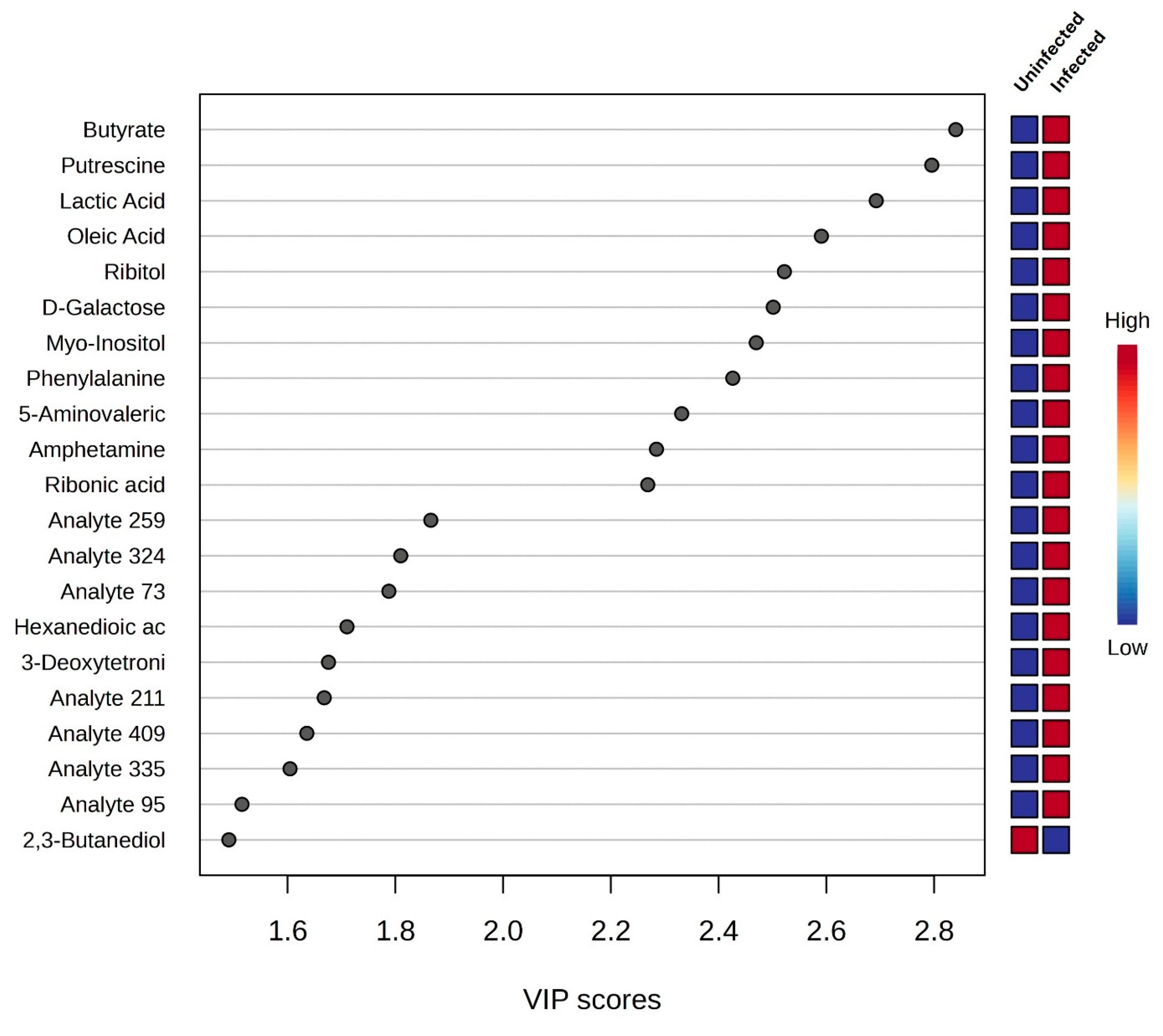
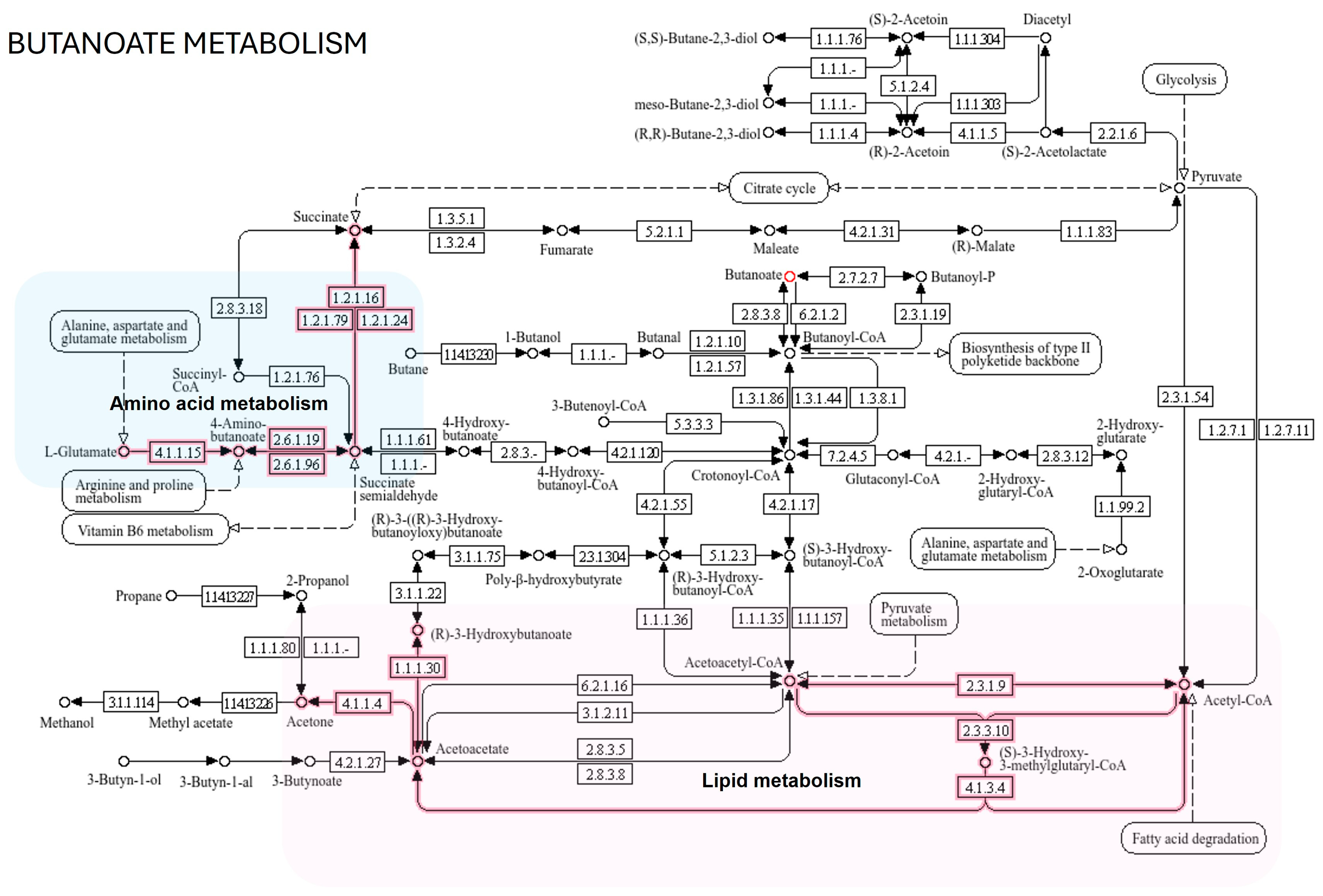
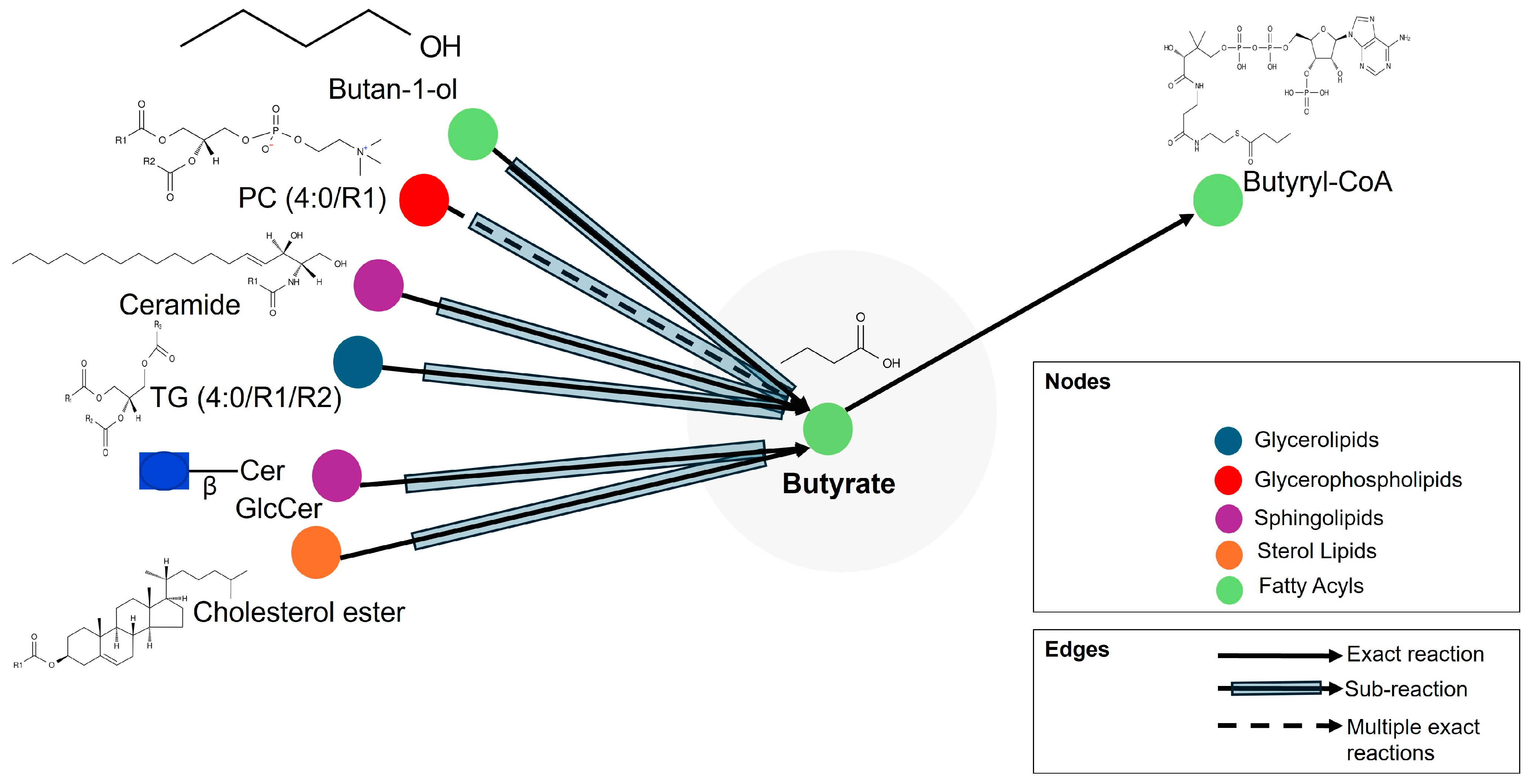
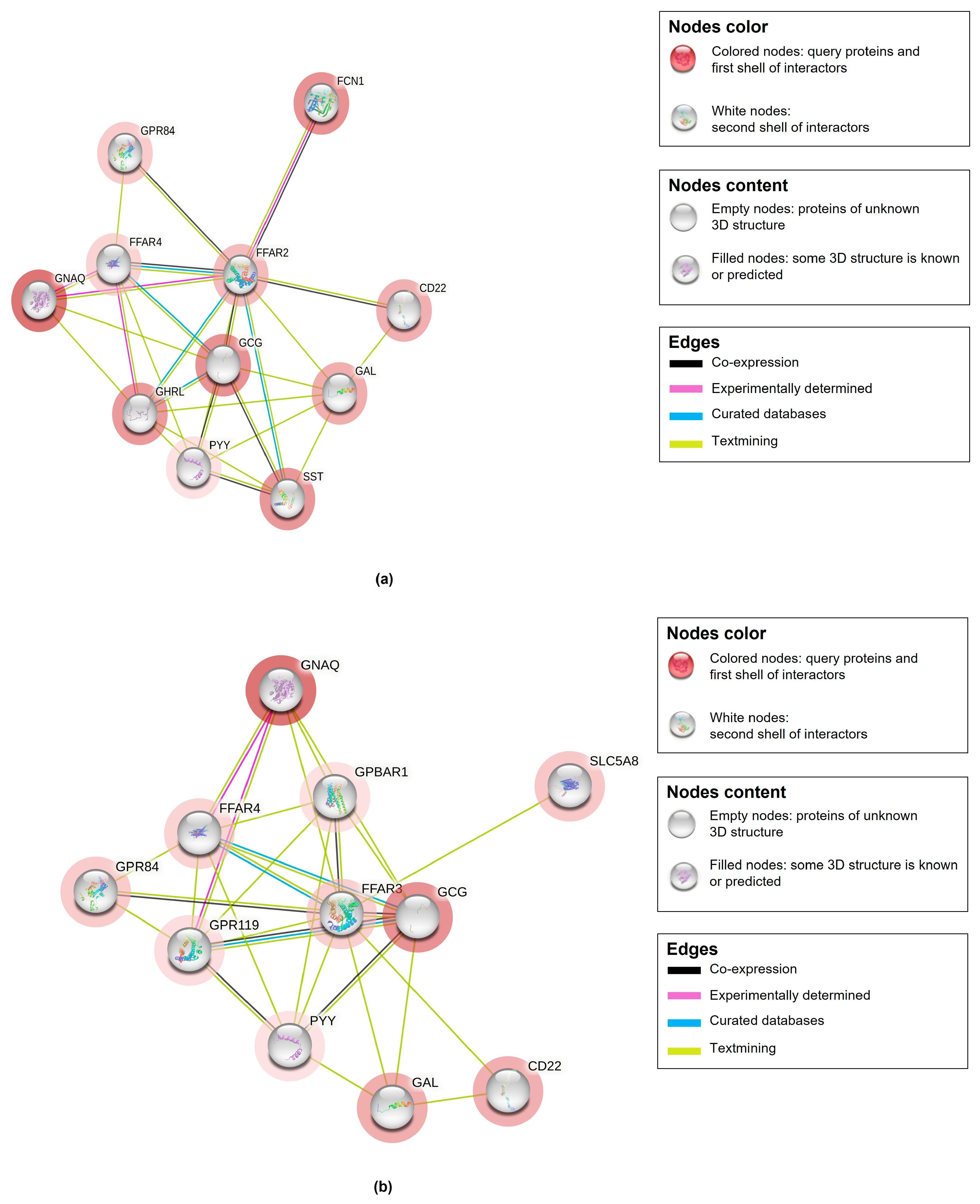
| Metabolic Molecules | Molecular Weight (g/mol) | Formula | Class | VIP | FC | p-Value Fisher’s Exact Test | FDR Critical Value | FDR q-Value |
|---|---|---|---|---|---|---|---|---|
| Butyrate | 88.1051 | C4H8O2 | Fatty acyls | 2.793 | 0.104 | 0.000 | 0.002 | 0.000 †† |
| Putrescine | 88.1515 | C4H12N2 | Organonitrogen compound | 2.749 | 0.071 | 0.000 | 0.005 | 0.000 †† |
| Lactic acid | 90.0779 | C3H6O3 | Hydroxy acid | 2.648 | 0.109 | 0.000 | 0.007 | 0.000 †† |
| Oleic acid | 282.4614 | C18H34O2 | Fatty acyls | 2.548 | 0.178 | 0.001 | 0.010 | 0.005 †† |
| Ribitol | 152.1458 | C5H12O5 | Organooxygen compound | 2.480 | 0.226 | 0.002 | 0.012 | 0.005 †† |
| D-Galactose | 180.1559 | C6H12O6 | Organooxygen compound | 2.460 | 0.186 | 0.002 | 0.014 | 0.005 †† |
| Myo-Inositol | 180.16 | C6H12O | Organooxygen compound | 2.429 | 0.194 | 0.002 | 0.017 | 0.005 †† |
| Phenylalanine | 165.1891 | C9H11NO2 | Carboxylic acid | 2.386 | 0.096 | 0.003 | 0.019 | 0.005 †† |
| 5-Aminovaleric acid | 117.1463 | C5H11NO2 | Carboxylic acid | 2.293 | 0.108 | 0.006 | 0.021 | 0.007 †† |
| Amphetamine | 135.2062 | C9H13N | Benzene | 2.247 | 0.257 | 0.008 | 0.024 | 0.013 †† |
| Ribonic acid | 166.1293 | C5H10O6 | Organooxygen compound | 2.231 | 0.468 | 0.009 | 0.026 | 0.015 †† |
| Analyte 259 UM159 | *** | *** | *** | 1.835 | 0.369 | 0.049 | 0.029 | 0.016 †† |
| Analyte 324 UM73 | *** | *** | *** | 1.780 | 0.378 | 0.058 | 0.031 | 0.079 *† |
| Analyte 73 UM116 | *** | *** | *** | 1.758 | 0.389 | 0.063 | 0.033 | 0.087 *† |
| Hexanedioic acid | 146.1412 | C6H10O4 | Fatty acyls | 1.682 | 0.230 | 0.078 | 0.036 | 0.088 *† |
| 3-Deoxytetronic acid | 120.1039 | C4H8O4 | Hydroxy acid | 1.648 | 0.484 | 0.086 | 0.038 | 0.091 *† |
| Analyte 211 UM110 | *** | *** | *** | 1.640 | 0.484 | 0.094 | 0.040 | 0.095 *† |
| Analyte 409 UM256 | *** | *** | *** | 1.608 | 0.333 | 0.095 | 0.043 | 0.095 *† |
| Analyte 335 UM74 | *** | *** | *** | 1.578 | 0.403 | 0.093 | 0.045 | 0.095 *† |
| Analyte 95 UM168 | *** | *** | *** | 1.490 | 0.356 | 0.069 | 0.048 | 0.095 *† |
| 2,3-Butanediol | 90.121 | C4H10O2 | Organooxygen compound | 1.466 | 4.297 | 0.002 | 0.050 | 0.095 *† |
| Pathway Name | Hitsa | p-Value | Holm pb | Impact Value |
|---|---|---|---|---|
| Galactose metabolism | 2 | 0.00426 | 0.341 | 0.105 |
| Phenylalanine, tyrosine, and tryptophan biosynthesis | 1 | 0.0155 | 1 | 0.0156 |
| Phenylalanine metabolism | 1 | 0.0308 | 1 | 0.0312 |
| Ascorbate and aldarate metabolism | 1 | 0.0346 | 1 | 0.0351 |
| Butanoate metabolism | 1 | 0.0542 | 1 | 0.0585 |
| Glutathione metabolism | 1 | 0.104 | 1 | 0.109 |
| Inositol phosphate metabolism | 1 | 0.112 | 1 | 0.117 |
| Arginine and proline metabolism | 1 | 0.133 | 1 | 0.14 |
| Biosynthesis of unsaturated fatty acids | 1 | 0.133 | 1 | 0.14 |
| Saliva-Secreting Glands Abundance in Homo sapiens * | Protein–Protein Interaction Network ** | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Proteins Interacting with Butyrate *** | Interaction Consistency Score | Coverage | Abundance Levels (ppm) | No. of Nodes | No. of Edges | Avg. Local Clustering Coefficient | Avg. Node Degree | Exp. No. of Edges | PPI Enrichment p-Value |
| Free fatty acid receptor 2 | 30.1 | 54% | 10,486 | 11 | 28 | 0.835 | 5.09 | 11 | 9.72 × 10−6 |
| Free fatty acid receptor 3 | 30.1 | 54% | 10,486 | 10 | 27 | 5.4 | 0.837 | 9 | 1.25 × 10−6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mametja, P.M.; Motshudi, M.C.; Naidoo, C.M.; Rakau, K.; Seheri, L.M.; Mkolo, N.M. Tapping into Metabolomics for Understanding Host and Rotavirus Group A Interactome. Life 2025, 15, 765. https://doi.org/10.3390/life15050765
Mametja PM, Motshudi MC, Naidoo CM, Rakau K, Seheri LM, Mkolo NM. Tapping into Metabolomics for Understanding Host and Rotavirus Group A Interactome. Life. 2025; 15(5):765. https://doi.org/10.3390/life15050765
Chicago/Turabian StyleMametja, Phiona Moloi, Mmei Cheryl Motshudi, Clarissa Marcelle Naidoo, Kebareng Rakau, Luyanda Mapaseka Seheri, and Nqobile Monate Mkolo. 2025. "Tapping into Metabolomics for Understanding Host and Rotavirus Group A Interactome" Life 15, no. 5: 765. https://doi.org/10.3390/life15050765
APA StyleMametja, P. M., Motshudi, M. C., Naidoo, C. M., Rakau, K., Seheri, L. M., & Mkolo, N. M. (2025). Tapping into Metabolomics for Understanding Host and Rotavirus Group A Interactome. Life, 15(5), 765. https://doi.org/10.3390/life15050765







