Multiscale Segmentation-Guided Diffusion Model for CBCT-to-CT Synthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Preparation
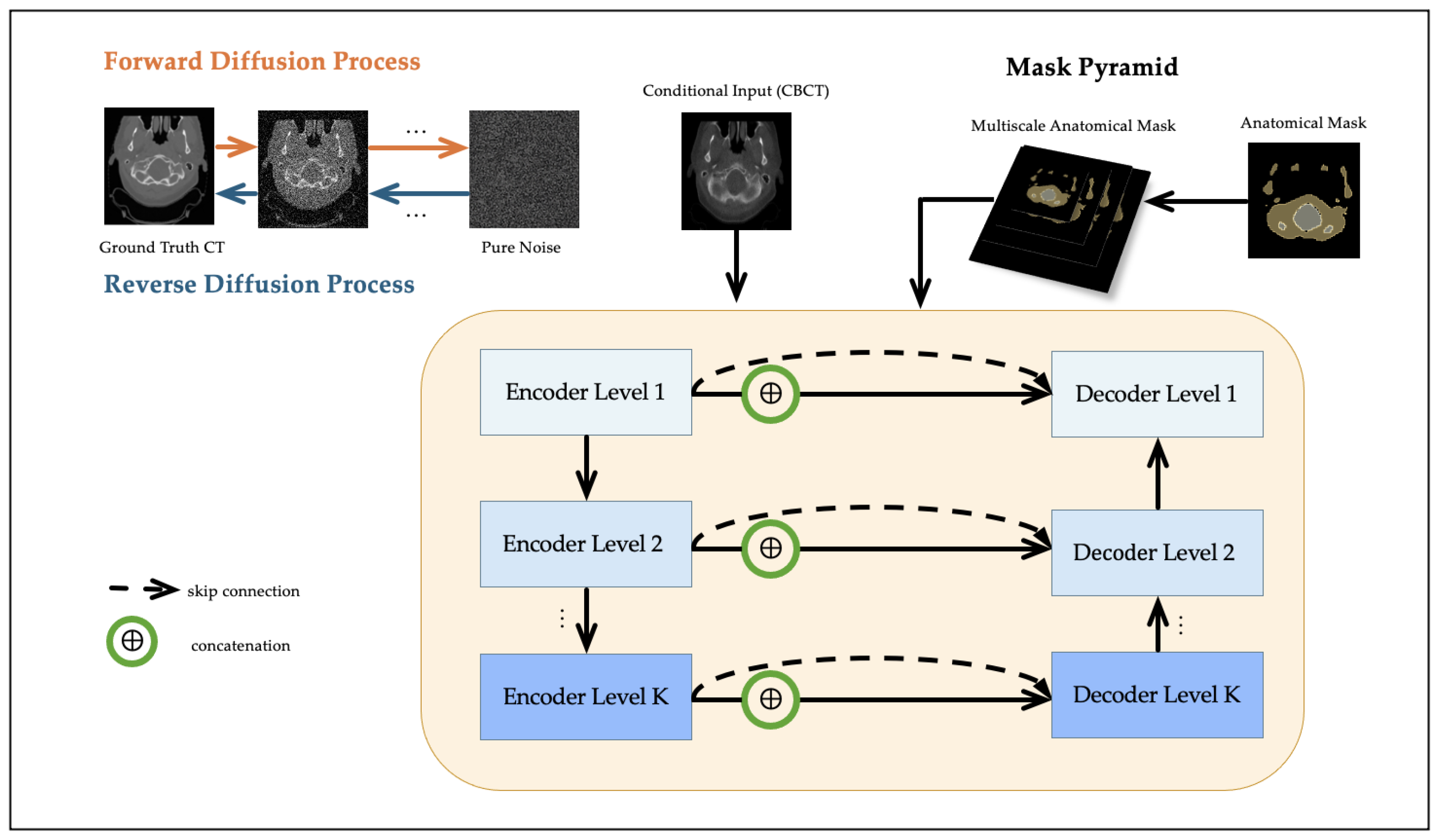
2.2. Conditional Diffusion Model
2.3. Multiscale Mask Pyramid Module
2.4. Multiscale Anatomical Loss
2.5. Evaluation Metrics
- Mean Absolute Error (MAE) measures the average voxel-wise intensity difference:
- Peak Signal-to-Noise Ratio (PSNR) is a logarithmic metric quantifying signal fidelity:where denotes the dynamic range of intensities. In our study, we set HU, corresponding to the intensity clipping range of HU.
- Structural Similarity Index (SSIM) evaluates structural agreement in local image patches:where x and y are local image patches from sCT and CT; , , and denote mean, variance, and covariance. Constants and are defined as and , where L is the dynamic range of the image. We set , corresponding to the intensity clipping range.
- HU Bias reflects the global intensity offset between sCT and reference CT, which is critical for dose calculation. A smaller HU bias (ideally close to 0) indicates better calibration of synthetic intensities:
- Gradient Root Mean Square Error (Grad-RMSE) is a common metric to assess anatomical sharpness and structural fidelity:where ∇ denotes the image gradient operator (e.g., Sobel or central difference), capturing spatial intensity changes. Lower Gradient RMSE indicates better preservation of anatomical boundaries, making it particularly important for downstream applications such as segmentation or dose calculation near organ interfaces.
- Dice Similarity Coefficient (DSC) is computed to quantify the structural agreement between anatomical segmentations derived from sCT and CT images. Specifically, we apply tools such as TotalSegmentator to extract segmentation masks from both domains and compute their overlap:where A and B denote the predicted and reference segmentation masks. A higher Dice score indicates improved anatomical consistency and potentially greater dosimetric reliability.
- To quantify tissue-wise accuracy, we also compute the MAE within three HU-defined anatomical regions: air (HU < ), soft tissue (HU between and 300), and bone (HU > 300). Let denote the set of pixels belonging to region r. The region-specific MAE is computed asThese region-specific errors provide a more clinically meaningful evaluation of synthetic CT accuracy, particularly in capturing air-tissue interfaces, soft tissue contrast, and bone density.
2.6. Implementation Details
3. Results
3.1. Experimental Setup
3.2. Ablation Study
- CBCT: Raw CBCT images directly compared with the ground-truth CT, without any synthesis.
- Baseline Diffusion [16]: A conditional DDPM with a UNet backbone, trained without anatomical supervision.
- SegGuided Diffusion (Single-scale, ) [23]: The baseline DDPM trained with a single binary anatomical mask derived from TotalSegmentator as structural guidance.
- Multiscale SegGuided Diffusion (): An extension of the SegGuided model that incorporates a multiscale mask pyramid by downsampling the original mask to K additional resolutions, aligned with the encoder. Here, K denotes the number of auxiliary mask levels beyond the full-resolution input .
- Multiscale SegGuided Diffusion with Multiscale Loss: The above model is further enhanced by a multiscale anatomical loss applied at each decoder resolution to enforce anatomical consistency across scales.
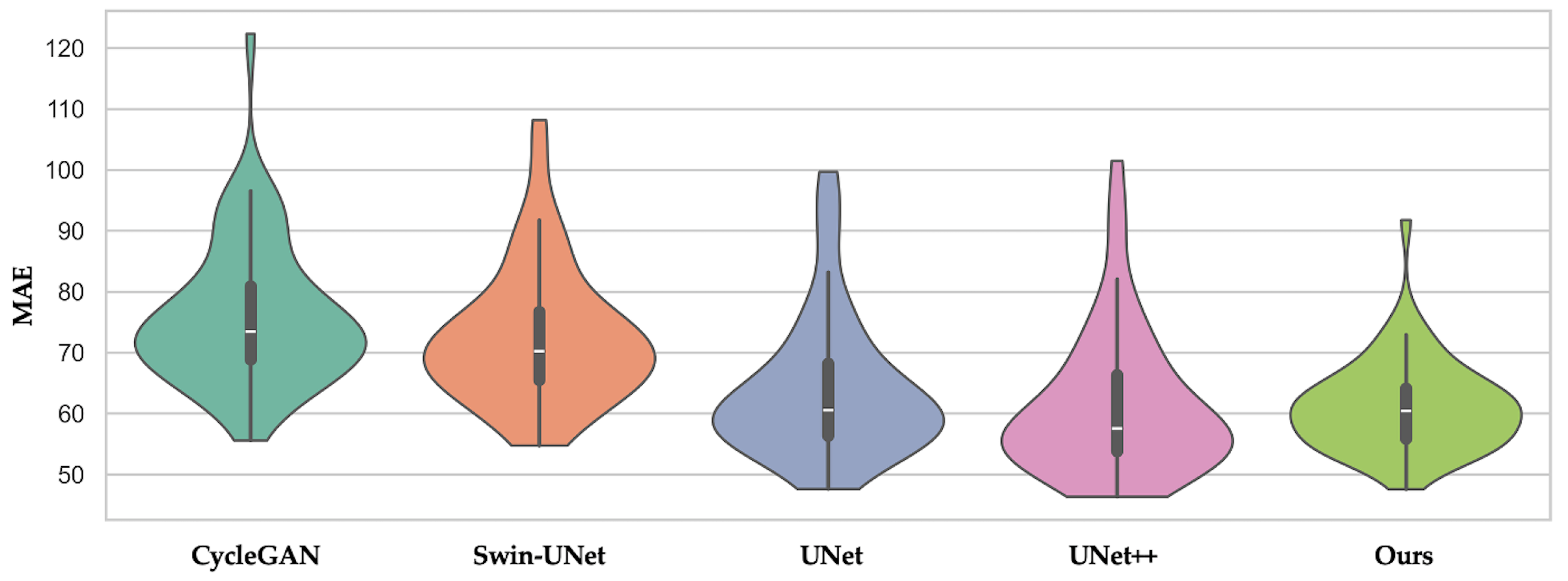
3.3. Quantitative Results
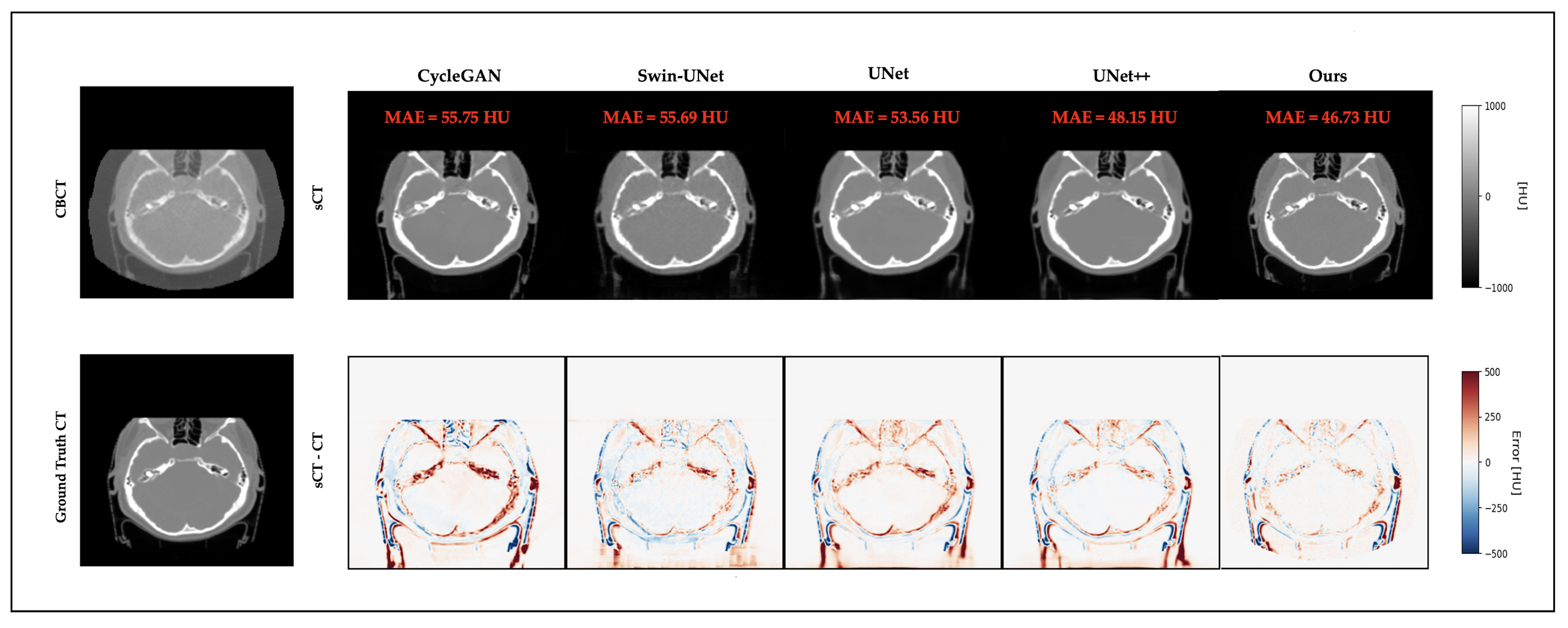
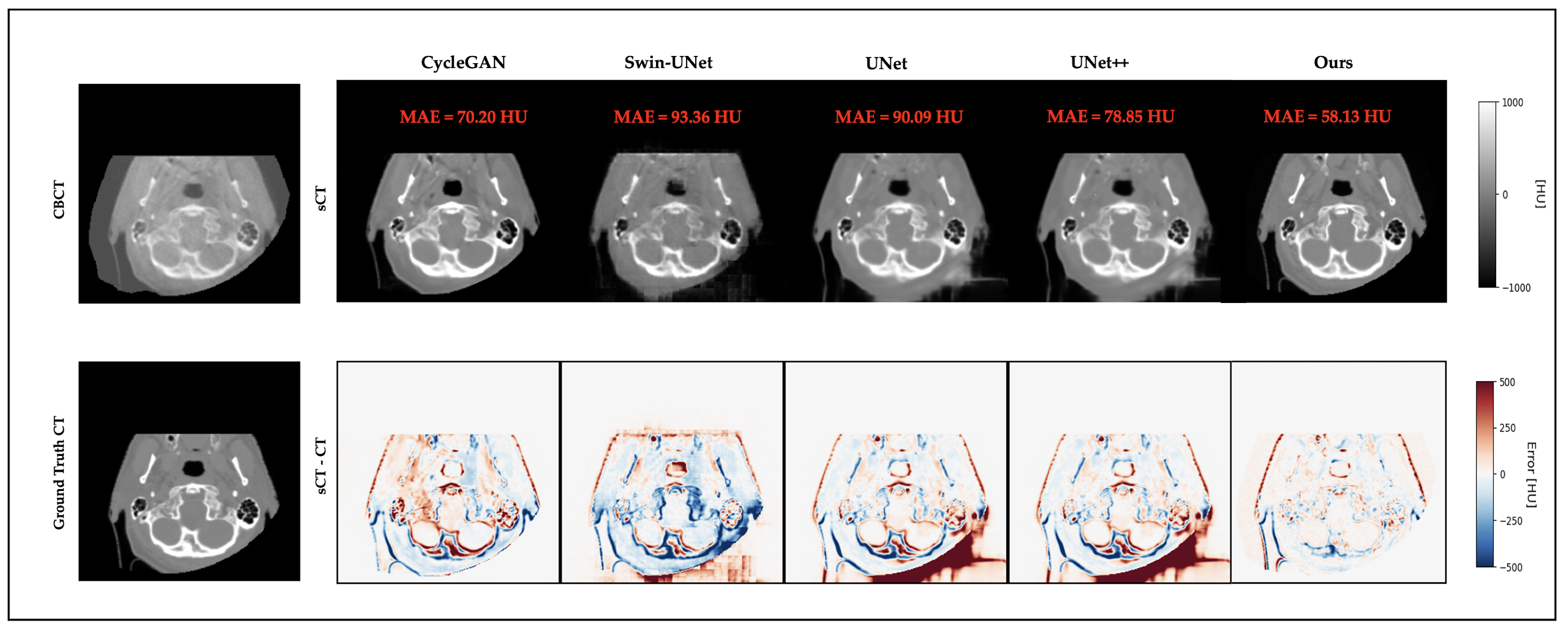
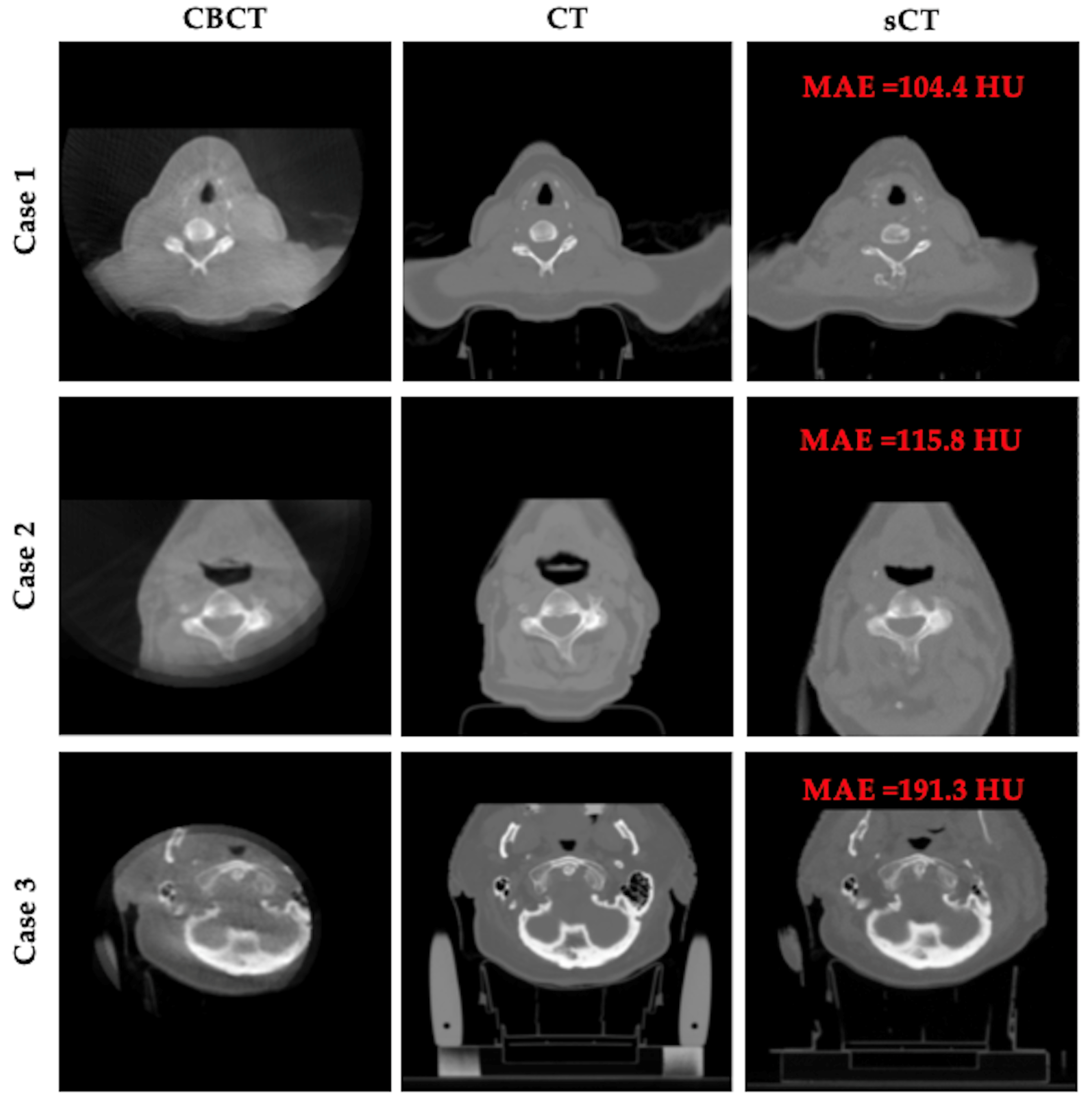
3.4. Qualitative Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CT | Computed tomography |
| sCT | Synthetic CT |
| CBCT | Cone-beam computed tomography |
| IGART | Image-guided adaptive radiotherapy |
| GAN | Generative adversarial networks |
| HU | Hounsfield units |
| DDPM | Denoising diffusion probabilistic model |
| MSE | Mean squared error |
| MAE | Mean absolute error |
| PSNR | Peak signal-to-noise ratio |
| SSIM | Structural similarity index |
| Grad-RMSE | Gradient mean squared error |
| DSC | Dice similarity coefficient |
References
- Lavrova, E.; Garrett, M.D.; Wang, Y.F.; Chin, C.; Elliston, C.; Savacool, M.; Price, M.; Kachnic, L.A.; Horowitz, D.P. Adaptive Radiation Therapy: A Review of CT-based Techniques. Radiol. Imaging Cancer 2023, 5. [Google Scholar] [CrossRef]
- Riou, O.; Prunaretty, J.; Michalet, M. Personalizing radiotherapy with adaptive radiotherapy: Interest and challenges. Cancer/Radiothérapie 2024, 28, 603–609. [Google Scholar] [CrossRef]
- Sherwani, M.K.; Gopalakrishnan, S. A Systematic Literature Review: Deep Learning Techniques for Synthetic Medical Image Generation and Their Applications in Radiotherapy. Front. Radiol. 2024, 4, 1385742. [Google Scholar] [CrossRef]
- Whitebird, R.R.; Solberg, L.I.; Bergdall, A.R.; López-Solano, N.; Smith-Bindman, R. Barriers to CT Dose Optimization: The Challenge of Organizational Change. Acad. Radiol. 2021, 28, 387–392. [Google Scholar] [CrossRef]
- Schulze, R.; Heil, U.; Groß, D.; Bruellmann, D.; Dranischnikow, E.; Schwanecke, U.; Schoemer, E. Artefacts in CBCT: A review. Dentomaxillofacial Radiol. 2011, 40, 265–273. [Google Scholar] [CrossRef]
- Altalib, A.; McGregor, S.; Li, C.; Perelli, A. Synthetic CT Image Generation From CBCT: A Systematic Review. IEEE Trans. Radiat. Plasma Med. Sci. 2025, 9, 691–707. [Google Scholar] [CrossRef]
- Hehakaya, C.; Van der Voort van Zyp, J.R.; Lagendijk, J.J.W.; Grobbee, D.E.; Verkooijen, H.M.; Moors, E.H.M. Problems and Promises of Introducing the Magnetic Resonance Imaging Linear Accelerator Into Routine Care: The Case of Prostate Cancer. Front. Oncol. 2020, 10, 1741. [Google Scholar] [CrossRef]
- Arnold, T.C.; Freeman, C.W.; Litt, B.; Stein, J.M. Low-field MRI: Clinical promise and challenges. J. Magn. Reson. Imaging 2022, 57, 25–44. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Schaal, D.; Curry, H.; Clark, R.; Magliari, A.; Kupelian, P.; Khuntia, D.; Beriwal, S. Review of cone beam computed tomography based online adaptive radiotherapy: Current trend and future direction. Radiat. Oncol. 2023, 18, 144. [Google Scholar] [CrossRef] [PubMed]
- Ghaznavi, H.; Maraghechi, B.; Zhang, H.; Zhu, T.; Laugeman, E.; Zhang, T.; Zhao, T.; Mazur, T.R.; Darafsheh, A. Quantitative Use of Cone-Beam Computed Tomography in Proton Therapy: Challenges and Opportunities. Phys. Med. Biol. 2025, 70, 09TR01. [Google Scholar] [CrossRef] [PubMed]
- Landry, G.; Kurz, C.; Thummerer, A. Perspectives for Using Artificial Intelligence Techniques in Radiation Therapy. Eur. Phys. J. Plus 2024, 139, 883. [Google Scholar] [CrossRef]
- Mastella, E.; Calderoni, F.; Manco, L.; Ferioli, M.; Medoro, S.; Turra, A.; Giganti, M.; Stefanelli, A. A Systematic Review of the Role of Artificial Intelligence in Automating Computed Tomography-Based Adaptive Radiotherapy for Head and Neck Cancer. Phys. Imaging Radiat. Oncol. 2025, 33, 100731. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.; Jain, A.; Abbeel, P. Denoising Diffusion Probabilistic Models. arXiv 2020. [Google Scholar] [CrossRef]
- Song, J.; Meng, C.; Ermon, S. Denoising Diffusion Implicit Models. arXiv 2022. [Google Scholar] [CrossRef]
- Chen, X.; Qiu, R.L.J.; Peng, J.; Shelton, J.W.; Chang, C.W.; Yang, X.; Kesarwala, A.H. CBCT-based Synthetic CT Image Generation Using a Diffusion Model for CBCT-guided Lung Radiotherapy. Med. Phys. 2024, 51, 8168–8178. [Google Scholar] [CrossRef]
- Peng, J.; Qiu, R.L.J.; Wynne, J.F.; Chang, C.W.; Pan, S.; Wang, T.; Roper, J.; Liu, T.; Patel, P.R.; Yu, D.S.; et al. CBCT-Based Synthetic CT Image Generation Using Conditional Denoising Diffusion Probabilistic Model. Med. Phys. 2024, 51, 1847–1859. [Google Scholar] [CrossRef]
- Fu, L.; Li, X.; Cai, X.; Miao, D.; Yao, Y.; Shen, Y. Energy-Guided Diffusion Model for CBCT-to-CT Synthesis. Comput. Med. Imaging Graph. 2024, 113, 102344. [Google Scholar] [CrossRef]
- Li, Y.; Shao, H.C.; Liang, X.; Chen, L.; Li, R.; Jiang, S.; Wang, J.; Zhang, Y. Zero-Shot Medical Image Translation via Frequency-Guided Diffusion Models. IEEE Trans. Med. Imaging 2023, 43, 980–993. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, L.; Wang, J.; Yang, X.; Zhou, H.; He, J.; Xie, Y.; Jiang, Y.; Sun, W.; Zhang, X.; et al. Texture-Preserving Diffusion Model for CBCT-to-CT Synthesis. Med. Image Anal. 2025, 99, 103362. [Google Scholar] [CrossRef]
- Kazerouni, A.; Aghdam, E.K.; Heidari, M.; Azad, R.; Fayyaz, M.; Hacihaliloglu, I.; Merhof, D. Diffusion models in medical imaging: A comprehensive survey. Med. Image Anal. 2023, 88, 102846. [Google Scholar] [CrossRef] [PubMed]
- Khader, F.; Müller-Franzes, G.; Tayebi Arasteh, S.; Han, T.; Haarburger, C.; Schulze-Hagen, M.; Schad, P.; Engelhardt, S.; Baessler, B.; Foersch, S.; et al. Denoising diffusion probabilistic models for 3D medical image generation. Sci. Rep. 2023, 13, 7303. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Chen, L.; Nguyen, D.; Zhou, Z.; Gu, X.; Yang, M.; Wang, J.; Jiang, S. Synthetic CT generation from CBCT images via deep learning. Med. Phys. 2020, 47, 1115–1125. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, Z.; Sun, Y.; Strittmatter, A.; Raj, A.; Allababidi, A.; Rink, J.S.; Zöllner, F.G. seg2med: A bridge from artificial anatomy to multimodal medical images. arXiv 2025. [Google Scholar] [CrossRef]
- Poch, D.V.; Estievenart, Y.; Zhalieva, E.; Patra, S.; Yaqub, M.; Taieb, S.B. Segmentation-Guided CT Synthesis with Pixel-Wise Conformal Uncertainty Bounds. arXiv 2025. [Google Scholar] [CrossRef]
- Sinha, A.; Dolz, J. Multi-scale self-guided attention for medical image segmentation. IEEE J. Biomed. Health Inform. 2020, 25, 121–130. [Google Scholar] [CrossRef]
- Rahman, M.M.; Munir, M.; Marculescu, R. Emcad: Efficient multi-scale convolutional attention decoding for medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 17–21 June 2024; pp. 11769–11779. [Google Scholar]
- Kolahi, S.G.; Chaharsooghi, S.K.; Khatibi, T.; Bozorgpour, A.; Azad, R.; Heidari, M.; Hacihaliloglu, I.; Merhof, D. MSA2Net: Multi-scale Adaptive Attention-guided Network for Medical Image Segmentation. arXiv 2024, arXiv:2407.21640. [Google Scholar]
- Thummerer, A.; van der Bijl, E.; Galapon, A., Jr.; Verhoeff, J.J.C.; Langendijk, J.A.; Both, S.; van den Berg, C.N.A.T.; Maspero, M. SynthRAD2023 Grand Challenge Dataset: Generating Synthetic CT for Radiotherapy. Med. Phys. 2023, 50, 4664–4674. [Google Scholar] [CrossRef]
- Wasserthal, J.; Breit, H.C.; Meyer, M.T.; Pradella, M.; Hinck, D.; Sauter, A.W.; Heye, T.; Boll, D.T.; Cyriac, J.; Yang, S.; et al. TotalSegmentator: Robust Segmentation of 104 Anatomic Structures in CT Images. Artif. Intell. 2023, 5, e230024. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A Self-Configuring Method for Deep Learning-Based Biomedical Image Segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired Image-to-Image Translation using Cycle-Consistent Adversarial Networks. arXiv 2020. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015. [Google Scholar] [CrossRef]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. arXiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-like Pure Transformer for Medical Image Segmentation. arXiv 2021. [Google Scholar] [CrossRef]
- Chen, J.; Ye, Z.; Zhang, R.; Li, H.; Fang, B.; Zhang, L.b.; Wang, W. Medical Image Translation with Deep Learning: Advances, Datasets and Perspectives. Med. Image Anal. 2025, 103, 103605. [Google Scholar] [CrossRef]
- Altalib, A.; Li, C.; Perelli, A. Conditional Diffusion Models for CT Image Synthesis from CBCT: A Systematic Review. arXiv 2025. [Google Scholar] [CrossRef]
- Chen, Y.; Konz, N.; Gu, H.; Dong, H.; Chen, Y.; Li, L.; Lee, J.; Mazurowski, M.A. ContourDiff: Unpaired Image-to-Image Translation with Structural Consistency for Medical Imaging. arXiv 2024. [Google Scholar] [CrossRef]
- Brioso, R.C.; Crespi, L.; Seghetto, A.; Dei, D.; Lambri, N.; Mancosu, P.; Scorsetti, M.; Loiacono, D. ARTInp: CBCT-to-CT Image Inpainting and Image Translation in Radiotherapy. arXiv 2025. [Google Scholar] [CrossRef]
- Zhou, X.; Wu, J.; Zhao, H.; Chen, L.; Zhang, S.; Wang, G. GLFC: Unified Global-Local Feature and Contrast Learning with Mamba-Enhanced UNet for Synthetic CT Generation from CBCT. arXiv 2025. [Google Scholar] [CrossRef]
| Method | MAE (↓) | PSNR (↑) | SSIM (↑) |
|---|---|---|---|
| CBCT | 507.32 ± 470.80 | 20.48 ± 7.49 | 0.65 ± 0.20 |
| Baseline Diffusion [16] | 69.78 ± 46.09 | 30.63 ± 3.47 | 0.87 ± 0.09 |
| SegGuided Diffusion () [23] | 64.15 ± 34.86 | 30.90 ± 3.32 | 0.88 ± 0.07 |
| Multiscale SegGuided Diffusion () | 65.06 ± 35.93 | 29.47 ± 4.05 | 0.88 ± 0.08 |
| Multiscale SegGuided Diffusion () | 63.1 ± 36.61 | 30.87 ± 4.15 | 0.88 ± 0.08 |
| Multiscale SegGuided Diffusion () | 62.69 ± 32.83 | 31.83 ± 3.34 | 0.89 ± 0.07 |
| Multiscale SegGuided Diffusion with Multiscale Loss () | 61.82 ± 30.59 | 32.05 ± 3.27 | 0.90 ± 0.05 |
| Method | MAE (↓) | PSNR (↑) | SSIM (↑) | HU Bias (↓) | Grad-RMSE (↓) | DSC (↑) | MAEair (↓) | MAEsoft (↓) | MAEbone (↓) |
|---|---|---|---|---|---|---|---|---|---|
| CBCT | 507.32 ± 470.80 | 20.48 ± 7.49 | 0.65 ± 0.20 | −219.36 ± 141.52 | 2631.38 ± 363.70 | 0.0001 ± 0.0003 | 962.54 ± 12.75 | 56.90 ± 3.01 | 852.62 ± 112.24 |
| CycleGAN | 75.61 ± 77.43 | 29.99 ± 3.80 | 0.87 ± 0.10 | −7.82 ± 9.77 | 1092.85 ± 171.46 | 0.84 ± 0.04 | 29.75 ± 12.68 | 61.09 ± 27.58 | 191.24 ± 86.50 |
| Swin-UNet | 73.13 ± 44.16 | 30.49 ± 3.21 | 0.87 ± 0.85 | −5.21 ± 10.09 | 980.44 ± 160.21 | 0.86 ± 0.03 | 32.13 ± 13.13 | 52.5 ± 19.41 | 165.83 ± 74.65 |
| UNet | 64.60 ± 40.07 | 31.18 ± 3.37 | 0.89 ± 0.78 | 3.42 ± 8.21 | 765.14 ± 295.74 | 0.94 ± 0.02 | 24.15 ± 12.71 | 41.83 ± 16.95 | 127.9 ± 64.12 |
| UNet++ | 62.04 ± 40.35 | 31.25 ± 3.52 | 0.89 ± 0.79 | 4.1 ± 7.9 | 730.56 ± 310.23 | 0.94 ± 0.02 | 23.1 ± 12.36 | 40.56 ± 15.93 | 124.65 ± 63.38 |
| Multiscale SegGuided Diffusion + Multiscale Loss (Ours) | 61.82 ± 30.59 | 32.05 ± 3.27 | 0.90 ± 0.05 | −0.72 ± 7.13 | 696.86 ± 341.59 | 0.95 ± 0.02 | 21.0 ± 12.49 | 34.84 ± 14.58 | 113.06 ± 62.07 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Luo, Y.; Hooshangnejad, H.; Zhang, R.; Feng, X.; Chen, Q.; Ngwa, W.; Ding, K. Multiscale Segmentation-Guided Diffusion Model for CBCT-to-CT Synthesis. Life 2025, 15, 1871. https://doi.org/10.3390/life15121871
Guo Y, Luo Y, Hooshangnejad H, Zhang R, Feng X, Chen Q, Ngwa W, Ding K. Multiscale Segmentation-Guided Diffusion Model for CBCT-to-CT Synthesis. Life. 2025; 15(12):1871. https://doi.org/10.3390/life15121871
Chicago/Turabian StyleGuo, Yike, Yi Luo, Hamed Hooshangnejad, Rui Zhang, Xue Feng, Quan Chen, Wilfred Ngwa, and Kai Ding. 2025. "Multiscale Segmentation-Guided Diffusion Model for CBCT-to-CT Synthesis" Life 15, no. 12: 1871. https://doi.org/10.3390/life15121871
APA StyleGuo, Y., Luo, Y., Hooshangnejad, H., Zhang, R., Feng, X., Chen, Q., Ngwa, W., & Ding, K. (2025). Multiscale Segmentation-Guided Diffusion Model for CBCT-to-CT Synthesis. Life, 15(12), 1871. https://doi.org/10.3390/life15121871








