Prediction of Early Diagnosis in Ovarian Cancer Patients Using Machine Learning Approaches with Boruta and Advanced Feature Selection
Abstract
1. Introduction
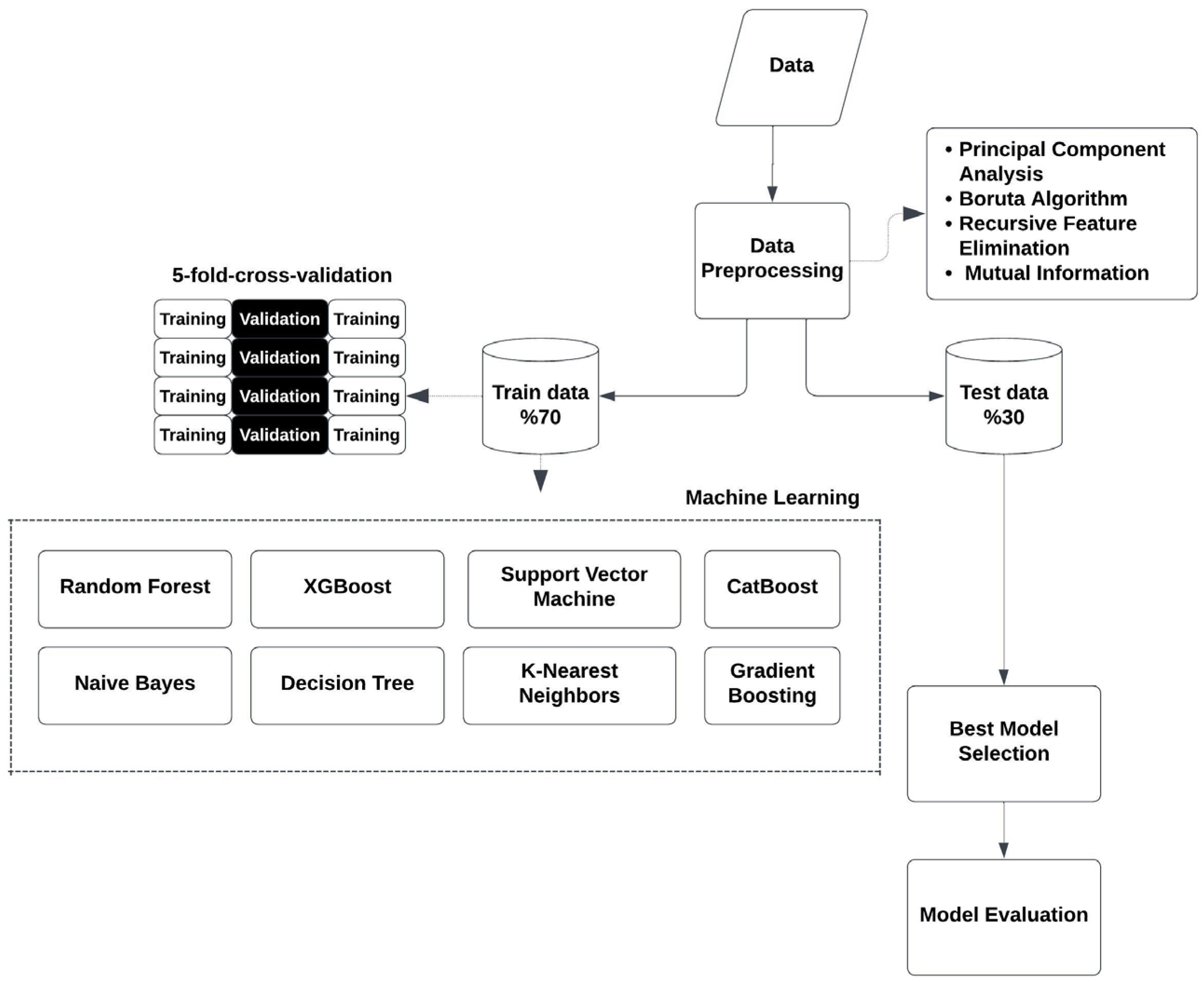
2. Materials and Methods
2.1. Data Preprocessing
2.2. Feature Selection and Dimensionality Reduction
2.3. Data Splitting
2.4. Classification Algorithms
2.5. Hyperparameter Tuning
2.6. Evaluation Metrics
- AUC (Area Under the Curve): This index assesses the ability of a model to perform discrimination between classes and has particular significance for unbalanced datasets [35].
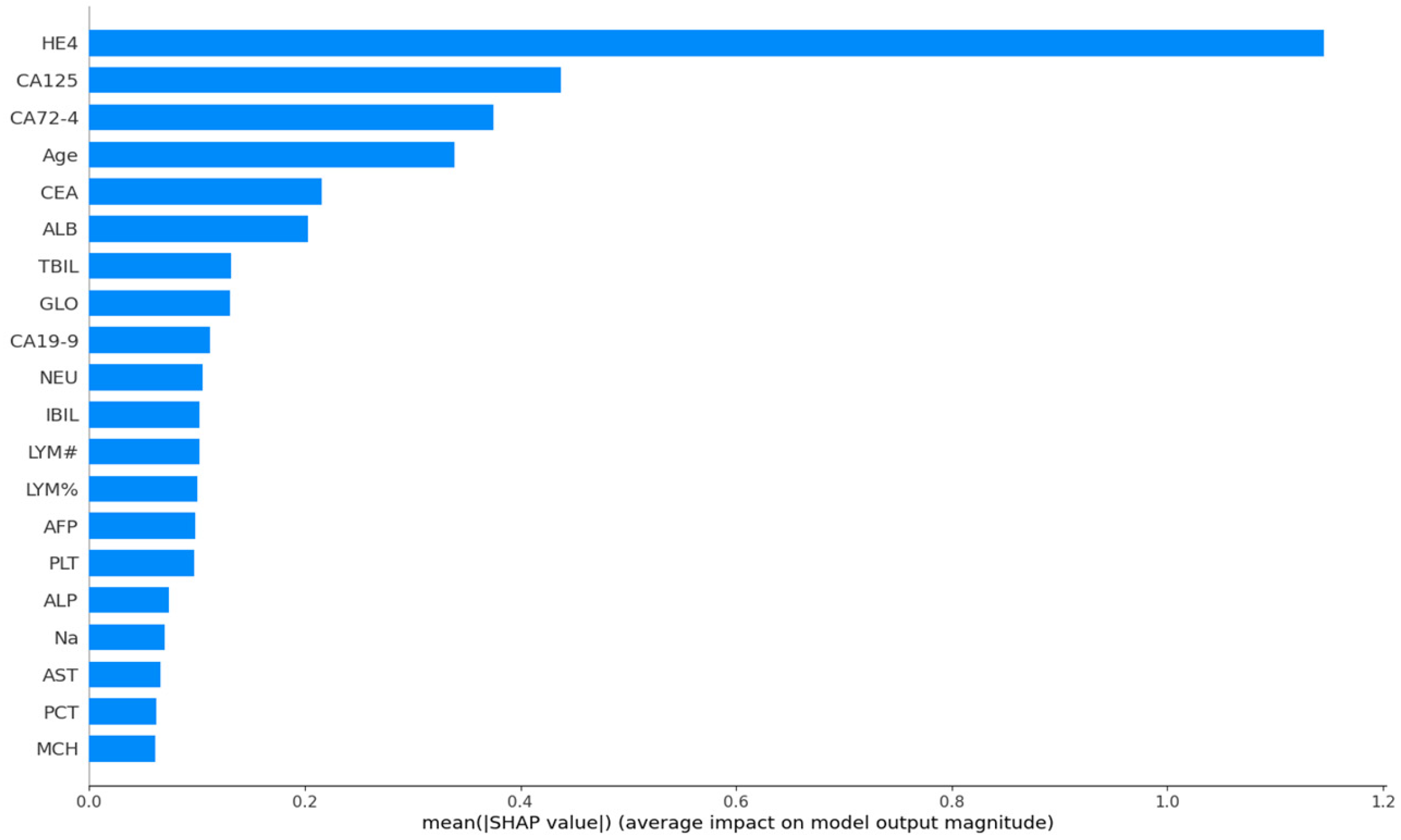
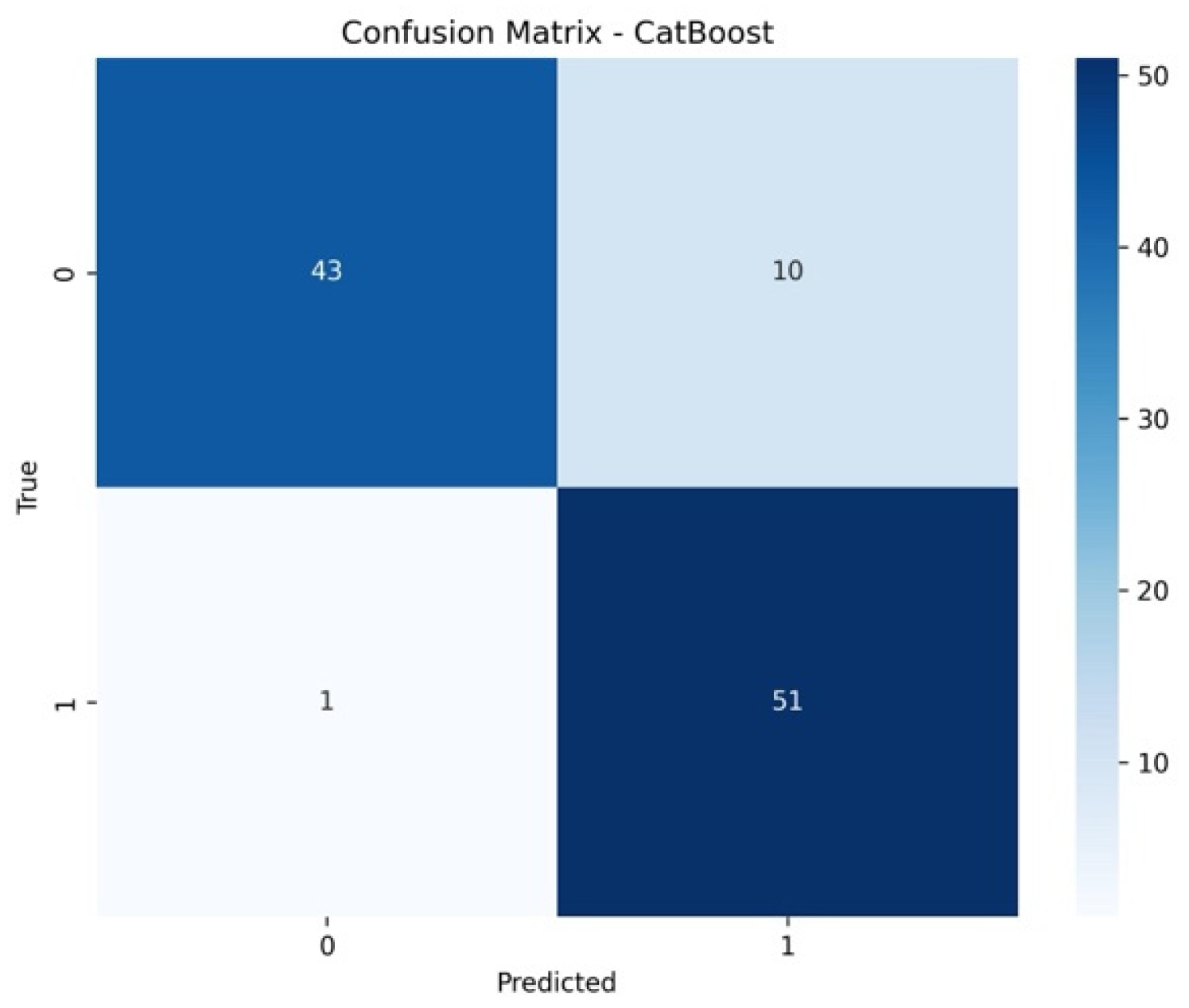
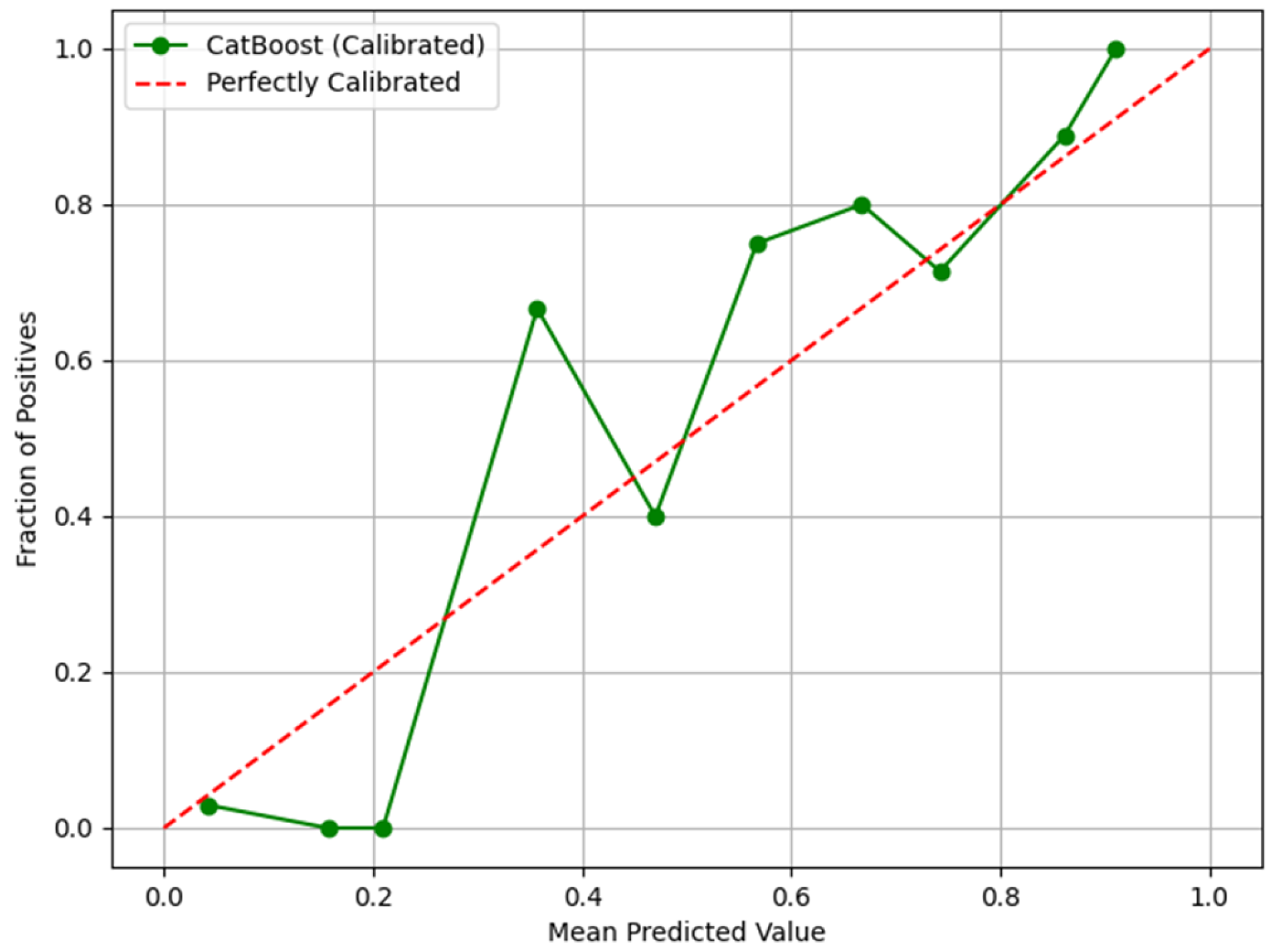
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PCA | Principal Component Analysis |
| RFE | Recursive Feature Elimination |
| MI | Mutual Information |
| SVM | Support Vector Machine |
| ANN | Artificial Neural Network |
| AUC | Area Under the Curve |
| TP | True Positive |
| TN | True Negative |
| FP | False Positive |
| FN | False Negative |
References
- Globocan. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020; Available online: https://gco.iarc.fr/today (accessed on 31 March 2025).
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87. [Google Scholar] [PubMed]
- Surveillance, Epidemiology, and End Results Cancer Stat Facts: Ovarian cancer. Available online: http://seer.cancer.gov/statfacts/html/ovary.html (accessed on 25 July 2017).
- American Cancer Society. Ovarian Cancer Survival Rates. Available online: https://www.cancer.org/cancer/ovarian-cancer/detection-diagnosis-staging/survival-rates.html (accessed on 31 March 2025).
- Fischerova, D.; Burgetova, A. Imaging techniques for the evaluation of ovarian cancer. Best. Pract. Res. Clin. Obstet. Gynaecol. 2014, 28, 697–720. [Google Scholar] [CrossRef] [PubMed]
- Wernick, M.N.; Yang, Y.; Brankov, J.G.; Yourganov, G.; Strother, S.C. Machine Learning in Medical Imaging. IEEE Signal Process. Mag. 2010, 27, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine Learning in Medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef]
- Ayyoubzadeh, S.M.; Ahmadi, M.; Yazdipour, A.B.; Ghorbani-Bidkorpeh, F.; Ahmadi, M. Prediction of ovarian cancer using artificial intelligence tools. Health Sci. Rep. 2024, 7, e2203. [Google Scholar] [CrossRef]
- Lu, M.; Fan, Z.; Xu, B.; Chen, L.; Zheng, X.; Li, J.; Znati, T.; Mi, Q.; Jiang, J. Using machine learning to predict ovarian cancer. Int. J. Med. Inform. 2020, 141, 104195. [Google Scholar] [CrossRef]
- Little, R.; Rubin, D. Statistical Analysis with Missing Data, 3rd ed.; Wiley: Hoboken, NJ, USA, 2019. [Google Scholar] [CrossRef]
- Schafer, J.L. Analysis of Incomplete Multivariate Data, 1st ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 1997. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Liu, H.; Motoda, H. Computational Methods of Feature Selection, 1st ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 2007. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal Component Analysis, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar] [CrossRef]
- Kursa, M.B.; Rudnicki, W.R. Feature selection with the Boruta package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene Selection for Cancer Classification using Support Vector Machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Beraha, M.; Metelli, A.; Papini, M.; Tirinzoni, A.; Restelli, M. Feature Selection via Mutual. Information: New Theoretical Insights. In Proceedings of the 2019 International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019; pp. 1–9. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD’ 16); Association for Computing Machinery: New York, NY, USA, 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Prokhorenkova, L.; Gusev, G.; Vorobev, A.; Dorogush, A.V.; Gulin, A. Catboost: Unbiased Boosting with Categorical Features. In Proceedings of the 32nd International Conference on Neural Information Processing Systems, Montréal, QC, Canada, 3–8 December 2018; pp. 6639–6649. [Google Scholar]
- Quinlan, J.R. Induction of decision trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Zhang, H. The optimality of Naive Bayes. In Proceedings of the Seventeenth International Florida Artificial Intelligence Research Society Conference (FLAIRS), Miami Beach, FL, USA, 17–19 May 2004; pp. 562–567. [Google Scholar]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar]
- Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Hutter, F.; Babic, D.; Hoos, H.H.; Hu, A.J. Boosting verification by automatic tuning of decision procedures. In Proceedings of the Formal Methods in Computer Aided Design (FMCAD’ 07); IEEE Computer Society: Washington, DC, USA, 2007; pp. 27–34. [Google Scholar]
- Hutter, F.; Hoos, H.H.; Leyton-Brown, K. Sequential Model-Based Optimization for General Algorithm Configuration. In Learning and Intelligent Optimization; Coello, C.A.C., Ed.; LION 2011. Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2011; Volume 6683. [Google Scholar] [CrossRef]
- Bartz-Beielstein, T.; Markon, S. Tuning search algorithms for real-world applications: A regression tree based approach. In Proceedings of the 2004 Congress on Evolutionary Computation, Portland, OR, USA, 19–23 June 2004; pp. 1111–1118. [Google Scholar]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Rainio, O.; Teuho, J.; Klén, R. Evaluation metrics and statistical tests for machine learning. Sci. Rep. 2024, 14, 6086. [Google Scholar] [CrossRef]
- Ferri, C.; Hernández-Orallo, J.; Modroiu, R. An experimental comparison of performance measures for classification. Pattern Recogn. Lett. 2009, 30, 27–38. [Google Scholar] [CrossRef]
- Fawcett, T. An introduction to ROC analysis. Pattern Recogn. Lett. 2006, 27, 861–874. [Google Scholar]
- Yang, L.R.; Yang, M.; Chen, L.L.; Shen, Y.L.; He, Y.; Meng, Z.T.; Wang, W.Q.; Li, F.; Liu, Z.J.; Li, L.H.; et al. Machine learning for epithelial ovarian cancer platinum resistance recurrence identification using routine clinical data. Front. Oncol. 2024, 14, 1457294. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Gu, S.; Yang, J.; Zhao, Y.; Sheng, J.; Cheng, S.; Xu, S.; Wu, Y.; Ma, M.; Luo, X.; et al. Comprehensive machine learning-based preoperative blood features predict the prognosis for ovarian cancer. BMC Cancer 2024, 24, 267. [Google Scholar] [CrossRef]
- Sheela Lavanya, J.M.; Subbulakshmi, P. Innovative approach towards early prediction of ovarian cancer: Machine learning-enabled XAI techniques. Heliyon 2024, 10, e29197. [Google Scholar] [CrossRef]
- Amniouel, S.; Yalamanchili, K.; Sankararaman, S.; Jafri, M.S. Evaluating Ovarian Cancer Chemotherapy Response Using Gene Expression Data and Machine Learning. BioMedInformatics 2024, 4, 1396–1424. [Google Scholar] [CrossRef]
- Paik, E.S.; Lee, J.W.; Park, J.Y.; Kim, J.H.; Kim, M.; Kim, T.J.; Choi, C.H.; Kim, B.G.; Bae, D.S.; Seo, S.W. Prediction of survival outcomes in patients with epithelial ovarian cancer using machine learning methods. J. Gynecol. Oncol. 2019, 30, e65. [Google Scholar] [CrossRef]
- Gui, T.; Cao, D.; Yang, J.; Wei, Z.; Xie, J.; Wang, W.; Xiang, Y.; Peng, P. Early prediction and risk stratification of ovarian cancer based on clinical data using machine learning approaches. J. Gynecol. Oncol. 2024, 36, e53. [Google Scholar] [CrossRef]
- Chen, Z.; Ouyang, H.; Sun, B.; Ding, J.; Zhang, Y.; Li, X. Utilizing explainable machine learning for progression-free survival prediction in high-grade serous ovarian cancer: Insights from a prospective cohort study. Int. J. Surg. 2025. [Google Scholar] [CrossRef]
- Piedimonte, S.; Mohamed, M.; Rosa, G.; Gerstl, B.; Vicus, D. Predicting Response to Treatment and Survival in Advanced Ovarian Cancer Using Machine Learning and Radiomics: A Systematic Review. Cancers 2025, 17, 336. [Google Scholar] [CrossRef]

| Variable | Description |
|---|---|
| AFP | Alpha-fetoprotein; a tumor marker primarily used to assess liver function and detect certain cancers. |
| AG | Albumin/Globulin ratio; a diagnostic indicator of liver function and protein balance. |
| Age | The age of the individual, typically used as a demographic variable. |
| ALB | Albumin; a protein produced by the liver, used to assess liver function and nutritional status. |
| ALP | Alkaline phosphatase; an enzyme related to liver, bone, and bile duct function. |
| ALT | Alanine aminotransferase; an enzyme that helps assess liver damage. |
| AST | Aspartate aminotransferase; an enzyme that is indicative of liver and heart function. |
| BASO# | Absolute basophil count; basophils are a type of white blood cell involved in immune responses, including allergies. |
| BASO% | Percentage of basophils among total white blood cells. |
| BUN | Blood urea nitrogen; a marker used to evaluate kidney function. |
| Ca | Calcium; a mineral essential for bone health, muscle function, and nerve signaling. |
| CA125 | Cancer antigen 125; a biomarker used to assess ovarian cancer. |
| CA19-9 | Cancer antigen 19-9; a marker used to assess pancreatic cancer. |
| CA72-4 | Cancer antigen 72-4; a tumor marker mainly used for gastric cancer. |
| CEA | Carcinoembryonic antigen; a protein often elevated in various cancers, particularly colorectal cancer. |
| CL | Chloride; an electrolyte that helps maintain fluid balance and acid–base status. |
| CO2CP | Carbon dioxide content; measures the blood’s bicarbonate concentration, important for assessing acid–base balance. |
| CREA | Creatinine; a waste product of muscle metabolism, commonly used to assess kidney function. |
| DBIL | Direct bilirubin; a form of bilirubin that is conjugated in the liver and used to assess liver function and jaundice. |
| EO# | Absolute eosinophil count; eosinophils are white blood cells involved in allergic responses and parasitic infections. |
| EO% | Percentage of eosinophils among total white blood cells. |
| GGT | Gamma-glutamyl transferase; an enzyme used to evaluate liver and biliary system disorders. |
| GLO | Globulin; a class of proteins that includes immunoglobulins, which play a role in immune function. |
| GLU | Glucose; a key source of energy for cells, its levels are used to assess metabolic function and diabetes. |
| HCT | Hematocrit; the proportion of blood that is composed of red blood cells, used to assess anemia or dehydration. |
| HE4 | Human epididymis protein 4; a biomarker for ovarian cancer detection. |
| HGB | Hemoglobin; a protein in red blood cells that carries oxygen from the lungs to the tissues. |
| IBIL | Indirect bilirubin; the unconjugated form of bilirubin, elevated in liver dysfunction and hemolysis. |
| K | Potassium; an essential electrolyte that regulates cell function, heart rhythm, and muscle contractions. |
| LYM# | Absolute lymphocyte count; lymphocytes are a subset of white blood cells that are critical for immune function. |
| LYM% | Percentage of lymphocytes among total white blood cells. |
| MCH | Mean corpuscular hemoglobin; a measure of the average amount of hemoglobin per red blood cell. |
| MCV | Mean corpuscular volume; the average volume of a red blood cell, used to classify anemia. |
| Mg | Magnesium; a mineral important for muscle and nerve function and enzymatic processes. |
| MONO# | Absolute monocyte count; monocytes are white blood cells involved in immune response and inflammation. |
| MONO% | Percentage of monocytes among total white blood cells. |
| MPV | Mean platelet volume; a measure of the size of platelets in the blood, used to assess platelet production and function. |
| Na | Sodium; an electrolyte that helps regulate fluid balance, blood pressure, and nerve function. |
| NEU | Neutrophils; the most abundant type of white blood cell, important for fighting bacterial infections. |
| PCT | Procalcitonin; a biomarker used to detect bacterial infections and assess sepsis. |
| PDW | Platelet distribution width; a measure of the variability in platelet size, useful for assessing platelet function. |
| PHOS | Phosphate; a mineral important for bone health and cellular energy production. |
| PLT | Platelets; cells involved in blood clotting and wound healing. |
| RBC | Red blood cells; cells responsible for oxygen transport in the body. |
| RDW | Red cell distribution width; a measure of the variability in red blood cell size, useful for diagnosing anemia. |
| TBIL | Total bilirubin; a combination of direct and indirect bilirubin, used to assess liver function and jaundice. |
| TP | Total protein; the sum of albumin and globulin in the blood, reflecting overall nutritional and liver status. |
| UA | Uric acid; a waste product of purine metabolism; elevated levels can indicate kidney dysfunction or gout. |
| Menopause | A binary categorical variable indicating whether the individual is postmenopausal. |
| TYPE | A binary categorical variable. |
| Model | Method | Accuracy | F1 | Precision | Recall | AUC |
|---|---|---|---|---|---|---|
| Random Forest | Boruta | 0.8667 | 0.8665 | 0.8689 | 0.8667 | 0.9426 |
| PCA | 0.7238 | 0.7228 | 0.72821 | 0.7238 | 0.8146 | |
| RFE | 0.8952 | 0.8952 | 0.8954 | 0.8952 | 0.9505 | |
| Mutual Information | 0.8571 | 0.8569 | 0.8605 | 0.8571 | 0.9307 | |
| XGBoost | Boruta | 0.8381 | 0.8375 | 0.8444 | 0.8381 | 0.9216 |
| PCA | 0.7429 | 0.7424 | 0.7453 | 0.7429 | 0.8320 | |
| RFE | 0.8381 | 0.8378 | 0.84133 | 0.8381 | 0.9508 | |
| Mutual Information | 0.8381 | 0.8378 | 0.8413 | 0.8381 | 0.9175 | |
| CatBoost | Boruta | 0.8952 | 0.8945 | 0.9073 | 0.8952 | 0.9502 |
| PCA | 0.7524 | 0.7511 | 0.7587 | 0.7524 | 0.8396 | |
| RFE | 0.8857 | 0.8856 | 0.8881 | 0.8857 | 0.9430 | |
| Mutual Information | 0.8857 | 0.8854 | 0.8909 | 0.8857 | 0.9296 | |
| Decision Tree | Boruta | 0.7905 | 0.7904 | 0.7909 | 0.7905 | 0.7896 |
| PCA | 0.6190 | 0.6186 | 0.6200 | 0.6190 | 0.6540 | |
| RFE | 0.8285 | 0.8284 | 0.8289 | 0.8285 | 0.8494 | |
| Mutual Information | 0.7904 | 0.7902 | 0.7923 | 0.7904 | 0.7908 | |
| K-Nearest Neighbors | Boruta | 0.8190 | 0.8190 | 0.8191 | 0.8190 | 0.8534 |
| PCA | 0.7428 | 0.7378 | 0.7650 | 0.7428 | 0.7821 | |
| RFE | 0.8285 | 0.8276 | 0.8366 | 0.8285 | 0.8798 | |
| Mutual Information | 0.8285 | 0.8283 | 0.8306 | 0.8285 | 0.8844 | |
| Naive Bayes | Boruta | 0.8000 | 0.7986 | 0.8093 | 0.8000 | 0.9023 |
| PCA | 0.7142 | 0.7079 | 0.7369 | 0.7142 | 0.7634 | |
| RFE | 0.8190 | 0.8164 | 0.8402 | 0.8190 | 0.9296 | |
| Mutual Information | 0.838095 | 0.836465 | 0.853893 | 0.838095 | 0.929245 | |
| Gradient Boosting | Boruta | 0.8476 | 0.8472 | 0.8523 | 0.8476 | 0.9183 |
| PCA | 0.7333 | 0.7319 | 0.7392 | 0.7333 | 0.8004 | |
| RFE | 0.8571 | 0.8565 | 0.8637 | 0.8571 | 0.9346 | |
| Mutual Information | 0.8476 | 0.8472 | 0.8523 | 0.8476 | 0.9174 | |
| SVM | Boruta | 0.8476 | 0.8474 | 0.8497 | 0.8476 | 0.8762 |
| PCA | 0.8000 | 0.7998 | 0.8010 | 0.8000 | 0.8534 | |
| RFE | 0.8761 | 0.8759 | 0.8797 | 0.8761 | 0.9154 | |
| Mutual Information | 0.8761 | 0.8753 | 0.8877 | 0.8761 | 0.8925 | |
| ANN | Boruta | 0.8476 | 0.8472 | 0.8524 | 0.8476 | 0.8828 |
| PCA | 0.7905 | 0.7845 | 0.8297 | 0.7905 | 0.8842 | |
| RFE | 0.7905 | 0.7894 | 0.7977 | 0.7905 | 0.8157 | |
| Mutual Information | 0.8095 | 0.8093 | 0.8115 | 0.8095 | 0.8330 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Öznacar, T.; Güler, T. Prediction of Early Diagnosis in Ovarian Cancer Patients Using Machine Learning Approaches with Boruta and Advanced Feature Selection. Life 2025, 15, 594. https://doi.org/10.3390/life15040594
Öznacar T, Güler T. Prediction of Early Diagnosis in Ovarian Cancer Patients Using Machine Learning Approaches with Boruta and Advanced Feature Selection. Life. 2025; 15(4):594. https://doi.org/10.3390/life15040594
Chicago/Turabian StyleÖznacar, Tuğçe, and Tunç Güler. 2025. "Prediction of Early Diagnosis in Ovarian Cancer Patients Using Machine Learning Approaches with Boruta and Advanced Feature Selection" Life 15, no. 4: 594. https://doi.org/10.3390/life15040594
APA StyleÖznacar, T., & Güler, T. (2025). Prediction of Early Diagnosis in Ovarian Cancer Patients Using Machine Learning Approaches with Boruta and Advanced Feature Selection. Life, 15(4), 594. https://doi.org/10.3390/life15040594






