Wild Emmer (Triticum turgidum ssp. dicoccoides) Diversity in Southern Turkey: Evaluation of SSR and Morphological Variations
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Agromorphological Variation in Wild Emmer Populations
3.2. Genetic Diversity Within and Between the Populations and Sub-Regions
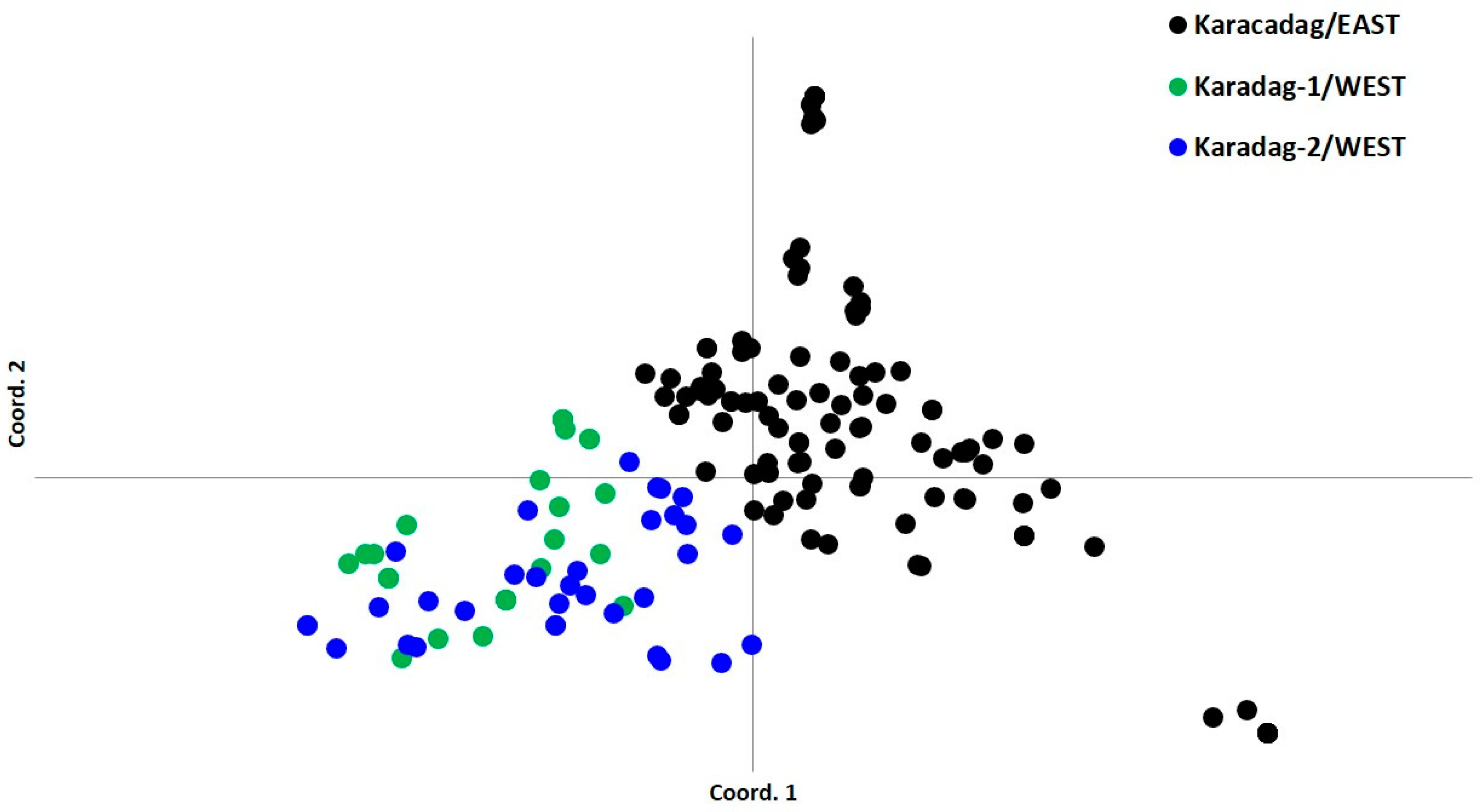
3.3. PCoA and Neighbor-Joining Grouping Patterns in Wild Emmer Populations
4. Discussion
4.1. Agromorphological Diversity
4.2. Genetic Diversity of In situ Populations
4.3. PCoA and Neighbor-Joining Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Snowdon, R.J.; Wittkop, B.; Chen, T.-W.; Stahl, A. Crop adaptation to climate change as a consequence of long-term breeding. Theor. Appl. Genet. 2021, 134, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Bai, S.; Li, H.; Sun, G.; Zhang, D.; Ma, F.; Zhao, X.; Nie, F.; Li, J.; Chen, L.; et al. Introgressing the Aegilops tauschii genome into wheat as a basis for cereal improvement. Nat. Plants 2021, 7, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Bohra, A.; Roorkiwal, M.; Barmukh, R.; Cowling, W.; Chitikineni, A.; Lam, H.-M.; Hickey, L.T.; Croser, J.; Edwards, D.; et al. Rapid delivery systems for future food security. Nat. Biotechnol. 2021, 39, 1179–1181. [Google Scholar] [CrossRef]
- Feng, K.W.; Nie, X.J.; Cui, L.C.; Deng, P.C.; Wang, M.X.; Song, W.N. Genome-Wide Identification and Characterization of Salinity Stress-Responsive miRNAs in Wild Emmer Wheat (Triticum turgidum ssp dicoccoides). Genes 2017, 8, 156. [Google Scholar] [CrossRef]
- Peng, J.H.; Sun, D.F.; Nevo, E. Wild emmer wheat, Triticum dicoccoides, occupies a pivotal position in wheat domestication process. Aust. J. Crop Sci. 2011, 5, 1127–1143. [Google Scholar]
- Dong, P.; Wei, Y.M.; Chen, G.Y.; Li, W.; Wang, J.R.; Nevo, E.; Zheng, Y.L. Sequence-related amplified polymorphism (SRAP) of wild emmer wheat (Triticum dicoccoides) in Israel and its ecological association. Biochem. Syst. Ecol. 2010, 38, 1–11. [Google Scholar] [CrossRef]
- Lack, H.W.; Van Slageren, M. The discovery, typification and rediscovery of wild emmer wheat, Triticum turgidum subsp. dicoccoides (Poaceae). Willdenowia 2020, 50, 207–216. [Google Scholar] [CrossRef]
- Ozkan, H.; Willcox, G.; Graner, A.; Salamini, F.; Kilian, B. Geographic distribution and domestication of wild emmer wheat (Triticum dicoccoides). Genet. Resour. Crop Evol. 2011, 58, 11–53. [Google Scholar] [CrossRef]
- Kantar, M.; Lucas, S.J.; Budak, H. miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 2011, 233, 471–484. [Google Scholar] [CrossRef]
- Shavrukov, Y.; Langridge, P.; Tester, M.; Nevo, E. Wide genetic diversity of salinity tolerance, sodium exclusion and growth in wild emmer wheat, Triticum dicoccoides. Breed. Sci. 2010, 60, 426–435. [Google Scholar] [CrossRef]
- Feng, K.; Cui, L.; Lv, S.; Bian, J.; Wang, M.; Song, W.; Nie, X. Comprehensive evaluating of wild and cultivated emmer wheat (Triticum turgidum L.) genotypes response to salt stress. Plant Growth Regul. 2017, 84, 261–273. [Google Scholar] [CrossRef]
- Soresi, D.; Bagnaresi, P.; Crescente, J.M.; Diaz, M.; Cattivelli, L.; Vanzetti, L.; Carrera, A. Genetic Characterization of a Fusarium Head Blight Resistance QTL from Triticum turgidum ssp. dicoccoides. Plant Mol. Biol. Rep. 2020, 39, 710–726. [Google Scholar] [CrossRef]
- Soresi, D.; Zappacosta, D.; Garayalde, A.; Irigoyen, I.; Basualdo, J.; Carrera, A. A Valuable QTL for Fusarium Head Blight Resistance from Triticum turgidum L. ssp dicoccoides has a Stable Expression in Durum Wheat Cultivars. Cereal Res. Commun. 2017, 45, 234–247. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, L.; Wang, C.Y.; Wang, Y.J.; Zhou, X.L.; Lv, S.K.; Liu, X.L.; Kang, Z.S.; Ji, W.Q. Molecular mapping and marker development for the Triticum dicoccoides-derived stripe rust resistance gene YrSM139-1B in bread wheat cv. Shaanmai 139. Theor. Appl. Genet. 2016, 129, 369–376. [Google Scholar] [CrossRef]
- Sela, H.; Ezrati, S.; Ben-Yehuda, P.; Manisterski, J.; Akhunov, E.; Dvorak, J.; Breiman, A.; Korol, A. Linkage disequilibrium and association analysis of stripe rust resistance in wild emmer wheat (Triticum turgidum ssp. dicoccoides) population in Israel. Theor. Appl. Genet. 2014, 127, 2453–2463. [Google Scholar] [CrossRef]
- Toktay, H.; Imren, M.; Elekcioglu, I.H.; Dababat, A.A. Evaluation of Turkish wild Emmers (Triticum dicoccoides Koern.) and wheat varieties for resistance to the root lesion nematodes (Pratylenchus thornei and Pratylenchus neglectus). Turk. Entomoloji Derg.-Turk. J. Entomol. 2015, 39, 219–227. [Google Scholar] [CrossRef]
- Saidou, M.; Wang, C.Y.; Alam, M.A.; Chen, C.H.; Ji, W.Q. Genetic Analysis of Powdery Mildew Resistance Gene Using ssr Markers in Common Wheat Originated from Wild Emmer (Triticum dicoccoides Thell). Turk. J. Field Crops 2016, 21, 10–15. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, D.Y.; Ouyang, S.H.; Xie, J.Z.; Wu, Q.H.; Wang, Z.Z.; Cui, Y.; Lu, P.; Zhang, D.; Liu, Z.J.; et al. Dynamic evolution of resistance gene analogs in the orthologous genomic regions of powdery mildew resistance gene MlIW170 in Triticum dicoccoides and Aegilops tauschii. Theor. Appl. Genet. 2015, 128, 1617–1629. [Google Scholar] [CrossRef]
- Qiu, L.N.; Liu, N.N.; Wang, H.F.; Shi, X.H.; Li, F.; Zhang, Q.; Wang, W.D.; Guo, W.L.; Hu, Z.R.; Li, H.J.; et al. Fine mapping of a powdery mildew resistance gene MlIW39 derived from wild emmer wheat (Triticum turgidum ssp. dicoccoides). Theor. Appl. Genet. 2021, 134, 2469–2479. [Google Scholar] [CrossRef]
- Ouyang, S.H.; Zhang, D.; Han, J.; Zhao, X.J.; Cui, Y.; Song, W.; Huo, N.X.; Liang, Y.; Xie, J.Z.; Wang, Z.Z.; et al. Fine Physical and Genetic Mapping of Powdery Mildew Resistance Gene MlIW172 Originating from Wild Emmer (Triticum dicoccoides). PLoS ONE 2014, 9, e100160. [Google Scholar] [CrossRef] [PubMed]
- Xue, F.; Ji, W.Q.; Wang, C.Y.; Zhang, H.; Yang, B.J. High-density mapping and marker development for the powdery mildew resistance gene PmAS846 derived from wild emmer wheat (Triticum turgidum var. dicoccoides). Theor. Appl. Genet. 2012, 124, 1549–1560. [Google Scholar] [CrossRef] [PubMed]
- Strejčková, B.; Mazzucotelli, E.; Čegan, R.; Milec, Z.; Brus, J.; Çakır, E.; Mastrangelo, A.M.; Özkan, H.; Šafář, J. Wild emmer wheat, the progenitor of modern bread wheat, exhibits great diversity in the VERNALIZATION1 gene. Front. Plant Sci. 2023, 13, 1106164. [Google Scholar] [CrossRef] [PubMed]
- Suneja, Y.; Gupta, A.K.; Bains, N.S. Stress Adaptive Plasticity: Aegilops tauschii and Triticum dicoccoides as Potential Donors of Drought Associated Morpho-Physiological Traits in Wheat. Front. Plant Sci. 2019, 10, 211. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Hu, X.; Islam, S.; She, M.Y.; Peng, Y.C.; Yu, Z.T.; Wylie, S.; Juhasz, A.; Dowla, M.; Yang, R.C.; et al. New insights into the evolution of wheat avenin-like proteins in wild emmer wheat (Triticum dicoccoides). Proc. Natl. Acad. Sci. USA 2018, 115, 13312–13317. [Google Scholar] [CrossRef]
- Tene, M.; Adhikari, E.; Cobo, N.; Jordan, K.W.; Matny, O.; del Blanco, I.A.; Roter, J.; Ezrati, S.; Govta, L.; Manisterski, J.; et al. GWAS for Stripe Rust Resistance in Wild Emmer Wheat (Triticum dicoccoides) Population: Obstacles and Solutions. Crops 2022, 2, 42–61. [Google Scholar] [CrossRef]
- Kilian, B.; Dempewolf, H.; Guarino, L.; Werner, P.; Coyne, C.; Warburton, M.L. Crop Science special issue: Adapting agriculture to climate change: A walk on the wild side. Crop Sci. 2021, 61, 32–36. [Google Scholar] [CrossRef]
- Bellil, I.; Hamdi, O.; Benbelkacem, A.; Khelifi, D. The Genetic Potential of a Germplasm of Interspecific Crosses between Durum Wheats (Triticum turgidum L. ssp. durum (Desf.) Husn.) and their Relatives (T. dicoccum Schubl. and T. polonicum L.) in Five Glutenin Loci. Cereal Res. Commun. 2019, 47, 678–688. [Google Scholar] [CrossRef]
- Rasheed, A.; Mujeeb-Kazi, A.; Ogbonnaya, F.C.; He, Z.; Rajaram, S. Wheat genetic resources in the post-genomics era: Promise and challenges. Ann. Bot. 2017, 121, 603–616. [Google Scholar] [CrossRef]
- Mujeeb-Kazi, A.; Kazi, A.G.; Dundas, I.; Rasheed, A.; Ogbonnaya, F.; Kishii, M.; Bonnett, D.; Wang, R.R.C.; Xu, S.; Chen, P.; et al. Chapter Four—Genetic Diversity for Wheat Improvement as a Conduit to Food Security. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: Cambridge, MA, USA, 2013; Volume 122, pp. 179–257. [Google Scholar]
- Nevo, E.; Golenberg, E.; Beiles, A.; Brown, A.H.D.; Zohary, D. Genetic Diversity and Environmental Associations of Wild Wheat, Triticum-Dicoccoides, in Israel. Theor. Appl. Genet. 1982, 62, 241–254. [Google Scholar] [CrossRef]
- Poyarkova, H.; Gerechteramitai, Z.K.; Genizi, A. 2 Variants of Wild Emmer (Triticum dicoccoides) Native to Israel—Morphology and Distribution. Can. J. Bot. 1991, 69, 2772–2789. [Google Scholar] [CrossRef]
- Chandrasekhar, K.; Nashef, K.; Ben-David, R. Agronomic and genetic characterization of wild emmer wheat (Triticum turgidum subsp. dicoccoides) introgression lines in a bread wheat genetic background. Genet. Resour. Crop Evol. 2017, 64, 1917–1926. [Google Scholar] [CrossRef]
- Arystanbekkyzy, M.; Nadeem, M.A.; Aktas, H.; Yeken, M.Z.; Zencirci, N.; Nawaz, M.A.; Ali, F.; Haider, M.S.; Tunc, K.; Chung, G.; et al. Phylogenetic and Taxonomic Relationship of Turkish Wild and Cultivated Emmer (Triticum turgidum ssp. dicoccoides) Revealed by iPBS-Retrotransposons Markers. Int. J. Agric. Biol. 2019, 21, 155–163. [Google Scholar] [CrossRef]
- Shizuka, T.; Mori, N.; Ozkan, H.; Ohta, S. Chloroplast DNA haplotype variation within two natural populations of wild emmer wheat (Triticum turgidum ssp. dicoccoides) in southern Turkey. Biotechnol. Biotechnol. Equip. 2015, 29, 423–430. [Google Scholar] [CrossRef]
- Ozbek, O.; Millet, E.; Anikster, Y.; Arslan, O.; Feldman, M. Comparison of the genetic structure of populations of wild emmer wheat, Triticum turgidum ssp dicoccoides, from Israel and Turkey revealed by AFLP analysis. Genet. Resour. Crop Evol. 2007, 54, 1587–1598. [Google Scholar] [CrossRef]
- Gerechteramitai, Z.K.; Sharp, E.L.; Reinhold, M. Temperature Sensitive Genes for Stripe Rust Resistance in Triticum-Dicoccoides Indigenous to Israel. Phytopathology 1981, 71, 218. [Google Scholar]
- Gerechteramitai, Z.K.; Vansilfhout, C.H.; Grama, A.; Kleitman, F. YR15—A New Gene for Resistance to Puccinia-Striiformis in Triticum dicoccoides sel. G-25. Euphytica 1989, 43, 187–190. [Google Scholar] [CrossRef]
- Nevo, E.; Beiles, A.; Gutterman, Y.; Storch, N.; Kaplan, D. Genetic-Resources of Wild Cereals in Israel and Vicinity. I. Phenotypic Variation Within and Between Populations of Wild Wheat, Triticum dicoccoides. Euphytica 1984, 33, 717–735. [Google Scholar] [CrossRef]
- Silfhout, C.H.; Grama, A.; Gerechter-Amitai, Z.K.; Kleitman, F. Resistance to yellow rust in Triticum dicoccoides. I. Crosses with susceptible Triticum durum. Neth. J. Plant Pathol. 1989, 95, 73–78. [Google Scholar] [CrossRef]
- Poyarkova, H. Morphology, Geography and Infraspecific Taxonomics of Triticum dicoccoides Korn—A Retrospective of 80 Years of Research. Euphytica 1988, 38, 11–23. [Google Scholar] [CrossRef]
- Nevo, E.; Krugman, T.; Beiles, A. Genetic-Resources for Salt Tolerance in the Wild Progenitors of Wheat (Triticum dicoccoides) and Barley (Hordeum spontaneum) in Israel. Plant Breed. 1993, 110, 338–341. [Google Scholar] [CrossRef]
- Kato, K.; Mori, Y.; Beiles, A.; Nevo, E. Geographical variation in heading traits in wild emmer wheat, Triticum dicoccoides. I. Variation in vernalization response and ecological differentiation. Theor. Appl. Genet. 1997, 95, 546–552. [Google Scholar] [CrossRef]
- Kato, K.; Tanizoe, C.; Beiles, A.; Nevo, E. Geographical variation in heading traits in wild emmer wheat, Triticum dicoccoides. II. Variation in heading date and adaptation to diverse eco-geographical conditions. Hereditas 1998, 128, 33–39. [Google Scholar] [CrossRef]
- Fahima, T.; Sun, G.L.; Beharav, A.; Krugman, T.; Beiles, A.; Nevo, E. RAPD polymorphism of wild emmer wheat populations, Triticum dicoccoides, in Israel. Theor. Appl. Genet. 1999, 98, 434–447. [Google Scholar] [CrossRef]
- Belyayev, A.; Raskina, O.; Korol, A.; Nevo, E. Coevolution of A and B genomes in allotetraploid Triticum dicoccoides. Genome 2000, 43, 1021–1026. [Google Scholar] [CrossRef]
- Li, Y.C.; Fahima, T.; Peng, J.H.; Roder, M.S.; Kirzhner, V.M.; Beiles, A.; Korol, A.B.; Nevo, E. Edaphic microsatellite DNA divergence in wild emmer wheat, Triticum dicoccoides, at a microsite: Tabigha, Israel. Theor. Appl. Genet. 2000, 101, 1029–1038. [Google Scholar] [CrossRef]
- Peng, J.H.; Fahima, T.; Roder, M.S.; Huang, Q.Y.; Dahan, A.; Li, Y.C.; Grama, A.; Nevo, E. High-density molecular map of chromosome region harboring stripe-rust resistance genes YrH52 and Yr15 derived from wild emmer wheat, Triticum dicoccoides. Genetica 2000, 109, 199–210. [Google Scholar] [CrossRef]
- Li, Y.C.; Krugman, T.; Fahima, T.; Beiles, A.; Korol, A.B.; Nevo, E. Spatiotemporal allozyme divergence caused by aridity stress in a natural population of wild wheat, Triticum dicoccoides, at the Ammiad microsite, Israel. Theor. Appl. Genet. 2001, 102, 853–864. [Google Scholar] [CrossRef]
- Fahima, T.; Roder, M.S.; Wendehake, K.; Kirzhner, V.M.; Nevo, E. Microsatellite polymorphism in natural populations of wild emmer wheat, Triticum dicoccoides, in Israel. Theor. Appl. Genet. 2002, 104, 17–29. [Google Scholar] [CrossRef]
- Li, Y.C.; Roder, M.S.; Fahima, T.; Kirzhner, V.M.; Beiles, A.; Korol, A.B.; Nevo, E. Climatic effects on microsatellite diversity in wild emmer wheat (Triticum dicoccoides) at the Yehudiyya microsite, Israel. Heredity 2002, 89, 127–132. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Stierschneider, M.; Steiner, B.; Lemmens, M.; Griesser, M.; Nevo, E.; Fahima, T. Variation for resistance to head blight caused by Fusarium graminearum in wild emmer (Triticum dicoccoides) originating from Israel. Euphytica 2003, 130, 17–23. [Google Scholar] [CrossRef]
- Li, Y.C.; Fahima, T.; Roder, M.S.; Kirzhner, V.M.; Beiles, A.; Korol, A.B.; Nevo, E. Genetic effects on microsatellite diversity in wild emmer wheat (Triticum dicoccoides) at the Yehudiyya microsite, Israel. Heredity 2003, 90, 150–156. [Google Scholar] [CrossRef]
- Xu, S.S.; Khan, K.; Klindworth, D.L.; Faris, J.D.; Nygard, G. Chromosomal location of genes for novel glutenin subunits and gliadins in wild emmer wheat (Triticum turgidum L. var. dicoccoides). Theor. Appl. Genet. 2004, 108, 1221–1228. [Google Scholar] [CrossRef]
- Anikster, Y.; Manisterski, J.; Long, D.L.; Leonard, K.J. Leaf rust and stem rust resistance in Triticum dicoccoides populations in Israel. Plant Dis. 2005, 89, 55–62. [Google Scholar] [CrossRef]
- Syouf, M.; Abu-Irmaileh, B.E.; Valkoun, J.; Bdour, S. Introgression from durum wheat landraces in wild emmer wheat (Triticum dicoccoides (Korn. ex Asch et Graebner) Schweinf). Genet. Resour. Crop Evol. 2006, 53, 1165–1172. [Google Scholar] [CrossRef]
- Hua, W.; Liu, Z.J.; Zhu, J.; Xie, C.J.; Yang, T.M.; Zhou, Y.L.; Duan, X.Y.; Sun, Q.X.; Liu, Z.Y. Identification and genetic mapping of pm42, a new recessive wheat powdery mildew resistance gene derived from wild emmer (Triticum turgidum var. dicoccoides). Theor. Appl. Genet. 2009, 119, 223–230. [Google Scholar] [CrossRef]
- Liu, Z.J.; Zhu, J.; Cui, Y.; Liang, Y.; Wu, H.B.; Song, W.; Liu, Q.; Yang, T.M.; Sun, Q.X.; Liu, Z.Y. Identification and comparative mapping of a powdery mildew resistance gene derived from wild emmer (Triticum turgidum var. dicoccoides) on chromosome 2BS. Theor. Appl. Genet. 2012, 124, 1041–1049. [Google Scholar] [CrossRef]
- Domb, K.; Keidar, D.; Yaakov, B.; Khasdan, V.; Kashkush, K. Transposable elements generate population-specific insertional patterns and allelic variation in genes of wild emmer wheat (Triticum turgidum ssp dicoccoides). BMC Plant Biol. 2017, 17, 175. [Google Scholar] [CrossRef]
- Vuorinen, A.L.; Kalendar, R.; Fahima, T.; Korpelainen, H.; Nevo, E.; Schulman, A.H. Retrotransposon-Based Genetic Diversity Assessment in Wild Emmer Wheat (Triticum turgidum ssp. dicoccoides). Agronomy 2018, 8, 107. [Google Scholar] [CrossRef]
- Salamini, F.; Ozkan, H.; Brandolini, A.; Schafer-Pregl, R.; Martin, W. Genetics and geography of wild cereal domestication in the near east. Nat. Rev. Genet. 2002, 3, 429–441. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Ozkan, H.; Kafkas, S.; Sertac Ozer, M.; Brandolini, A. Genetic relationships among South-East Turkey wild barley populations and sampling strategies of Hordeum spontaneum. Theor. Appl. Genet. 2005, 112, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef]
- Gonzalez-Paleo, L.; Ravetta, D.A. Allocation patterns and phenology in wild and selected accessions of annual and perennial Physaria (Lesquerella, Brassicaceae). Euphytica 2012, 186, 289–302. [Google Scholar] [CrossRef]
- Feuillet, C.; Langridge, P.; Waugh, R. Cereal breeding takes a walk on the wild side. Trends Genet. 2007, 24, 24–32. [Google Scholar] [CrossRef]
- Ortiz, R.; Sayre, K.D.; Govaerts, B.; Gupta, R.; Subbarao, G.V.; Ban, T.; Hodson, D.; Dixon, J.A.; Ortiz-Monasterio, J.I.; Reynolds, M. Climate change: Can wheat beat the heat? Agric. Ecosyst. Environ. 2008, 126, 46–58. [Google Scholar] [CrossRef]
- El Haddad, N.; Kabbaj, H.; Zaïm, M.; El Hassouni, K.; Tidiane Sall, A.; Azouz, M.; Ortiz, R.; Baum, M.; Amri, A.; Gamba, F.; et al. Crop wild relatives in durum wheat breeding: Drift or thrift? Crop Sci. 2021, 61, 37–54. [Google Scholar] [CrossRef]
- Castaneda-Alvarez, N.P.; Khoury, C.K.; Achicanoy, H.A.; Bernau, V.; Dempewolf, H.; Eastwood, R.J.; Guarino, L.; Harker, R.H.; Jarvis, A.; Maxted, N.; et al. Global conservation priorities for crop wild relatives. Nat. Plants 2016, 2, 16022. [Google Scholar] [CrossRef]
- Harlan, J.R.; Wet, J.M.J. Toward a Rational Classification of Cultivated Plants. Taxon 1971, 20, 509–517. [Google Scholar] [CrossRef]
- Chhuneja, P.; Arora, J.K.; Kaur, P.; Kaur, S.; Singh, K. Characterization of wild emmer wheat Triticum dicoccoides germplasm for vernalization alleles. J. Plant Biochem. Biotechnol. 2015, 24, 249–253. [Google Scholar] [CrossRef]
- Peng, J.H.; Sun, D.; Nevo, E. Domestication evolution, genetics and genomics in wheat. Mol. Breed. 2011, 28, 281–301. [Google Scholar] [CrossRef]
- Nevo, E. Genetic resources of wild emmer, Triticum dicoccoides, for wheat improvement in the third millennium. Isr. J. Plant Sci. 2001, 49, S77–S91. [Google Scholar] [CrossRef]
- Schmidt, K. Göbekli Tepe; Arkeoloji ve Sanat Yayınları: Istanbul, Turkey, 2007. [Google Scholar]
- Dietrich, O.; Heun, M.; Notroff, J.; Schmidt, K.; Zarnkow, M. The role of cult and feasting in the emergence of Neolithic communities. New evidence from Göbekli Tepe, south-eastern Turkey. Antiquity 2012, 86, 674–695. [Google Scholar] [CrossRef]
- Jorgensen, C.; Luo, M.-C.; Ramasamy, R.; Dawson, M.; Gill, B.S.; Korol, A.B.; Distelfeld, A.; Dvorak, J. A High-Density Genetic Map of Wild Emmer Wheat from the Karaca Dağ Region Provides New Evidence on the Structure and Evolution of Wheat Chromosomes. Front. Plant Sci. 2017, 8, 1798. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Zhu, K.Y.; Dong, L.L.; Liang, Y.; Li, G.Q.; Fang, T.L.; Guo, G.H.; Wu, Q.H.; Xie, J.Z.; Chen, Y.X.; et al. Wheat powdery mildew resistance gene Pm64 derived from wild emmer (Triticum turgidum var. dicoccoides) is tightly linked in repulsion with stripe rust resistance gene Yr5. Crop J. 2019, 7, 761–770. [Google Scholar] [CrossRef]
- Ahmadi, H.; Nazarian, F. The inheritance and chromosomal location of morphological traits in wild wheat, Triticum turgidum L. ssp. dicoccoides. Euphytica 2007, 158, 103–108. [Google Scholar] [CrossRef]
- Rawale, K.S.; Khan, M.A.; Gill, K.S. The novel function of the Ph1 gene to differentiate homologs from homoeologs evolved in Triticum turgidum ssp. dicoccoides via a dramatic meiosis-specific increase in the expression of the 5B copy of the C-Ph1 gene. Chromosoma 2019, 128, 561–570. [Google Scholar] [CrossRef]
- Negisho, K.; Shibru, S.; Pillen, K.; Ordon, F.; Wehner, G. Genetic diversity of Ethiopian durum wheat landraces. PLoS ONE 2021, 16, e0247016. [Google Scholar] [CrossRef]
- Teklu, Y.; Hammer, K.; Röder, M.S. Simple sequence repeats marker polymorphism in emmer wheat (Triticum dicoccon Schrank): Analysis of genetic diversity and differentiation. Genet. Resour. Crop Evol. 2007, 54, 543–554. [Google Scholar] [CrossRef]
- Harlan, J.R.; Zohary, D. Distribution of wild wheats and barley. Science 1966, 153, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
- Zohary, D.; Hopf, M. Domestication of Plants in the Old World: The Origin and Spread of Cultivated Plants in West Asia, Europe and the Nile Valley; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Hegde, S.G.; Valkoun, J.; Waines, J.G. Genetic diversity in wild wheats and goat grass. Theor. Appl. Genet. 2000, 101, 309–316. [Google Scholar] [CrossRef]
- Lu, H.; Bernardo, R. Molecular marker diversity among current and historical maize inbreds. Theor. Appl. Genet. 2001, 103, 613–617. [Google Scholar] [CrossRef]
- Ozbek, O.; Millet, E.; Anikster, Y.; Arslan, O.; Feldman, M. Spatio-temporal genetic variation in populations of wild emmer wheat, Triticum turgidum ssp dicoccoides, as revealed by AFLP analysis. Theor. Appl. Genet. 2007, 115, 19–26. [Google Scholar] [CrossRef]

| Collection No | Collection Locality | Zone | Altitude (m) | Latitude (°N) | Longitude (°E) |
|---|---|---|---|---|---|
| 1 | 24.5 km SW from Diyarbakır to Ovadag | East1 | 780 | 37°47′38″ | 40°12′14″ |
| 2 | 12.9 km NW from Ovadag to Pirinçlik | East1 | 1007 | 37°47′31″ | 39°57′18″ |
| 3 | 18.5 km NW from Ovadag to Pirinçlik | East1 | 920 | 37°49′17″ | 39°59′34″ |
| 4 | 20.1 km SW from Pirinçlik | East1 | 1080 | 37°52′02″ | 39°51′05″ |
| 5 | 20 km SW from Pirinçlik | East1 | 1260 | 37°50′40″ | 39°47′58″ |
| 6 | 2.9 km NE from Karabahçe to Pirinçlik | East1 | 1300 | 37°49′12″ | 39°46′29″ |
| 7 | 41.2 km SW from Pirinçlik | East1 | 1250 | 37°46′42″ | 39°44′50″ |
| 8 | 6.3 km N from Karabahçe (42.9 km W from Diyarbakır to Siverek) | East1 | 1070 | 37°50′21″ | 39°43′23″ |
| 9 | 4.6 km SW from Karabahçe | East1 | 1180 | 37°46′19″ | 39°44′03″ |
| 10 | 17.9 km SW from Karabahçe | East1 | 1160 | 37°44′29″ | 39°42′50″ |
| 11 | 21.7 km SW from Karabahçe | East1 | 1235 | 37°42′51″ | 39°44′03″ |
| 12 | 37.9 km SW from Karabahçe | East1 | 1170 | 37°39′49″ | 39°42′49″ |
| 13 | 37.9 km SW from Karabahçe | East1 | 1180 | 37°36′27″ | 39°43′41″ |
| 14 | 41.6 km SW from Karabahçe | East1 | 1170 | 37°35′08″ | 39°44′36″ |
| 15 | 48.7 km SW from Karabahçe | East1 | 1030 | 37°33′09″ | 39°42′06″ |
| 16 | 27.6 km SW from Karacadag (69.6 km SW from Karabahçe) | East1 | 950 | 37°37′40″ | 39°33′40″ |
| 17 | 30.2 km SW from Çermik to Siverek | East1 | 800 | 38°00′56″ | 39°22′11″ |
| 18 | Siverek Karakeçi Road Azemi Village | East1 | 733 | 37°36′51″ | 39°20′12″ |
| 19 | Karakeçi road | East1 | 737 | 37°33′27″ | 39°20′35″ |
| 20 | Karakeçi grassland | East1 | 758 | 37°32′22″ | 39°21′52″ |
| 21 | 5 km from Siverek to Siverek Hilvan Road | East1 | 645 | 37°42′23″ | 39°16′34″ |
| 22 | 72 km SE from Turkoglu SE (W of Karadag) | West1 | 800(853) | 37°19′46″ | 37°16′29″ |
| 23 | 72 km SE from Turkoglu SE (W of Karadag) | West1 | 800(853) | 37°19′46″ | 37°16′29″ |
| 24 | 34 km ESE from Narlı (WSW of Karadag) | West1 | 840 (877) | 37°18′53″ | 37°15′41″ |
| 25 | 34 km ESE from Narlı (WSW of Karadag) | West1 | 780 (813) | 37°20′12″ | 37°17′53″ |
| 26 | 39 km ESE from Narlı (SW of Karadag) | West1 | 760 (793) | 37°17′06″ | 37°17′39″ |
| 27 | 39 km ESE from Narlı (SW of Karadag) | West1 | 760 (793) | 37°17′06″ | 37°17′39″ |
| 28 | Between Kahramanmaraş Kelleş village and Yiğitce village | West1 | 791 | 37°20′25″ | 37°17′54″ |
| 29 | Between Gaziantep Tekirsin village and Dundarlı village | West1 | 882 | 37°15′20″ | 37°23′26″ |
| 30 | 37 km NE from Kilis to Gaziantep | West2 | 830 | 37°20′19″ | 37°16′50″ |
| 31 | 39 km NE from Kilis to Gaziantep | West2 | 920 | 37°19′50″ | 37°18′51″ |
| 32 | 41 km NE from Kilis to Gaziantep | West2 | 770 | 37°24′23″ | 37°25′47″ |
| 33 | 42 km NE from Kilis to Gaziantep | West2 | 750 | 37°24′58″ | 37°24′50″ |
| 34 | 58 km NE from Kilis to Gaziantep | West2 | 720 | 37°16′01″ | 37°30′52″ |
| 35 | 59 km NE from Kilis to Gaziantep | West2 | 770 | 37°15′33″ | 37°29′03″ |
| 36 | 21 km NE from Kilis to Gaziantep | West2 | 620 | 36°45′52″ | 37°15′04″ |
| 37 | 24 km NE from Kilis to Gaziantep | West2 | 700 | 36°52′20″ | 37°12′12″ |
| 38 | 25 km NE from Kilis to Gaziantep | West2 | 830 | 36°33′25″ | 37°11′57″ |
| Name | Ch | Motif | Forward Primer Sequence | Reverse Primer Sequence |
| cfa2219 | 1A | (GT)21 | TCTGCCGAGTCACTTCATTG | GACAAGGCCAGTCCAAAAGA |
| wmc312 | 1A | (GA)10 | TGTGCCCGCTGGTGCGAAG | CCGACGCAGGTGAGCGAAG |
| wmc658 | 2A | ---- | CTCATCGTCCTCCTCCACTTTG | GCCATCCGTTGACTTGAGGTTA |
| wmc313 | 4A | (CA)18 | GCAGTCTAATTATCTGCTGGCG | GGGTCCTTGTCTACTCATGTCT |
| wmc110 | 5A | (GT)11 | GCAGATGAGTTGAGTTGGATTG | GTACTTGGAAACTGTGTTTGGG |
| cfa2190 | 5A | (TC)31 | CAGTCTGCAATCCACTTTGC | AAAAGGAAACTAAAGCGATGGA |
| wmc626 | 1B | ---- | AGCCCATAAACATCCAACACGG | AGGTGGGCTTGGTTACGCTCTC |
| gwm498 | 1B | ---- | GGTGGTATGGACTATGGACACT | GGTGGTATGGACTATGGACACT |
| wmc128 | 1B | (GA)10 | CGGACAGCTACTGCTCTCCTTA | CTGTTGCTTGCTCTGCACCCTT |
| wmc149 | 2B | (CT)24 | ACAGACTTGGTTGGTGCCGAGC | ATGGGCGGGGGTGTAGAGTTTG |
| wmc332 | 2B | (CT)12 | CATTTACAAAGCGCATGAAGCC | GAAAACTTTGGGAACAAGAGCA |
| gwm335 | 5B | --- | CGTACTCCACTCCACACGG | CGGTCCAAGTGCTACCTTTC |
| gwm630 | 6B | (GT)16 | GTGCCTGTGCCATCGTC | CGAAAGTAACAGCGCAGTGA |
| gwm146 | 7B | --- | CCAAAAAAACTGCCTGCATG | CTCTGGCATTGCTCCTTGG |
| wmc76 | 7B | (GT)19 | CTTCAGAGCCTCTTTCTCTACA | CTGCTTCACTTGCTGATCTTTG |
| gwm333 | 7B | (GA)19 | GCCCGGTCATGTAAAACG | TTTCAGTTTGCGTTAAGCTTTG |
| Traits | Sum of Squares | Mean Square |
|---|---|---|
| Plant height | 990.53 | 495.269 * |
| Heading date | 750.785 | 375.393 *** |
| Peduncle length | 1155.930 | 577.695 *** |
| Flag leaf area | 262.141 | 131.071 **** |
| Auriculas length | 0.927 | 0.463 ns |
| Auriculas width | 2444.0 | 1222.0 ns |
| Spike length | 5387.0 | 2694.0 * |
| Spike number | 137.496 | 68.748 **** |
| Length of the uppermost awn in the spikelet | 822.418 | 411.209 ns |
| Awn length in the fourth flower | 1474.411 | 737.205 ns |
| Length of the lowermost awn in the spikelet | 1236.780 | 618.390 ns |
| Spikelet length | 13.883 | 6941.0 * |
| Spikelet width | 2467.0 | 1233.0 ns |
| Lemma length | 18.152 | 9076.0 ** |
| Lemma width | 0.263 | 0.132 ns |
| Palea length | 7100.0 | 3550.0 * |
| Palea width | 0.239 | 0.119 ns |
| Glume length | 7473.0 | 3736.0 * |
| Glume hull thickness | 6745.0 | 3373.0 * |
| Glume height | 0.481 | 0.240 ns |
| Anther length | 4556.0 | 2278.0 **** |
| Anther width | 0.068 | 0.034 * |
| Maturation | 673.340 | 366.670 * |
| Traits | Whole Collections | Karacadag/EAST | Karadag-1/WEST | Karadag-2/WEST |
|---|---|---|---|---|
| Plant height (cm) | 126.95 ±18.45 | 126.35 ± 19.68 | 127.64 ± 16.27 | 128.31 ± 16.28 |
| Heading date (day) | 166.06 ± 8.36 | 164.37 ± 9.12 | 170.38 ± 7.08 | 167.49 ± 4.58 |
| Peduncle length(cm) | 38.95 ± 6.85 | 39.17 ± 6.83 | 35.09 ± 7.09 | 41.48 ± 5.26 |
| Flag leaf area (cm2) | 16.38 ± 6.18 | 15.66 ± 6.33 | 17.76 ± 5.76 | 17.55 ± 4.65 |
| Auriculas length (mm) | 4.52 ± 0.64 | 4.50 ± 0.68 | 4.45 ± 0.39 | 4.65 ± 0.65 |
| Auriculas width(mm) | 5.42 ± 0.90 | 5.43 ± 0.88 | 5.68 ± 0.87 | 5.19 ± 0.91 |
| Spike length (cm) | 9.18 ± 1.16 | 9.28 ± 1.23 | 9.11 ± 0.91 | 8.91 ± 1.08 |
| Spikelet number | 20.02 ± 2.49 | 20.69 ± 2.62 | 18.89 ± 1.75 | 18.80 ± 1.67 |
| Length of the uppermost awn in the spikelet (mm) | 76.37 ± 18.84 | 74.91 ± 19.56 | 80.76 ± 15.78 | 77.40 ± 18.71 |
| Awn length in the fourth flower(mm) | 95.97 ± 19.73 | 93.68 ± 20.45 | 100.23 ± 17.34 | 98.85 ± 18.51 |
| Length of the lowermost awn in the spikelet (mm) | 59.49 ± 22.21 | 56.35 ± 22.72 | 68.41 ± 18.75 | 62.11 ± 21.37 |
| Spikelet length (mm) | 15.72 ± 1.44 | 15.50 ± 1.47 | 16.13 ± 1.49 | 16.07 ± 1.17 |
| Spikelet width (mm) | 4.67 ± 0.82 | 4.74 ± 0.86 | 4.38 ± 0.68 | 4.71 ± 0.72 |
| Lemma length (mm) | 13.46 ± 1.66 | 13.26 ± 1.87 | 13.95 ± 1.30 | 13.71 ± 0.91 |
| Lemma width (mm) | 2.53 ± 0.35 | 2.50 ± 0.35 | 2.51 ± 0.35 | 2.62 ± 0.37 |
| Palea length (mm) | 12.49 ± 1.12 | 12.32 ± 1.19 | 12.88 ± 1.02 | 12.70 ± 0.86 |
| Palea width (mm) | 1.81 ± 0.51 | 1.77 ± 0.26 | 1.73 ± 0.26 | 2.03 ± 1.01 |
| Glume length (mm) | 12.27 ± 1.12 | 12.09 ± 1.14 | 12.59 ± 1.04 | 12.61 ± 1.04 |
| Glume hull thickness (mm) | 0.24 ± 0.09 | 0.26 ± 0.09 | 0.22 ± 0.09 | 0.20 ± 0.06 |
| Glume height (mm) | 2.45 ± 0.35 | 2.43 ± 0.33 | 2.45 ± 0.35 | 2.55 ± 0.40 |
| Anther length (mm) | 3.73 ± 0.50 | 3.62 ± 0.50 | 3.80 ± 0.43 | 4.10 ± 0.42 |
| Anther width (mm) | 0.58 ± 0.10 | 0.59 ± 0.10 | 0.53 ± 0.09 | 0.58 ± 0.11 |
| Maturation (day) | 198.12 ± 4.56 | 97.18 ± 4.97 | 199.48 ± 4.03 | 199.86 ± 2.48 |
| Source | df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Among Pops | 2 | 439.551 | 219.776 | 4.429 | 16 |
| Within Pops | 166 | 3803.206 | 22.911 | 22.911 | 84 |
| Total | 168 | 4242.757 | 27.340 | 100 |
| Population | Karacadag/EAST | Karadag-1/WEST | Karadag-2/WEST |
|---|---|---|---|
| Karacadag/EAST | --- | 0.518 | 0.539 |
| Karadag-1/WEST | 0.484 | --- | 0.214 |
| Karadag-2/WEST | 0.463 | 0.788 | --- |
| Pops | N | Na | Ne | I | He | uHe |
|---|---|---|---|---|---|---|
| Karacadag/EAST | 108 | 9.938 ± 1.871 | 5.470 ± 0.869 | 1.692 ± 0.177 | 0.725 ± 0.046 | 0.729± 0.046 |
| Karadag-1/WEST | 28 | 3.938 ± 0.470 | 2.747 ± 0.308 | 1.030 ± 0.118 | 0.561 ± 0.051 | 0.572 ± 0.052 |
| Karadag-2/WEST | 33 | 6.125 ± 0.861 | 4.135 ± 0.644 | 1.348 ± 0.181 | 0.621 ± 0.070 | 0.632 ± 0.071 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Çakır, E.; Alsaleh, A.; Bektas, H.; Özkan, H. Wild Emmer (Triticum turgidum ssp. dicoccoides) Diversity in Southern Turkey: Evaluation of SSR and Morphological Variations. Life 2025, 15, 203. https://doi.org/10.3390/life15020203
Çakır E, Alsaleh A, Bektas H, Özkan H. Wild Emmer (Triticum turgidum ssp. dicoccoides) Diversity in Southern Turkey: Evaluation of SSR and Morphological Variations. Life. 2025; 15(2):203. https://doi.org/10.3390/life15020203
Chicago/Turabian StyleÇakır, Esra, Ahmad Alsaleh, Harun Bektas, and Hakan Özkan. 2025. "Wild Emmer (Triticum turgidum ssp. dicoccoides) Diversity in Southern Turkey: Evaluation of SSR and Morphological Variations" Life 15, no. 2: 203. https://doi.org/10.3390/life15020203
APA StyleÇakır, E., Alsaleh, A., Bektas, H., & Özkan, H. (2025). Wild Emmer (Triticum turgidum ssp. dicoccoides) Diversity in Southern Turkey: Evaluation of SSR and Morphological Variations. Life, 15(2), 203. https://doi.org/10.3390/life15020203






