Exploring Childhood Lower Urinary Tract Symptoms (LUTS), Urinary Tract Infections (UTIs) and the Microbiome—A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Inclusion Criteria
2.2. Exclusion Criteria
2.3. Study Selection and Screening
2.4. Data Extraction
2.5. Risk of Bias Assessment
3. Results
3.1. Study Characteristics and Patient Group Distribution
3.2. Predominant Bacteria by Sample Type
3.2.1. Stool Samples
Urinary Tract Infection (UTI)
Voiding Dysfunction (VD)
3.2.2. Urine Samples
Urinary Tract Infection (UTI)
Vesicoureteral Reflux (VUR)
| Publication Year | First Author | Patient Sex Male: Female (n:n) | Type of Sample | Total n | Groups | n per Group | Mean Patient Age | Predominant Bacteria | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phylum | Class | Order | Family | Genus | Species | ||||||||
| 2018 | Paalanne [22] | 30:76 | stool | 106 | UTI | 37 | 20.3 months | Bacteroidetes, Firmicutes | Bacteroides,Enterobacter | Escherichia coli, Bacteroides fragilis, Bacteroides uniformis | |||
| Control | 69 | 21.8 months | Bacteroidetes, Firmicutes | Peptostreptococcaceae | Bacteroides | Bacteroides fragilis | |||||||
| 2020 | Forster [23] | 19:15 | urine | 34 | UTI | 11 | 11 years | Enterobacteriaceae | Klebsiella, Staphylococcus | ||||
| ASB | 19 | 8.8 years | Enterobacteriaceae | ||||||||||
| Control | 4 | 15 years | Enterobacteriaceae, Neisseriaceae | Staphylococcus | |||||||||
| 2020 | Kinneman [24] | 26:59 | urine | 85 | UTI | 9 | 382 days | Firmicutes, Proteobacteria | Clostridia, Bacteroidia, Gammaproteobacteria, Actinobacteria, Betaproteobacteria | Clostridiales, Bacteroidales, Enterobacteriales, Burkholderiales, Actinomycetales | Tissierellaceae, Prevotellaceae, Veillonellaceae, Enterobacteriaceae, Comamonadacea | Prevotella, Peptoniphilus, Escherichia, Veillonella, Finegoldia | |
| Control | 76 | ||||||||||||
| 2021 | Vitko [25] | 12:37 | urine | 49 | VUR | 20 | 4.8 years | Dorea, Escherichia | |||||
| 13 | 3.8 years | ||||||||||||
| controls | 16 | 10.2 years | Prevotella,Lactobacillus | ||||||||||
| 2022 | Akarken [26] | 20:29 | stool | 49 | VD | 25 | 8.26 years | Fusobacterium nucleatum, Clostridium difficile,Bacteriodes clarus | |||||
| Control | 24 | 8.00 years | Roseburia intestinalis | ||||||||||
| 2023 | Cole [27] | 0:33 | urine | 33 | BBD | 25 | 8.0 years | Porphyromonas, Varibaculum, Ezakiella, Campylobacter, Corynebacterium, Dialister, Streptococcus, Escherichia, Lagierella, Schaalia, Lawsonella, Peptoniphilus, Anaerococcus, Lactobacillus, Fenollaria, Finegoldia | |||||
| Control | 8 | 6.3 years | Peptoniphilus, Anaerococcus, Lactobacillus, Fenollaria, Finegoldia | ||||||||||
| 2023 | Urakami [28] | 42:37 | Stool | 79 | UTI | 28 | 5 months | Actinobacceriota, Actinobacteria | Bacilli | Bifidobacteriales, Enterobacteriales | Bifidobacteriaceae, Enterobacteriaceae | Escherichia, Shigella | Escherichia coli |
| Control | 51 | 5 months | Bacteroidiota | Bacteroidia | Negativicutes, Bacteroidales, Veillonellases, Selenomonadales | Bacteroidaceae, Veillonellaceae | Veilonella, Bacteroides | ||||||
| 2024 | Kelly [29] | Male | urine | 33 | Healthy | 13 | 40.1 months | Peptoniphillus, Ezakiella, Sphingomonas, Ralstonia | |||||
| Female | 20 | Prevotella, Anaerococcus, Shaalia | Prevotella timonensis, Schaalia turincensis,Anaerococcus lactolyticus | ||||||||||
| 13:20 | 33 | 0 UTI or Unknown (excluded from analysis) | 5 | ||||||||||
| History of 1 UTI | 10 | ||||||||||||
| History of 2 UTIs | 8 | ||||||||||||
| History of 3+ UTIs | 10 | Proteobacteria DECREASED: Bacteriodetes | DECREASED: Enterococcus, Lawsonella, Corynebacterium | ||||||||||
| 2024 | Luyang Hong [30] | 74:77 | stool | 151 | Gram-positive UTI | 53 | 29.49 weeks | Gammaproteobacteria, Bacilli | Enterococcaceae | Enterococcus faecalis | |||
| Gram-negative UTI | Gammaproteobacteria, Bacilli | Enterobacteriaceae | Klebsiella, Escherichia | Escherichia coli, Klebsiella aerogenes,Klebsiella pneumoniae, Enterobacter cloacae | |||||||||
| Control | 98 | 30.24 weeks | Clostridia | ||||||||||
Bladder-Bowel Dysfunction (BBD)
3.3. Microbiome Diversity by Sample Type
3.3.1. Stool Samples
Alpha Diversity
Beta Diversity
3.3.2. Urine Samples
Alpha Diversity
Beta Diversity
4. Discussion
4.1. Urinary Tract Infections (UTIs)
4.2. Prior Antibiotic Exposure
4.3. Bladder-Bowel-Dysfunction (BBD)
4.4. Sex-Based Differences in the Urinary Microbiome
4.5. The Role of the Gut Microbiome
4.6. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANS | Autonomic nervous system |
| BBD | Bladder-bowel dysfunction |
| CNS | Central nervous system |
| LUTS | Lower urinary tract symptoms |
| NE | Nocturnal enuresis |
| NGS | Next-generation sequencing |
| PICOS | Patient, Intervention, Comparison, Outcome, Study type |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| QUADAS | Quality Assessment of Diagnostic Accuracy Studies |
| QUADOMICS | Adaptation of the QUADAS, studies on the diagnostic accuracy of ‘-omics’-based technologies |
| rRNA | ribosomal RNA |
| SMD | Standardized mean difference |
| UTIs | Urinary tract infections |
| VD | Voiding dysfunction |
| VDSS | Voiding dysfunction symptom score |
| VUR | Vesicoureteral reflux |
Appendix A
| Section and Topic | Item # | Checklist Item | Location Where Item Is Reported |
|---|---|---|---|
| TITLE | |||
| Title | 1 | Identify the report as a systematic review. | p. 1 |
| ABSTRACT | |||
| Abstract | 2 | See the PRISMA 2020 for abstracts checklist. | p. 1 |
| INTRODUCTION | |||
| Rationale | 3 | Describe the rationale for the review in the context of existing knowledge. | p. 1 |
| Objectives | 4 | Provide an explicit statement of the objective(s) or question(s) the review addresses. | p. 2 |
| METHODS | |||
| Eligibility criteria | 5 | Specify the inclusion and exclusion criteria for the review and how studies were grouped for the syntheses. | pp. 2–3 |
| Information sources | 6 | Specify all databases, registers, websites, organizations, reference lists, and other sources searched or consulted to identify studies. Specify the date when each source was last searched or consulted. | p. 3 |
| Search strategy | 7 | Present the full search strategies for all databases, registers, and websites, including any filters and limits used. | pp. 3–4 |
| Selection process | 8 | Specify the methods used to decide whether a study met the inclusion criteria of the review, including how many reviewers screened each record and each report retrieved, whether they worked independently, and, if applicable, details of automation tools used in the process. | pp. 4–5 |
| Data collection process | 9 | Specify the methods used to collect data from reports, including how many reviewers collected data from each report, whether they worked independently, any processes for obtaining or confirming data from study investigators, and, if applicable, details of automation tools used in the process. | pp. 4–5 |
| Data items | 10a | List and define all outcomes for which data were sought. Specify whether all results that were compatible with each outcome domain in each study were sought (e.g., for all measures, time points, analyses), and if not, the methods used to decide which results to collect. | p. 5 |
| 10b | List and define all other variables for which data were sought (e.g., participant and intervention characteristics, funding sources). Describe any assumptions made about any missing or unclear information. | p. 5 | |
| Study risk of bias assessment | 11 | Specify the methods used to assess risk of bias in the included studies, including details of the tool(s) used, how many reviewers assessed each study, and whether they worked independently, and, if applicable, details of automation tools used in the process. | p. 5 |
| Effect measures | 12 | Specify for each outcome the effect measure(s) (e.g., risk ratio, mean difference) used in the synthesis or presentation of results. | pp. 2–3 and p. 5 |
| Synthesis methods | 13a | Describe the processes used to decide which studies were eligible for each synthesis (e.g., tabulating the study intervention characteristics and comparing against the planned groups for each synthesis (item #5). | pp. 3–5 |
| 13b | Describe any methods required to prepare the data for presentation or synthesis, such as handling of missing summary statistics or data conversions. | N/A | |
| 13c | Describe any methods used to tabulate or visually display the results of individual studies and syntheses. | N/A | |
| 13d | Describe any methods used to synthesize results and provide a rationale for the choice(s). If meta-analysis was performed, describe the model(s), method(s) to identify the presence and extent of statistical heterogeneity, and software package(s) used. | N/A | |
| 13e | Describe any methods used to explore possible causes of heterogeneity among study results (e.g., subgroup analysis, meta-regression). | N/A | |
| 13f | Describe any sensitivity analyses conducted to assess the robustness of the synthesized results. | N/A | |
| Reporting bias assessment | 14 | Describe any methods used to assess the risk of bias due to missing results in a synthesis (arising from reporting biases). | N/A |
| Certainty assessment | 15 | Describe any methods used to assess certainty (or confidence) in the body of evidence for an outcome. | N/A |
| RESULTS | |||
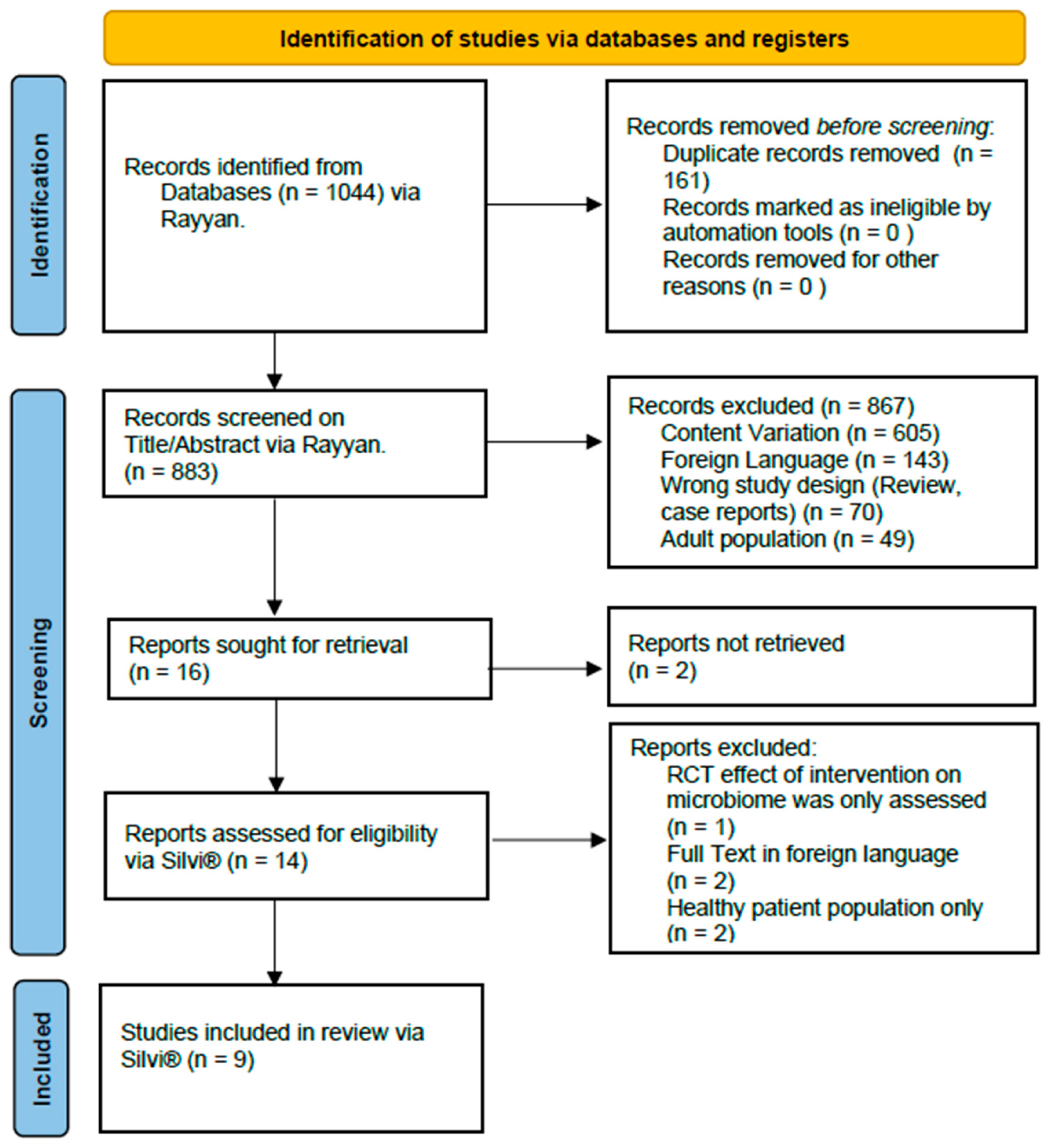
| Study selection | 16a | Describe the results of the search and selection process, from the number of records identified in the search to the number of studies included in the review, ideally using a flow diagram. | pp. 5–8 |
| 16b | Cite studies that might appear to meet the inclusion criteria, but which were excluded, and explain why they were excluded. | pp. 5–8 | |
| Study characteristics | 17 | Cite each included study and present its characteristics. | pp. 7–8 |
| Risk of bias in studies | 18 | Present assessments of the risk of bias for each included study. | Appendix A3 |
| Results of individual studies | 19 | For all outcomes, present, for each study: (a) summary statistics for each group (where appropriate) and (b) an effect estimate and its precision (e.g., confidence/credible interval), ideally using structured tables or plots. | pp. 5–15 |
| Results of syntheses | 20a | For each synthesis, briefly summarize the characteristics and risk of bias among contributing studies. | Appendix A3 |
| 20b | Present the results of all statistical syntheses conducted. If meta-analysis was done, present for each the summary estimate and its precision (e.g., confidence/credible interval) and measures of statistical heterogeneity. If comparing groups, describe the direction of the effect. | N/A | |
| 20c | Present the results of all investigations of possible causes of heterogeneity among study results. | N/A | |
| 20d | Present results of all sensitivity analyses conducted to assess the robustness of the synthesized results. | N/A | |
| Reporting biases | 21 | Present assessments of risk of bias due to missing results (arising from reporting biases) for each synthesis assessed. | N/A |
| Certainty of evidence | 22 | Present assessments of certainty (or confidence) in the body of evidence for each outcome assessed. | N/A |
| DISCUSSION | |||
| Discussion | 23a | Provide a general interpretation of the results in the context of other evidence. | pp. 15–17 |
| 23b | Discuss any limitations of the evidence included in the review. | p. 17 | |
| 23c | Discuss any limitations of the review processes used. | p. 17 | |
| 23d | Discuss implications of the results for practice, policy, and future research. | p. 17 | |
| OTHER INFORMATION | |||
| Registration and protocol | 24a | Provide registration information for the review, including the register name and registration number, or state that the review was not registered. | p. 2 |
| 24b | Indicate where the review protocol can be accessed, or state that a protocol was not prepared. | p. 2 | |
| 24c | Describe and explain any amendments to the information provided at registration or in the protocol. | N/A | |
| Support | 25 | Describe sources of financial or non-financial support for the review, and the role of the funders or sponsors in the review. | p. 18 |
| Competing interests | 26 | Declare any competing interests of review authors. | p. 18 |
| Availability of data, code and other materials | 27 | Report which of the following are publicly available and where they can be found: template data collection forms; data extracted from included studies; data used for all analyses; analytic code; any other materials used in the review. | N/A |
| Items | Possible Answers: Yes/No/Unclear/Not Applicable |
|---|---|
| 1. Were selection criteria clearly described? 2. Was the spectrum of patients representative of patients who will receive the test in practice? 3. Was the type of sample fully described? 4. Were the procedures and timing of biological sample collection with respect to clinical factors described with enough detail? 5. Were handling and pre-analytical procedures reported in sufficient detail and similar for the whole sample? And, if differences in procedures were reported, was their effect on the results assessed? 6. Is the time period between the reference standard and the index test short enough to reasonably guarantee that the target condition did not change between the two tests? 7. Is the reference standard likely to correctly classify the target condition? 8. Did the whole sample or a random selection of the sample receive verification using a reference standard of diagnosis? 9. Did patients receive the same reference standard regardless of the result of the index test? 10. Was the execution of the index test described in sufficient detail to permit replication of the test? 11. Was the execution of the reference standard described in sufficient detail to permit its replication? 12. Were the index test results interpreted without knowledge of the results of the reference standard? 13. Were the reference standard results interpreted without knowledge of the results of the index test? 14. Were the same clinical data available when test results were interpreted as would be available when the test is used in practice? 15. Were uninterpretable/intermediate test results reported? 16. Is it likely that the presence of overfitting was avoided? |
| Article | Observer | QUADOMICS Checklist | QUADOMICS-Score | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 1 | 0 | U | N/A | Total | ||
| Paalanne et al. (2018) [22] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | U | U | 1 | 0 | U | 12 | 1 | 3 | 0 | 12 |
| 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | U | 1 | 1 | 1 | 1 | 15 | 0 | 1 | 0 | 15 | |
| Final | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | U | 1 | 1 | 0 | 0 | 13 | 2 | 1 | 0 | 13 | |
| Forster et al. (2020) [23] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | U | U | 1 | 0 | U | 12 | 1 | 3 | 0 | 12 |
| 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | U | 1 | 1 | 1 | 1 | 15 | 0 | 1 | 0 | 15 | |
| Final | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | U | 1 | 1 | 0 | 0 | 13 | 2 | 1 | 0 | 13 | |
| Kinneman et al. (2020) [24] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | U | U | 1 | 0 | U | 12 | 1 | 3 | 0 | 12 |
| 2 | 1 | 1 | 1 | 1 | U | 1 | 1 | 0 | 1 | 1 | U | 1 | 1 | 0 | U | 1 | 11 | 2 | 3 | 0 | 11 | |
| Final | 1 | 1 | 1 | 1 | U | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 12 | 3 | 1 | 0 | 12 | |
| Vitko et al. (2021) [25] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | U | U | 1 | 0 | U | 12 | 1 | 3 | 0 | 12 |
| 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 16 | 0 | 0 | 0 | 16 | |
| Final | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 14 | 2 | 0 | 0 | 14 | |
| Akarken et al. (2022) [26] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | U | U | 1 | 0 | 0 | 12 | 1 | 3 | 0 | 12 |
| 2 | 1 | 1 | 1 | 1 | U | 1 | 1 | 0 | 1 | 1 | U | 1 | 1 | 0 | U | 1 | 11 | 2 | 3 | 0 | 11 | |
| Final | 1 | 1 | 1 | 1 | U | 1 | 1 | 1 | 1 | 1 | 0 | U | 1 | 1 | 1 | 0 | 12 | 2 | 2 | 0 | 12 | |
| Urakami et al. (2023) [28] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | U | U | 1 | 0 | 1 | 13 | 1 | 2 | 0 | 13 |
| 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | U | 1 | 1 | 1 | 1 | 15 | 0 | 1 | 0 | 15 | |
| Final | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | U | U | 1 | 0 | 1 | 13 | 1 | 2 | 0 | 13 | |
| Cole et al. (2023) [27] | 1 | 1 | U | 1 | 1 | 1 | U | 1 | 1 | 1 | 1 | 1 | U | U | 1 | 1 | 1 | 12 | 0 | 4 | 0 | 12 |
| 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 16 | 0 | 0 | 0 | 16 | |
| Final | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 16 | 0 | 0 | 0 | 16 | |
| Kelly et al. (2024) [29] | 1 | 1 | U | 1 | 1 | 1 | U | U | U | U | 1 | U | U | U | 1 | 1 | 1 | 8 | 0 | 8 | 0 | 8 |
| 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 16 | 0 | 0 | 0 | 16 | |
| Final | 1 | 1 | 1 | 1 | 1 | 1 | U | 1 | U | 1 | U | 1 | 1 | 1 | 1 | 1 | 13 | 0 | 3 | 0 | 13 | |
| Luyang Hong et al. (2024) [30] | 1 | 1 | 1 | 1 | 1 | 1 | U | 1 | 1 | 1 | 1 | 1 | U | U | 1 | 1 | 1 | 13 | 0 | 3 | 0 | 13 |
| 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | U | 1 | 1 | 1 | 1 | 15 | 0 | 1 | 0 | 15 | |
| Final | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | U | 1 | 1 | 1 | 1 | 15 | 0 | 1 | 0 | 15 | |
References
- Van den Ende, M.; Joinson, C.; Sinha, S.; Verbakel, I.; Ochoa, D.C.; Lazar, J.; Baird, A.; Selai, C.; Van Huele, A.; Bou Kheir, G.; et al. Navigating the Waters of LUTS from Childhood to Puberty—NOPIA meeting (ICI-RS 2024). 2025. J. Pediatr. Urol. 2025. submitted. [Google Scholar]
- Austin, P.F.; Bauer, S.B.; Bower, W.; Chase, J.; Franco, I.; Hoebeke, P.; Rittig, S.; Walle, J.V.; von Gontard, A.; Wright, A.; et al. The standardization of terminology of lower urinary tract function in children and adolescents: Update report from the standardization committee of the International Children’s Continence Society. Neurourol. Urodyn. 2016, 35, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, N.; Morone, N.E.; Bost, J.E.; Farrell, M.H. Prevalence of Urinary Tract Infection in Childhood. Pediatr. Infect. Dis. J. 2008, 27, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Hoebeke, P.; Van Laecke, E.; Van Camp, C.; Raes, A.; Van De Walle, J. One thousand video-urodynamic studies in children with non-neurogenic bladder sphincter dysfunction. BJU Int. 2001, 87, 575–580. [Google Scholar] [CrossRef]
- Bauer, S.B. Special considerations of the overactive bladder in children. Urology 2002, 60, 43–48. [Google Scholar] [CrossRef]
- Brubaker, L.; Wolfe, A.J. The female urinary microbiota, urinary health and common urinary disorders. Ann. Transl. Med. 2017, 5, 34. [Google Scholar] [CrossRef]
- Aragón, I.M.; Herrera-Imbroda, B.; Queipo-Ortuño, M.I.; Castillo, E.; Del Moral, J.S.-G.; Gómez-Millán, J.; Yucel, G.; Lara, M.F. The Urinary Tract Microbiome in Health and Disease. Eur. Urol. Focus 2018, 4, 128–138. [Google Scholar] [CrossRef]
- Davidson, G.L.; Cooke, A.C.; Johnson, C.N.; Quinn, J.L. The gut microbiome as a driver of individual variation in cognition and functional behaviour. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170286. [Google Scholar] [CrossRef]
- Reasoner, S.A.; Flores, V.; Van Horn, G.; Morales, G.; Peard, L.M.; Abelson, B.; Manuel, C.; Lee, J.; Baker, B.; Williams, T.; et al. Survey of the infant male urobiome and genomic analysis of Actinotignum spp. npj Biofilms Microbiomes 2023, 9, 91. [Google Scholar] [CrossRef]
- Wolfe, A.J.; Toh, E.; Shibata, N.; Rong, R.; Kenton, K.; FitzGerald, M.; Mueller, E.R.; Schreckenberger, P.; Dong, Q.; Nelson, D.E.; et al. Evidence of Uncultivated Bacteria in the Adult Female Bladder. J. Clin. Microbiol. 2012, 50, 1376–1383. [Google Scholar] [CrossRef]
- Shade, A. Diversity is the question, not the answer. ISME J. 2016, 11, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Jayalath, S.; Magana-Arachchi, D. Dysbiosis of the Human Urinary Microbiome and its Association to Diseases Affecting the Urinary System. Indian J. Microbiol. 2022, 62, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Pearce, M.M.; Hilt, E.E.; Rosenfeld, A.B.; Zilliox, M.J.; Thomas-White, K.; Fok, C.; Kliethermes, S.; Schreckenberger, P.C.; Brubaker, L.; Gai, X.; et al. The Female Urinary Microbiome: A Comparison of Women with and without Urgency Urinary Incontinence. mBio 2014, 5, e01283-14. [Google Scholar] [CrossRef] [PubMed]
- Brubaker, L.; Nager, C.W.; Richter, H.E.; Visco, A.; Nygaard, I.; Barber, M.D.; Schaffer, J.; Meikle, S.; Wallace, D.; Shibata, N.; et al. Urinary bacteria in adult women with urgency urinary incontinence. Int. Urogynecology J. 2014, 25, 1179–1184. [Google Scholar] [CrossRef]
- Price, T.K.; Wolff, B.; Halverson, T.; Limeira, R.; Brubaker, L.; Dong, Q.; Mueller, E.R.; Wolfe, A.J. Temporal Dynamics of the Adult Female Lower Urinary Tract Microbiota. mBio 2020, 11, e00475-20. [Google Scholar] [CrossRef]
- Panicker, J.N.; Marcelissen, T.; von Gontard, A.; Vrijens, D.; Abrams, P.; Wyndaele, M. Bladder-bowel interactions: Do we understand pelvic organ cross-sensitization? International Consultation on Incontinence Research Society (ICI-RS) 2018. Neurourol. Urodyn. 2019, 38, S25–S34. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Int. J. Surg. 2021, 88, 105906. [Google Scholar] [CrossRef]
- Holm-Larsen, T.; Ende, M.V.D.; Wendelboe, H.G.; Verbakel, I.; Kheir, G.B.; Hervé, F.; Everaert, K. The cost of lifelong LUTS—A systematic literature review. Neurourol. Urodyn. 2024, 43, 1058–1065. [Google Scholar] [CrossRef]
- Miller, S.A.; Forrest, J.L. Enhancing your practice through evidence-based decision making: PICO, learning how to ask good questions. J. Evid. Based Dent. Pract. 2001, 1, 136–141. [Google Scholar] [CrossRef]
- Cramond, F.; O’Mara-Eves, A.; Doran-Constant, L.; Rice, A.S.; Macleod, M.; Thomas, J. The development and evaluation of an online application to assist in the extraction of data from graphs for use in systematic reviews. Wellcome Open Res. 2019, 3, 157. [Google Scholar] [CrossRef]
- Lumbreras, B.; Porta, M.; Márquez, S.; Pollán, M.; Parker, L.A.; Hernández-Aguado, I. QUADOMICS: An adaptation of the Quality Assessment of Diagnostic Accuracy Assessment (QUADAS) for the evaluation of the methodological quality of studies on the diagnostic accuracy of ‘-omics’-based technologies. Clin. Biochem. 2008, 41, 1316–1325. [Google Scholar] [CrossRef] [PubMed]
- Paalanne, N.; Husso, A.; Salo, J.; Pieviläinen, O.; Tejesvi, M.V.; Koivusaari, P.; Pirttilä, A.M.; Pokka, T.; Mattila, S.; Jyrkäs, J.; et al. Intestinal microbiome as a risk factor for urinary tract infections in children. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1881–1891. [Google Scholar] [CrossRef] [PubMed]
- Forster, C.S.; Panchapakesan, K.; Stroud, C.; Banerjee, P.; Gordish-Dressman, H.; Hsieh, M.H. A cross-sectional analysis of the urine microbiome of children with neuropathic bladders. J. Pediatr. Urol. 2020, 16, 593.e1–593.e8. [Google Scholar] [CrossRef] [PubMed]
- Kinneman, L.D.; Zhu, W.; Wong, W.S.; Clemency, N.M.; Provenzano, M.M.; Vilboux, T.; Jane’t, K.; Seo-Mayer, P.; Levorson, R.; Kou, M.; et al. Assessment of the Urinary Microbiome in Children Younger Than 48 Months. Pediatr. Infect. Dis. J. 2020, 39, 565–570. [Google Scholar] [CrossRef]
- Vitko, D.; McQuaid, J.W.; Gheinani, A.H.; Hasegawa, K.; DiMartino, S.; Davis, K.H.; Chung, C.Y.; Petrosino, J.F.; Adam, R.M.; Mansbach, J.M.; et al. Urinary Tract Infections in Children with Vesicoureteral Reflux Are Accompanied by Alterations in Urinary Microbiota and Metabolome Profiles. Eur. Urol. 2022, 81, 151–154. [Google Scholar] [CrossRef]
- Akarken, I.; Tarhan, H.; Şener, G.; Deliktas, H.; Cengiz, N.; Şahin, H. Is there a difference in fecal microbiota of children with and without voiding dysfunction? Archivio Italiano di Urologia e Andrologia 2022, 94, 455–458. [Google Scholar] [CrossRef]
- Cole, E.B.; Khemmani, M.; Liu, H.; Halverson, T.M.; Noronha, M.F.; Forster, C.S.; Wolfe, A.J.; Shaikh, N. Urogenital urobiome of healthy children does not differ from that of children with bladder and bowel dysfunction. J. Pediatr. Urol. 2023, 19, 368.e1–368.e8. [Google Scholar] [CrossRef]
- Urakami, C.; Yamanouchi, S.; Kimata, T.; Tsuji, S.; Akagawa, S.; Kino, J.; Akagawa, Y.; Kato, S.; Araki, A.; Kaneko, K. Abnormal Development of Microbiota May Be a Risk Factor for Febrile Urinary Tract Infection in Infancy. Microorganisms 2023, 11, 2574. [Google Scholar] [CrossRef]
- Kelly, M.S.; Dahl, E.M.; Jeries, L.M.; Sysoeva, T.A.; Karstens, L. Characterization of pediatric urinary microbiome at species-level resolution indicates variation due to sex, age, and urologic history. J. Pediatr. Urol. 2024, 20, 884–893. [Google Scholar] [CrossRef]
- Hong, L.; Huang, Y.; Han, J.; Li, S.; Zhang, L.; Zhou, Q.; Cao, X.; Yu, W.; Guo, X.; Yang, Y.; et al. Pathogen-specific alterations in intestinal microbiota precede urinary tract infections in preterm infants: A longitudinal case-control study. Gut Microbes 2024, 16, 2333413. [Google Scholar] [CrossRef]
- Bossa, L.; Kline, K.; McDougald, D.; Lee, B.B.; Rice, S.A. Urinary catheter-associated microbiota change in accordance with treatment and infection status. PLoS ONE 2017, 12, e0177633. [Google Scholar] [CrossRef]
- Rund, S.A.; Rohde, H.; Sonnenborn, U.; Oelschlaeger, T.A. Antagonistic effects of probiotic Escherichia coli Nissle 1917 on EHEC strains of serotype O104:H4 and O157:H7. Int. J. Med. Microbiol. 2013, 303, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; Relman, D.A. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PLoS Biol. 2008, 6, e280. [Google Scholar] [CrossRef]
- Mulder, M.; Radjabzadeh, D.; Hassing, R.J.; Heeringa, J.; Uitterlinden, A.G.; Kraaij, R.; Stricker, B.H.; Verbon, A. The effect of antimicrobial drug use on the composition of the genitourinary microbiota in an elderly population. BMC Microbiol. 2019, 19, 9. [Google Scholar] [CrossRef]
- Storm, D.W.; Copp, H.L.; Halverson, T.M.; Du, J.; Juhr, D.; Wolfe, A.J. A Child’s urine is not sterile: A pilot study evaluating the Pediatric Urinary Microbiome. J. Pediatr. Urol. 2022, 18, 383–392. [Google Scholar] [CrossRef]
- Brubaker, L.; Putonti, C.; Dong, Q.; Wolfe, A.J. The human urobiome. Mamm. Genome 2021, 32, 232–238. [Google Scholar] [CrossRef]
- Fredsgaard, L.; Thorsteinsson, K.; Bundgaard-Nielsen, C.; Ammitzbøll, N.; Leutscher, P.; Chai, Q.; Jensen, A.-M.; Sørensen, S.; Pedersen, L.M.; Hagstrøm, S.; et al. Description of the voided urinary microbiota in asymptomatic prepubertal children—A pilot study. J. Pediatr. Urol. 2021, 17, 545.e1–545.e8. [Google Scholar] [CrossRef]
- Kassiri, B.; Shrestha, E.; Kasprenski, M.; Antonescu, C.; Florea, L.D.; Sfanos, K.S.; Wang, M.-H. A Prospective Study of the Urinary and Gastrointestinal Microbiome in Prepubertal Males. Urology 2019, 131, 204–210. [Google Scholar] [CrossRef]
- Akagawa, S.; Akagawa, Y.; Yamanouchi, S.; Kimata, T.; Tsuji, S.; Kaneko, K. Development of the gut microbiota and dysbiosis in children. Biosci. Microbiota Food Health 2021, 40, 12–18. [Google Scholar] [CrossRef]
- Carabotti, M.; Scirocco, A.; Maselli, M.A.; Severi, C. The gut-brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015, 28, 203–209. [Google Scholar]
- Yang, N.J.; Chiu, I.M. Bacterial Signaling to the Nervous System through Toxins and Metabolites. J. Mol. Biol. 2017, 429, 587–605. [Google Scholar] [CrossRef]
| Database | Search String(s) |
|---|---|
| PubMed | (“child”[MeSH Terms] OR “pediatrics”[MeSH Terms] OR “Infant, Newborn”[MeSH Terms] OR child*[Title/abstract] OR schoolchild*[Title/abstract] OR infan*[Title/abstract] OR adolescen*[Title/abstract] OR pediatri*[Title/abstract] OR paediatr*[Title/abstract] OR neonat*[Title/abstract] OR boy[Title/abstract] OR boys[Title/abstract] OR boyhood[Title/abstract] OR girl[Title/abstract] OR girls[Title/abstract] OR girlhood[Title/abstract] OR youth[Title/abstract] OR youths[Title/abstract] OR baby[Title/abstract] OR babies[Title/abstract] OR toddler*[Title/abstract] OR teen[Title/abstract] OR teens[Title/abstract] OR teenager*[Title/abstract] OR newborn*[Title/abstract] OR postneonat*[Title/abstract] OR postnat*[Title/abstract] OR perinat*[Title/abstract] OR puberty[Title/abstract] OR preschool*[Title/abstract] OR suckling*[Title/abstract] OR picu[Title/abstract] OR nicu[Title/abstract]) AND (“Urine/microbiology”[Mesh Terms] OR “microbiota”[MeSH Terms] OR “gastrointestinal microbiome”[MeSH Terms] OR “urine microbiome”[Title/abstract] OR (“gastrointestinal”[Title/abstract] AND “microbiome”[Title/abstract]) OR “gastrointestinal microbiome”[Title/abstract] OR (“gut”[Title/abstract] AND “microbiome”[Title/abstract]) OR “gut microbiome”[Title/abstract] OR “microbiota”[Title/abstract] OR “urinary microbiota”[Title/abstract] OR “gut microbiota”[Title/abstract] OR “urine microbiome”[Title/abstract] OR “urine microbiota”[Title/abstract]) AND (“Lower Urinary Tract Symptoms”[MeSH Terms] OR “Nocturia”[MeSH Terms] OR “urinary bladder diseases”[MeSH Terms] OR “Urinary Incontinence”[MeSH Terms] OR “Nocturnal Enuresis”[MeSH Terms] OR “urinary tract infections”[MeSH Terms] OR “pyelonephritis”[MeSH Terms] OR “cystitis”[MeSH Terms] OR “Lower Urinary Tract Symptoms”[Title/abstract] OR “luts”[Title/abstract] OR “Nocturia”[Title/abstract] OR “OAB”[Title/abstract] OR “overactive bladder”[Title/abstract] OR “bed wetting”[Title/abstract] OR “urological symptoms”[Title/abstract] OR “urinary disorders”[Title/abstract] OR “Urinary urge incontinence”[Title/abstract] OR “lower urinary tract dysfunction”[Title/abstract] OR “lower urinary tract problems”[Title/abstract] OR “urinary urgency”[Title/abstract] OR “urinary frequency”[Title/abstract] OR “voiding dysfunction”[Title/abstract] OR (“urinary”[Title/abstract] AND “tract”[Title/abstract] AND “infections”[Title/abstract]) OR “urinary tract infections”[Title/abstract] OR “urinary tract infection”[Title/abstract] OR “UTI”[Title/abstract] OR “UTIs”[Title/abstract] OR “UTIs”[Title/abstract] OR “bacteriuria”[Title/abstract] OR “pyelonephritis”[Title/abstract] OR “cystitis”[Title/abstract] OR “pyuria”[Title/abstract]) |
| Embase | (‘child’/mj OR ‘pediatrics’/mj OR ‘newborn’/mj OR ‘child*’:ti,ab,kw OR ‘schoolchild*’:ti,ab,kw OR ‘infan*’:ti,ab,kw OR ‘adolescen*’:ti,ab,kw OR ‘pediatri*’:ti,ab,kw OR ‘paediatr*’:ti,ab,kw OR ‘neonat*’:ti,ab,kw OR ‘boy’:ti,ab,kw OR ‘boys’:ti,ab,kw OR ‘boyhood’:ti,ab,kw OR ‘girl’:ti,ab,kw OR ‘girls’:ti,ab,kw OR ‘girlhood’:ti,ab,kw OR ‘youth’:ti,ab,kw OR ‘youths’:ti,ab,kw OR ‘baby’:ti,ab,kw OR ‘babies’:ti,ab,kw OR ‘toddler*’:ti,ab,kw OR ‘teen’:ti,ab,kw OR ‘teens’:ti,ab,kw OR ‘teenager*’:ti,ab,kw OR ‘newborn*’:ti,ab,kw OR ‘postneonat*’:ti,ab,kw OR ‘postnat*’:ti,ab,kw OR ‘perinat*’:ti,ab,kw OR ‘puberty’:ti,ab,kw OR ‘preschool*’:ti,ab,kw OR ‘suckling*’:ti,ab,kw OR ‘picu’:ti,ab,kw OR ‘nicu’:ti,ab,kw) AND (‘urine’/mj AND ‘microbiology’/de OR ‘microflora’/mj OR ‘intestine flora’/mj OR (‘gastrointestinal’:ti,ab,kw AND ‘microbiome’:ti,ab,kw) OR ‘gastrointestinal microbiome’:ti,ab,kw OR (‘gut’:ti,ab,kw AND ‘microbiome’:ti,ab,kw) OR ‘gut microbiome’:ti,ab,kw OR ‘microbiota’:ti,ab,kw OR ‘urinary microbiota’:ti,ab,kw OR ‘gut microbiota’:ti,ab,kw OR ‘urine microbiome’:ti,ab,kw OR ‘urine microbiota’:ti,ab,kw) AND (‘lower urinary tract symptom’/mj OR ‘nocturia’/mj OR ‘bladder disease’/mj OR ‘urine incontinence’/mj OR ‘nocturnal enuresis’/mj OR ‘urinary tract infection’/mj OR ‘pyelonephritis’/mj OR ‘cystitis’/mj OR ‘lower urinary tract symptoms’:ti,ab,kw OR ‘luts’:ti,ab,kw OR ‘nocturia’:ti,ab,kw OR ‘oab’:ti,ab,kw OR ‘overactive bladder’:ti,ab,kw OR ‘bed wetting’:ti,ab,kw OR ‘urological symptoms’:ti,ab,kw OR ‘urinary disorders’:ti,ab,kw OR ‘urinary urge incontinence’:ti,ab,kw OR ‘lower urinary tract dysfunction’:ti,ab,kw OR ‘lower urinary tract problems’:ti,ab,kw OR ‘urinary urgency’:ti,ab,kw OR ‘urinary frequency’:ti,ab,kw OR ‘voiding dysfunction’:ti,ab,kw OR (‘urinary’:ti,ab,kw AND ‘tract’:ti,ab,kw AND ‘infections’:ti,ab,kw) OR ‘urinary tract infections’:ti,ab,kw OR ‘urinary tract infection’:ti,ab,kw OR ‘uti’:ti,ab,kw OR ‘utis’:ti,ab,kw OR ‘uti‘s’:ti,ab,kw OR ‘bacteriuria’:ti,ab,kw OR ‘pyelonephritis’:ti,ab,kw OR ‘cystitis’:ti,ab,kw OR ‘pyuria’:ti,ab,kw) |
| CINAHL/Ebsco HOST | (((MH “Child”) OR (MH “Pediatrics”) OR (MH “Infant, Newborn”) OR (child*) OR (schoolchild*) OR (infan*) OR (adolescen*) OR (pediatri*) OR (paediatr*) OR (neonat*) OR (boy) OR (boys) OR (boyhood) OR (girl) OR (girls) OR (girlhood) OR (youth) OR (youths) OR (baby) OR (babies) OR (toddler*) OR (teen) OR (teens) OR (teenager*) OR (newborn*) OR (postneonat*) OR (postnat*) OR (perinat*) OR (puberty) OR (preschool*) OR (suckling*) OR (picu) OR (nicu))) AND (((MH “Urine/Microbiology”) OR (MH “Microbiota”) OR (MH “Gastrointestinal Microbiome”) OR (urine microbiome) OR (gastrointestinal AND microbiome) OR (gastrointestinal microbiome) OR (gut AND microbiome) OR (gut microbiome) OR (microbiota) OR (urinary microbiota) OR (gut microbiota) OR (urine microbiome) OR (urine microbiota))) AND (((MH “Lower Urinary Tract Symptoms”) OR (MH “Nocturia”) OR (MH “Urinary Bladder Diseases”) OR (MH “Urinary Incontinence”) OR (MH “Nocturnal Enuresis”) OR (MH “Urinary Tract Infections”) OR (MH “Pyelonephritis”) OR (MH “Cystitis”) OR (Lower Urinary Tract Symptoms) OR (LUTS) OR (Nocturia) OR (OAB) OR (overactive bladder) OR (bed wetting) OR (urological symptoms) OR (urinary disorders) OR (Urinary urge incontinence) OR (lower urinary tract dysfunction) OR (lower urinary tract problems) OR (urinary urgency) OR (urinary frequency) OR (voiding dysfunction) OR (urinary AND tract AND infections) OR (urinary tract infections) OR (urinary tract infection) OR (UTI) OR (UTIs) OR (UTIs) OR (bacteriuria) OR (pyelonephritis) OR (cystitis) OR (pyuria))) |
| Publication Year | First Author | Study Type | Retrospective vs. Prospective | Method of Microbiome Analysis | Patient Sex Male: Female (n:n) | Type of Sample | Total n | Groups | n per Group | Mean Patient Age |
|---|---|---|---|---|---|---|---|---|---|---|
| 2018 | Paalanne [22] | case-control | prospective | 16S Ribosomal RNA sequencing | 30:76 | stool | 106 | UTI | 37 | 20.3 months |
| Control | 69 | 21.8 months | ||||||||
| 2020 | Forster [23] | cross-sectional | retrospective | 16S Ribosomal RNA sequencing | 19:15 | urine | 34 | UTI | 11 | 11 years |
| ASB | 19 | 8.8 years | ||||||||
| Control | 4 | 15 years | ||||||||
| 2020 | Kinneman [24] | cross-sectional | prospective | 16S Ribosomal RNA sequencing | 26:59 | urine | 85 | UTI | 9 | 382 days |
| Control | 76 | |||||||||
| 2022 | Vitko [25] | case-control | prospective | 16S Ribosomal RNA sequencing | 12:37 | urine | 49 | VUR without Renal scarring | 20 | 4.8 years |
| VUR with Renal scarring | 13 | 3.8 years | ||||||||
| controls | 16 | 10.2 years | ||||||||
| 2022 | Akarken [26] | cross-sectional | retrospective | 16S Ribosomal RNA sequencing | 20:29 | stool | 49 | Voiding dysfunction | 25 | 8.26 years |
| Control | 24 | 8.00 years | ||||||||
| 2023 | Cole [27] | case-control | prospective | 16S Ribosomal RNA sequencing | 0:33 | urine | 33 | Bladder-Bowel Dysfunction (BBD) | 25 | 8.0 years |
| Control | 8 | 6.3 years | ||||||||
| 2023 | Urakami [28] | cross-sectional | prospective | 16S Ribosomal RNA sequencing | 42:37 | Stool | 79 | UTI | 28 | 5 months |
| Control | 51 | 5 months | ||||||||
| 2024 | Kelly [29] | cross-sectional | prospective | 16S Ribosomal RNA sequencing | 13:20 | urine | 33 | No UTI or Unknown (excluded for analysis) | 5 | 40.1 months |
| History of 1 UTI | 10 | |||||||||
| History of 2 UTIs | 8 | |||||||||
| History of 3+ UTIs | 10 | |||||||||
| 2024 | L. Hong [30] | Case-control | prospective | 16S Ribosomal RNA sequencing | 74:77 | stool | 151 | UTI | 53 | 29.49 weeks |
| Control | 98 | 30.24 weeks |
| Publication Year | First Author | Patient Sex Male:Female (n:n) | Type of Sample | Total n | Groups | n per Group | Mean Patient Age | Alpha Diversity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chao1-Index | SMD | Shannon-Waver | SMD | Inverse Simpson | SMD | Pielou | SMD | ||||||||
| 2018 | Paalanne [22] | 30:76 | stool | 106 | UTI | 37 | 20.3 months | 1040 (SD 540.5) | −0.02 | 5.9 (SD 1.61) | −0.13 | ||||
| Control | 69 | 21.8 months | 1050 (SD 485.0) | 6.09 (SD 1.37) | |||||||||||
| 2020 | Forster [23] | 19:15 | urine | 34 | UTI | 11 | 11 years | 311.38 (SD 140.75) | 0.13 1 | 1.65 (SD 0.44) | −0.23 1 | ||||
| ASB | 19 | 8.8 years | 156,77 (SD 138.24) | 1.54 2 | 1.34 (SD 1.35) | 0.14 2 | |||||||||
| Control | 4 | 15 years | 140.34 (SD 100.16) | 1.11 3 | 1.82 (SD 0.98) | 0.37 3 | |||||||||
| 2020 | Kinneman [24] | 26:59 | urine | 85 | UTI | 9 | 382 days | 1.65 (SD 0.44) | 3.33 | ||||||
| Control | 76 | 3.80 (SD 1.58) | |||||||||||||
| 2021 | Vitko [25] | 12:37 | urine | 49 | VUR | 20 | 4.8 years | Not Reported | |||||||
| 13 | 3.8 years | ||||||||||||||
| controls | 16 | 10.2 years | |||||||||||||
| 2022 | Akarken [26] | 20:29 | stool | 49 | VD | 25 | 8.26 years | Not Reported | |||||||
| Control | 24 | 8.00 years | |||||||||||||
| 2023 | Cole [27] | 0:33 | urine | 33 | BBD | 25 | 8.0 years | 139.03 (SD 81.25) | −0.41 | 2.51 (SD 1.68) | −0.71 | ||||
| Control | 8 | 6.3 years | 170.57 (SD 67.70) | 3.52 (SD 0.20) | |||||||||||
| 2023 | Urakami [28] | 42:37 | Stool | 79 | UTI | 28 | 5 months | 42.5 (IQR 33.5–48.5) | 1.4 | 3.0 (IQR 2.7–3.5) | 0.77 | ||||
| Control | 51 | 5 months | 97 (IQR 69.5–132.0) | 3.7 (IQR 3.2–4.6) | |||||||||||
| 2024 | Kelly [29] | Male | urine | 33 | Healthy | 13 | 40.1 months | 1.75 (SD 0.94) | 0.91 | 4.30 (SD 2.71) | 0.87 | 0.65 (SD 0.19) | 0.57 | ||
| Female | 20 | 2.37 (SD 0.43) | 7.66 (SD 4.46) | 0.73 (SD 0.10) | |||||||||||
| 13:20 | 33 | 0 UTI or Unknown (excluded for analysis) | 5 | / | / | ||||||||||
| History of 1 UTI | 10 | 2.58 (SD 0.40) | 0.58 4 | 8.64 (SD 4.34) | 0.32 4 | 0.83 (SD 0.04) | 2.38 4 | ||||||||
| History of 2 UTIs | 8 | 2.31 (SD 0.55) | 0.78 5 | 7.34 (SD 3.65) | 1.14 5 | 0.70 (SD 0.07) | 0.68 5 | ||||||||
| History of 3+ UTIs | 10 | 1.62 (SD 1.07) | 1.19 6 | 3.9 (SD 2.43) | 1.35 6 | 0.53 (SD 0.32) | 1.29 6 | ||||||||
| 2024 | Luyang Hong [30] | 74:77 | stool | 151 | Gram-positive UTI | 53 | 29.49 weeks | Only in the figure | |||||||
| Gram-negative UTI | Only in the figure | ||||||||||||||
| Control | 98 | 30.24 weeks | Only in the figure | ||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van den Ende, M.; Van de Steen, L.; Everaert, K.; Hervé, F.; Bou Kheir, G. Exploring Childhood Lower Urinary Tract Symptoms (LUTS), Urinary Tract Infections (UTIs) and the Microbiome—A Systematic Review. Life 2025, 15, 730. https://doi.org/10.3390/life15050730
Van den Ende M, Van de Steen L, Everaert K, Hervé F, Bou Kheir G. Exploring Childhood Lower Urinary Tract Symptoms (LUTS), Urinary Tract Infections (UTIs) and the Microbiome—A Systematic Review. Life. 2025; 15(5):730. https://doi.org/10.3390/life15050730
Chicago/Turabian StyleVan den Ende, Mauro, Laure Van de Steen, Karel Everaert, François Hervé, and George Bou Kheir. 2025. "Exploring Childhood Lower Urinary Tract Symptoms (LUTS), Urinary Tract Infections (UTIs) and the Microbiome—A Systematic Review" Life 15, no. 5: 730. https://doi.org/10.3390/life15050730
APA StyleVan den Ende, M., Van de Steen, L., Everaert, K., Hervé, F., & Bou Kheir, G. (2025). Exploring Childhood Lower Urinary Tract Symptoms (LUTS), Urinary Tract Infections (UTIs) and the Microbiome—A Systematic Review. Life, 15(5), 730. https://doi.org/10.3390/life15050730







