Global DNA Methylation and DNA Methyltransferase Status Among Cigarette Smokers in Saudi
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants and Samples
2.2. Quantification of Global DNA Methylation
2.3. Determination of ELISA
2.4. Statistical Analysis
3. Results
3.1. Socio-Demographic Data of the Participants
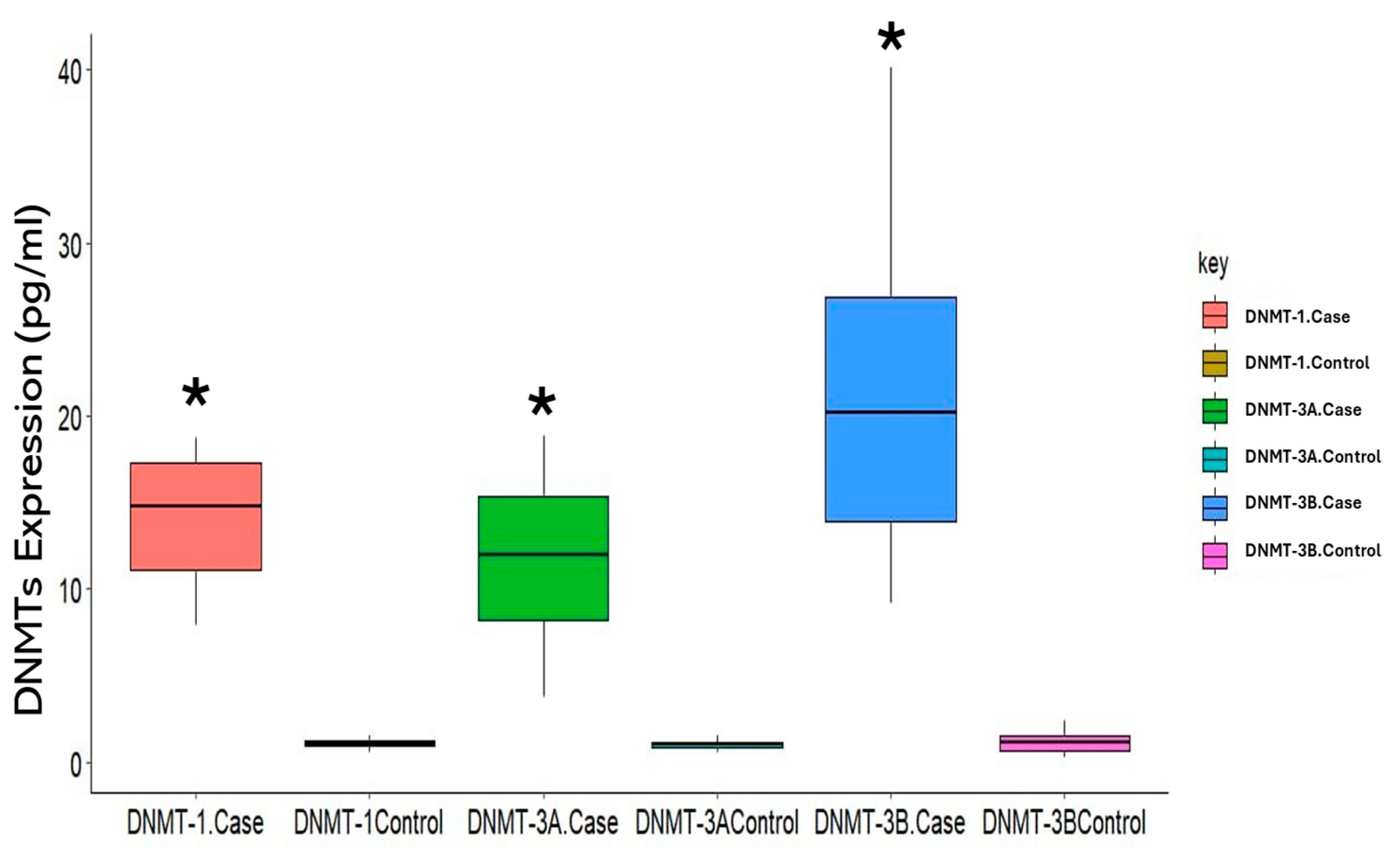
3.2. Expression of DNMTs in Cigarette Smokers
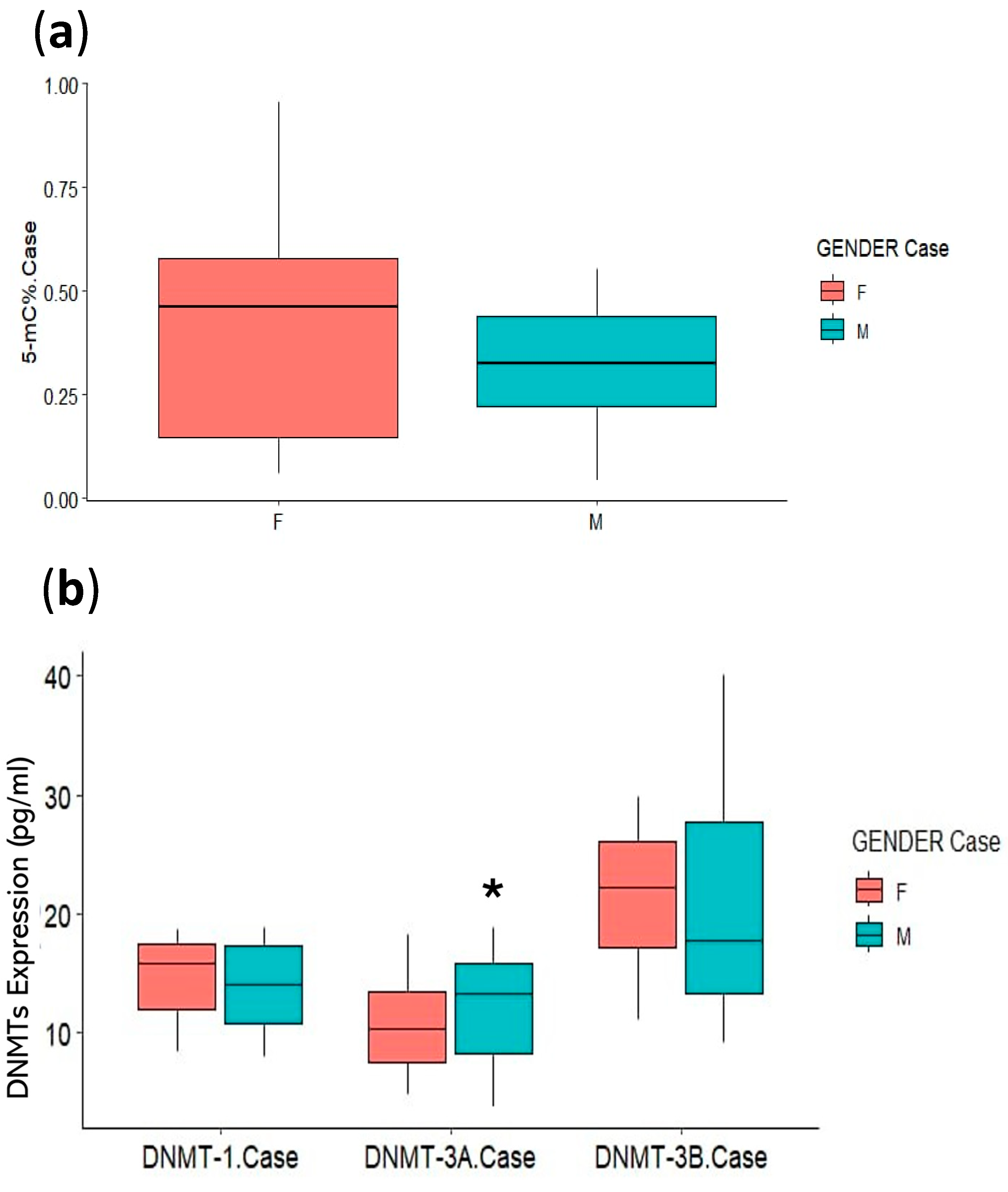
3.3. Association of Clinical Parameters with DNA Methylation Status
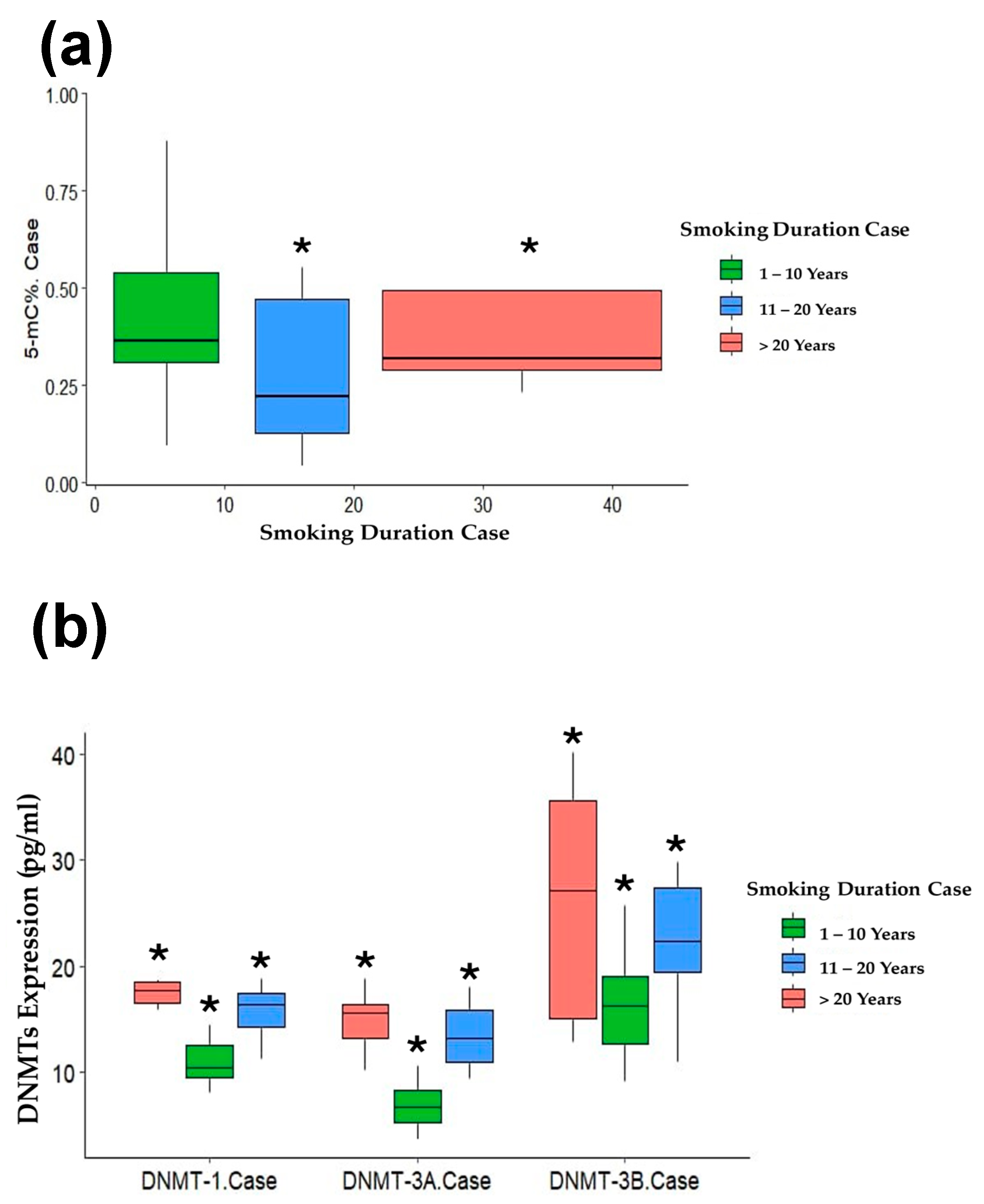
3.4. Association of Smoking Duration with DNA Methylation Status
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chang, J.T.; Anic, G.M.; Rostron, B.L.; Tanwar, M.; Chang, C.M. Cigarette Smoking Reduction and Health Risks: A Systematic Review and Meta-analysis. Nicotine Tob. Res. Off. J. Soc. Res. Nicotine Tob. 2021, 23, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.N. Tar level of cigarettes smoked and risk of smoking-related diseases. Inhal. Toxicol. 2018, 30, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, I.; Kabbarah, A.; Tasji, A.; Hawsawi, K.; Almutairi, R.; Almehmadi, J.; Almuylibi, L.; Bayrkdar, M.; Alherz, A.; Alzahrani, K. Association Between Tobacco Smoking and Prevalence of CVD in Young Adults in Saudi Arabia: Cross-Sectional Study. Arch. Pharm. Pract. 2024, 15, 1–6. [Google Scholar]
- Schabath, M.B.; Cote, M.L. Cancer progress and priorities: Lung cancer. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1563–1579. [Google Scholar] [CrossRef]
- Moradi-Lakeh, M.; El Bcheraoui, C.; Tuffaha, M.; Daoud, F.; Al Saeedi, M.; Basulaiman, M.; Memish, Z.A.; AlMazroa, M.A.; Al Rabeeah, A.A.; Mokdad, A.H. Tobacco consumption in the Kingdom of Saudi Arabia, 2013: Findings from a national survey. BMC Public Health 2015, 15, 611. [Google Scholar] [CrossRef]
- Monshi, S.S.; Arbaein, T.J.; Alzhrani, A.A.; Alzahrani, A.M.; Alharbi, K.K.; Alfahmi, A.; Alqahtani, M.Z.; Alzahrani, A.H.; Elkhobby, A.A.; Almazrou, A.M. Factors associated with the desire to quit tobacco smoking in Saudi Arabia: Evidence from the 2019 Global Adult Tobacco Survey. Tob. Induc. Dis. 2023, 21, 33. [Google Scholar] [CrossRef]
- Mao, Y.; Huang, P.; Wang, Y.; Wang, M.; Li, M.D.; Yang, Z. Genome-wide methylation and expression analyses reveal the epigenetic landscape of immune-related diseases for tobacco smoking. Clin. Epigenetics 2021, 13, 215. [Google Scholar] [CrossRef]
- Prijić, Ž.; Igić, R. Cigarette smoking and medical students. J. BUON 2021, 26, 1709–1718. [Google Scholar]
- Xie, Z.; Rahman, I.; Goniewicz, M.L.; Li, D. Perspectives on epigenetics alterations associated with smoking and vaping. Function 2021, 2, zqab022. [Google Scholar] [CrossRef]
- Fang, F.; Andersen, A.M.; Philibert, R.; Hancock, D.B. Epigenetic biomarkers for smoking cessation. Addict. Neurosci. 2023, 6, 100079. [Google Scholar] [CrossRef]
- Mattei, A.L.; Bailly, N.; Meissner, A. DNA methylation: A historical perspective. Trends Genet. 2022, 38, 676–707. [Google Scholar] [CrossRef] [PubMed]
- Kausar, S.; Abbas, M.N.; Cui, H. A review on the DNA methyltransferase family of insects: Aspect and prospects. Int. J. Biol. Macromol. 2021, 186, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Jan, Z.; Ahmed, W.S.; Biswas, K.H.; Jithesh, P.V. Identification of a potential DNA methyltransferase (DNMT) inhibitor. J. Biomol. Struct. Dyn. 2024, 42, 4730–4744. [Google Scholar] [CrossRef] [PubMed]
- Domingo-Relloso, A.; Joehanes, R.; Rodriguez-Hernandez, Z.; Lahousse, L.; Haack, K.; Fallin, M.D.; Herreros-Martinez, M.; Umans, J.G.; Best, L.G.; Huan, T.; et al. Smoking, blood DNA methylation sites and lung cancer risk. Environ. Pollut. 2023, 334, 122153. [Google Scholar] [CrossRef]
- Zhang, H.; Kalla, R.; Chen, J.; Zhao, J.; Zhou, X.; Adams, A.; Noble, A.; Ventham, N.T.; Wellens, J.; Ho, G.T.; et al. Altered DNA methylation within DNMT3A, AHRR, LTA/TNF loci mediates the effect of smoking on inflammatory bowel disease. Nat. Commun. 2024, 15, 595. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, L.; Yang, H.; Wu, X.; Luo, X.; Shen, J.; Xiao, Z.; Zhao, Y.; Du, F.; Chen, Y. Dysregulation of immunity by cigarette smoking promotes inflammation and cancer: A review. Environ. Pollut. 2023, 339, 122730. [Google Scholar] [CrossRef]
- Siemelink, M.A.; van der Laan, S.W.; Haitjema, S.; van Koeverden, I.D.; Schaap, J.; Wesseling, M.; de Jager, S.C.A.; Mokry, M.; van Iterson, M.; Dekkers, K.F.; et al. Smoking is Associated to DNA Methylation in Atherosclerotic Carotid Lesions. Circulation. Genom. Precis. Med. 2018, 11, e002030. [Google Scholar] [CrossRef]
- Avram, G.-E.; Marcu, A.; Moatar, A.; Samoila, C.; Podariu, A.; Seclaman, E.; Marian, C. Changes in global DNA methylation and hydroxymethylation in oral mucosa according to tobacco smoke exposure. J. Int. Med. Res. 2020, 48, 0300060520954677. [Google Scholar] [CrossRef]
- van Dongen, J.; Willemsen, G.; de Geus, E.J.; Boomsma, D.I.; Neale, M.C.; Consortium, B. Effects of smoking on genome-wide DNA methylation profiles: A study of discordant and concordant monozygotic twin pairs. eLife 2023, 12, e83286. [Google Scholar] [CrossRef]
- Mukhopadhyay, P.; Greene, R.M.; Pisano, M.M. Cigarette smoke induces proteasomal-mediated degradation of DNA methyltransferases and methyl CpG-/CpG domain-binding proteins in embryonic orofacial cells. Reprod. Toxicol. 2015, 58, 140–148. [Google Scholar] [CrossRef]
- Liu, F.; Killian, J.K.; Yang, M.; Walker, R.L.; Hong, J.A.; Zhang, M.; Davis, S.; Zhang, Y.; Hussain, M.; Xi, S.; et al. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene 2010, 29, 3650–3664. [Google Scholar] [CrossRef]
- Lee, K.W.; Pausova, Z. Cigarette smoking and DNA methylation. Front. Genet. 2013, 4, 132. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, Y.; Li, J.; Sun, X.; Wang, L.-P.; Ji, W.-Y. The tobacco-specific carcinogen NNK induces DNA methyltransferase 1 accumulation in laryngeal carcinoma. Oral Oncol. 2012, 48, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Caliri, A.W.; Tommasi, S.; Besaratinia, A. Relationships among smoking, oxidative stress, inflammation, macromolecular damage, and cancer. Mutat. Res./Rev. Mutat. Res. 2021, 787, 108365. [Google Scholar] [CrossRef]
- Belinsky, S.A.; Palmisano, W.A.; Gilliland, F.D.; Crooks, L.A.; Divine, K.K.; Winters, S.A.; Grimes, M.J.; Harms, H.J.; Tellez, C.S.; Smith, T.M. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res. 2002, 62, 2370–2377. [Google Scholar]
- Min, B.; Park, J.S.; Jeong, Y.S.; Jeon, K.; Kang, Y.K. Dnmt1 binds and represses genomic retroelements via DNA methylation in mouse early embryos. Nucleic Acids Res. 2020, 48, 8431–8444. [Google Scholar] [CrossRef]
- Rhee, I.; Bachman, K.E.; Park, B.H.; Jair, K.-W.; Yen, R.-W.C.; Schuebel, K.E.; Cui, H.; Feinberg, A.P.; Lengauer, C.; Kinzler, K.W. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 2002, 416, 552–556. [Google Scholar] [CrossRef]
- Fatemi, M.; Hermann, A.; Gowher, H.; Jeltsch, A. Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur. J. Biochem. 2002, 269, 4981–4984. [Google Scholar] [CrossRef]
- Tennis, M.A.; Vanscoyk, M.M.; Wilson, L.A.; Kelley, N.; Winn, R.A. Methylation of Wnt7a is modulated by DNMT1 and cigarette smoke condensate in non-small cell lung cancer. PLoS ONE 2012, 7, e32921. [Google Scholar] [CrossRef]
- Naessens, T.; Morias, Y.; Hamrud, E.; Gehrmann, U.; Budida, R.; Mattsson, J.; Baker, T.; Skogberg, G.; Israelsson, E.; Thörn, K. Human lung conventional dendritic cells orchestrate lymphoid neogenesis during chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2020, 202, 535–548. [Google Scholar] [CrossRef]
- Zhang, X.; Ulm, A.; Somineni, H.K.; Oh, S.; Weirauch, M.T.; Zhang, H.-X.; Chen, X.; Lehn, M.A.; Janssen, E.M.; Ji, H. DNA methylation dynamics during ex vivo differentiation and maturation of human dendritic cells. Epigenetics Chromatin 2014, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Zhou, B.; Luo, Z.Z.; Yu, S.C.; Tang, B. Cigarette smoke extract promotes DNA methyltransferase 3a expression in dendritic cells, inducing Th-17/Treg imbalance via the c-Jun/allograft inflammatory factor 1 axis. Kaohsiung J. Med. Sci. 2021, 37, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Husni, R.E.; Shiba-Ishii, A.; Iiyama, S.; Shiozawa, T.; Kim, Y.; Nakagawa, T.; Sato, T.; Kano, J.; Minami, Y.; Noguchi, M. DNMT3a expression pattern and its prognostic value in lung adenocarcinoma. Lung Cancer 2016, 97, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Statistics, E.G.C. GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2020, 70, 313. [Google Scholar]
- Dugué, P.-A.; Jung, C.-H.; Joo, J.E.; Wang, X.; Wong, E.M.; Makalic, E.; Schmidt, D.F.; Baglietto, L.; Severi, G.; Southey, M.C. Smoking and blood DNA methylation: An epigenome-wide association study and assessment of reversibility. Epigenetics 2020, 15, 358–368. [Google Scholar] [CrossRef]
- Huang, C.-C.; Lai, C.-Y.; Lin, I.-H.; Tsai, C.-H.; Tsai, S.-M.; Lam, K.-L.; Wang, J.-Y.; Chen, C.-C.; Wong, R.-H. Joint effects of cigarette smoking and green tea consumption with miR-29b and DNMT3B mRNA expression in the development of lung cancer. Genes 2022, 13, 836. [Google Scholar] [CrossRef]
- Huang, C.-C.; Lai, C.-Y.; Tsai, C.-H.; Wang, J.-Y.; Wong, R.-H. Combined effects of cigarette smoking, DNA methyltransferase 3B genetic polymorphism, and DNA damage on lung cancer. BMC Cancer 2021, 21, 1066. [Google Scholar] [CrossRef]
- Saito, Y.; Kanai, Y.; Sakamoto, M.; Saito, H.; Ishii, H.; Hirohashi, S. Overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, associated with DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 10060–10065. [Google Scholar] [CrossRef]

| Nonsmokers (Control) n = 36 | Smokers (Case) n = 36 | |||
|---|---|---|---|---|
| Personal Characteristics | No. | % | No. | % |
| Age (years) (Median-25th-75th quartiles) | 33.00 (26.25–36.50) | 37.00 (32.25–39.75) | ||
| Gender | ||||
| 10 | 27.8 | 21 | 58.3 |
| 26 | 72.2 | 15 | 41.7 |
| Nationality | Saudi individuals only | |||
| Duration of smoking (years) | 15 (7–20) | |||
| 13 | 36.1 | ||
| 15 | 41.7 | ||
| 8 | 22.2 | ||
| Smoking type | Cigarettes only, excluding water pipes or electronic cigarettes | |||
| Laboratory parameters | ||||
| 97.4 (88.7–100.7) | |||
| 183.5 (177.1–200.5) | |||
| 138.9 (129–150.4) | |||
| Parameters | Nonsmokers (n = 36) | Smokers (n = 36) | (95% CI) | p-Value | ||
|---|---|---|---|---|---|---|
| Median | 25–75 Quartiles | Median | 25–75 Quartiles | |||
| % 5 mC | 0.549 | (0.301–0.699) | 0.331 | (0.192–0.506) | (0.314–0.068) | <0.001 * |
| DMT1 (pg/mL) | 7.433 | (6.383–8.339) | 15.445 | (12.421–17.483) | (11.96–14.47) | <0.001 * |
| DNMT3A (pg/mL) | 6.827 | (6.100–8.151) | 10.412 | (7.861–14.606) | (9.34–11.99) | 0.003 * |
| DNMT3B (pg/mL) | 15.244 | (10.647–20.465) | 20.630 | (14.429–27.433) | (17.52–23.52) | 0.012 * |
| Variable | % 5 mC | DNMT1 | DNMT3A | DNMT3B | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r-Value | (95% CI) | p-Value | r-Value | (95% CI) | p-Value | r-Value | (95% CI) | p-Value | r-Value | (95% CI) | p-Value | |
| Age | 0.149 | (−0.373−0.301) | 0.384 | 0.108 | (0.156–0.696) | 0.530 | 0.103 | (0.042–0.632) | 0.552 | −0.067 | (0.227–0.72) | 0.699 |
| Glu (mg/dL) | −0.282 | (−0.566 −0.061) | 0.095 | 0.246 | (−0.100–0.539) | 0.148 | 0.290 | (−0.052–0.572) | 0.086 | 0.046 | (−0.296–0.377) | 0.791 |
| Chol (mg/dL) | 0.120 | (−0.227–0.440) | 0.485 | 0.232 | (−0.115–0.528) | 0.174 | 0.163 | (−0.184–0.475) | 0.342 | 0.070 | (−0.274–0.398) | 0.687 |
| TG (mg/dL) | 0.034 | (−0.307–0.367) | 0.843 | −0.018 | (−0.353−0.321) | 0.916 | 0.051 | (−0.292–0.382) | 0.768 | −0.012 | (−0.348–0.327) | 0.945 |
| Gender (Smokers) a | p-Value | Exp(B) | 95% CI for Exp(B) | |
|---|---|---|---|---|
| Lower Bound | Upper Bound | |||
| Intercept | 0.140 | |||
| % 5 mC | 0.058 | 75.838 | 0.868 | 6628.110 |
| DNMT1 | 0.162 | 0.791 | 0.570 | 1.098 |
| DNMT3A | 0.004 † | 1.529 | 1.149 | 2.035 |
| DNMT3B | 0.997 | 1.000 | 0.874 | 1.144 |
| Parameters | 1–10 Years n = 13 | 11–20 Years n = 15 | >20 Years n = 8 | |||
|---|---|---|---|---|---|---|
| r-Value | p-Value | r-Value | p-Value | r-Value | p-Value | |
| % 5 mC | −0.084 | 0.101 | −0.203 | 0.031* | −0.854 | 0.040 * |
| DMT1 | 0.085 | 0.001 * | 0.319 | 0.002* | 0.033 | 0.002 * |
| DNMT3A | 0.124 | 0.001 * | 0.361 | 0.028* | 0.431 | <0.001 * |
| DNMT3B | 0.148 | 0.049 * | 0.177 | 0.021* | 0.553 | 0.015 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
AlAmeer, A.; Sultan, S. Global DNA Methylation and DNA Methyltransferase Status Among Cigarette Smokers in Saudi. Life 2025, 15, 171. https://doi.org/10.3390/life15020171
AlAmeer A, Sultan S. Global DNA Methylation and DNA Methyltransferase Status Among Cigarette Smokers in Saudi. Life. 2025; 15(2):171. https://doi.org/10.3390/life15020171
Chicago/Turabian StyleAlAmeer, Areej, and Samar Sultan. 2025. "Global DNA Methylation and DNA Methyltransferase Status Among Cigarette Smokers in Saudi" Life 15, no. 2: 171. https://doi.org/10.3390/life15020171
APA StyleAlAmeer, A., & Sultan, S. (2025). Global DNA Methylation and DNA Methyltransferase Status Among Cigarette Smokers in Saudi. Life, 15(2), 171. https://doi.org/10.3390/life15020171






