Vaginal Microbiome and Functional Pathway Alterations in Preterm Premature Rupture of Membranes Revealed by 16S rRNA Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Vaginal Sampling
2.3. Genomic DNA Extraction and Amplicon Sequencing
2.4. Bioinformatic Analysis
2.5. Statistical Analysis
3. Results
3.1. Clinical Characteristics of Study Participants
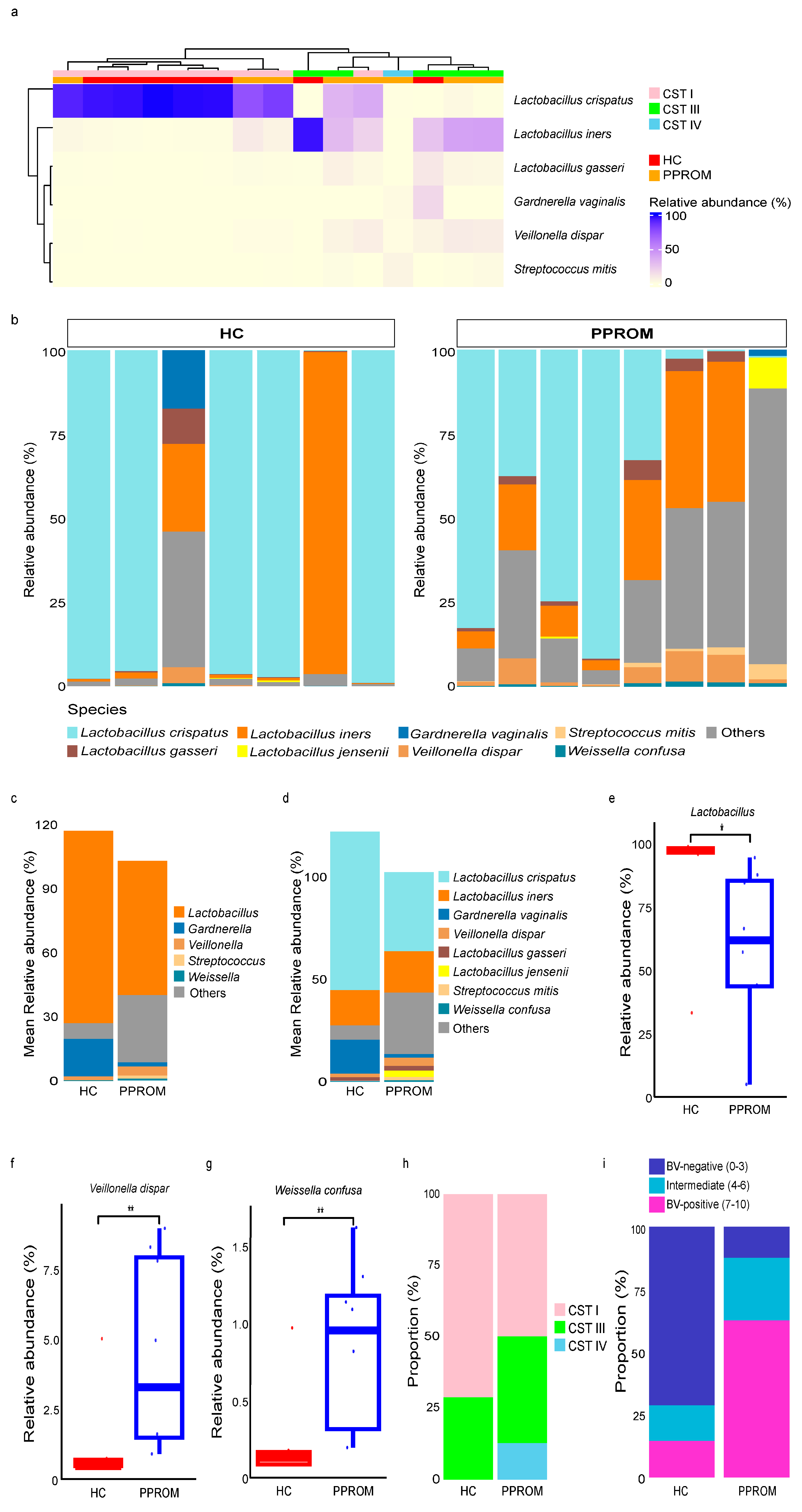
3.2. Hierarchical Clustering Analysis of the Vaginal Samples for HC and PPROM Groups
3.3. Community State Types (CSTs) and Correlation with Dysbiosis
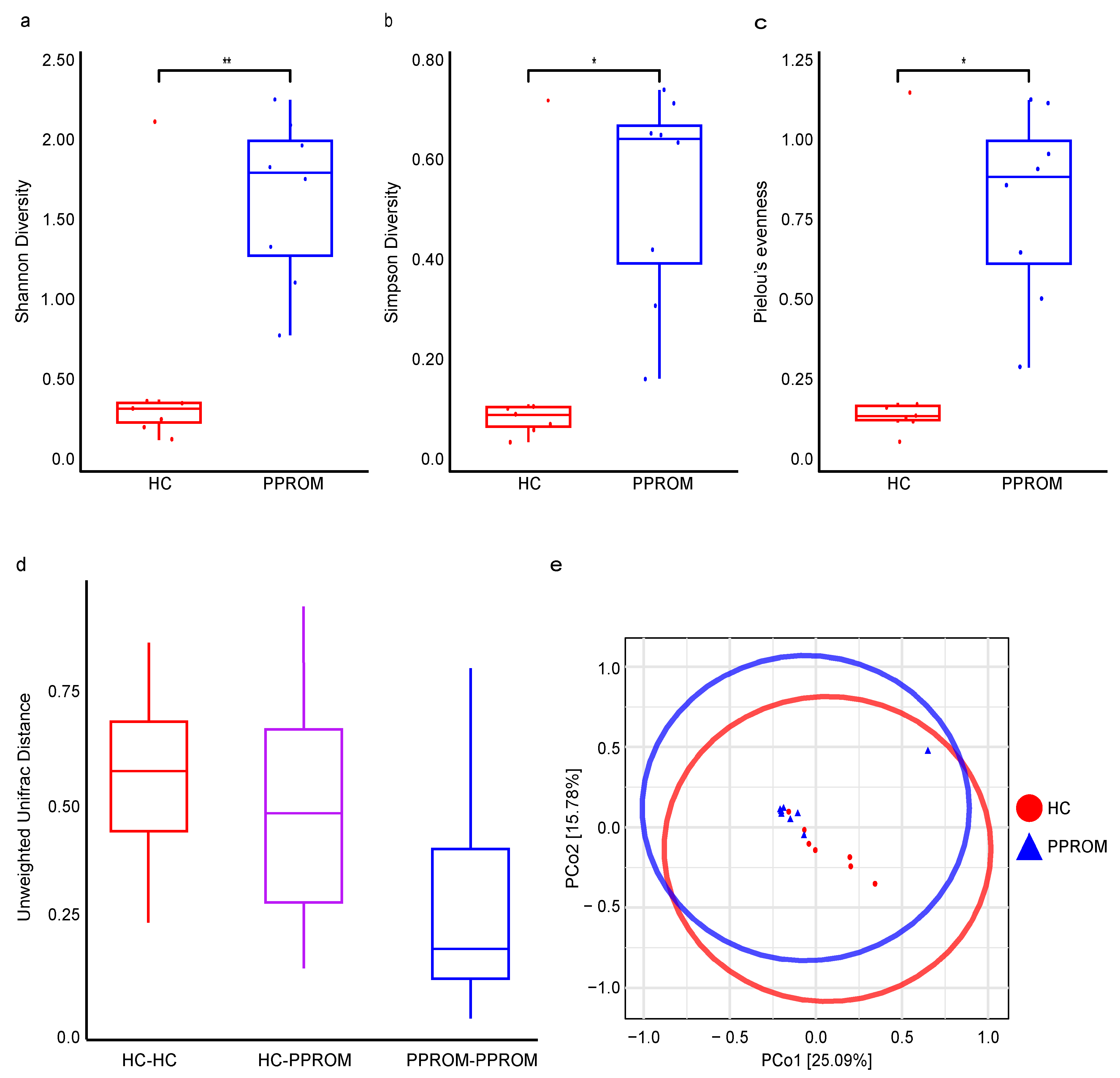
3.4. Alpha and Beta Diversity
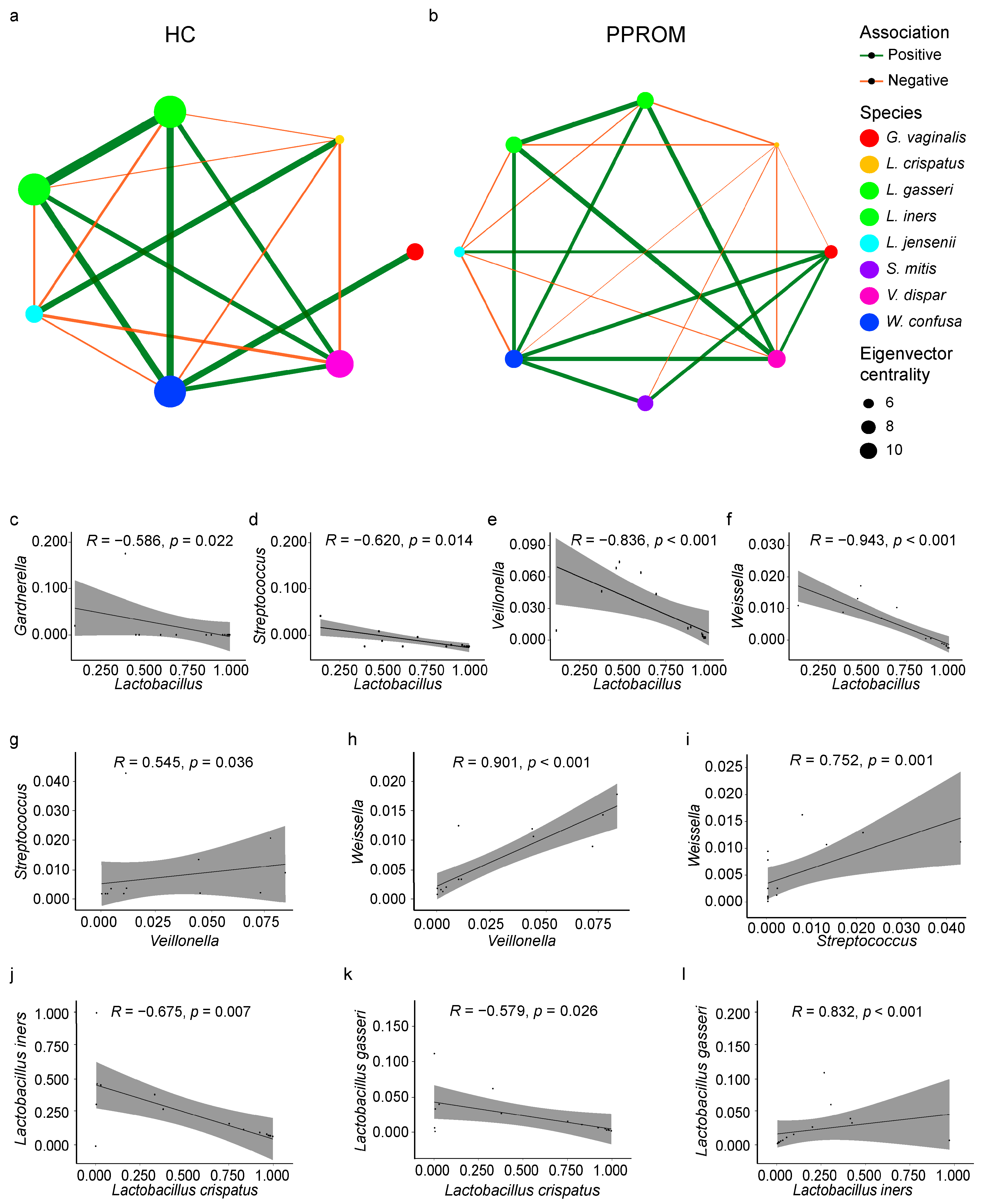
3.5. Co-Occurrence and Correlation Within the Vaginal Microbiome
3.6. Distinct Functional Pathways Between HC and PPROM Groups
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Parry, S.; Strauss, J.F., 3rd. Premature rupture of the fetal membranes. N. Engl. J. Med. 1998, 338, 663–670. [Google Scholar] [CrossRef]
- Sorrenti, S.; Di Mascio, D.; Khalil, A.; D’Antonio, F.; Rizzo, G.; Zullo, F.; D’Alberti, E.; D’Ambrosio, V.; Mappa, I.; Muzii, L.; et al. Outcome of prelabor rupture of membranes before or at the limit of viability: Systematic review and meta-analysis. Am. J. Obstet. Gynecol. MFM 2024, 6, 101370. [Google Scholar] [CrossRef]
- Fettweis, J.M.; Serrano, M.G.; Brooks, J.P.; Edwards, D.J.; Girerd, P.H.; Parikh, H.I.; Huang, B.; Arodz, T.J.; Edupuganti, L.; Glascock, A.L.; et al. The vaginal microbiome and preterm birth. Nat. Med. 2019, 25, 1012–1021. [Google Scholar] [CrossRef]
- DiGiulio, D.B.; Callahan, B.J.; McMurdie, P.J.; Costello, E.K.; Lyell, D.J.; Robaczewska, A.; Sun, C.L.; Goltsman, D.S.; Wong, R.J.; Shaw, G.; et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl. Acad. Sci. USA 2015, 112, 11060–11065. [Google Scholar] [CrossRef]
- Brown, R.G.; Marchesi, J.R.; Lee, Y.S.; Smith, A.; Lehne, B.; Kindinger, L.M.; Terzidou, V.; Holmes, E.; Nicholson, J.K.; Bennett, P.R.; et al. Vaginal dysbiosis increases risk of preterm fetal membrane rupture, neonatal sepsis and is exacerbated by erythromycin. BMC Med. 2018, 16, 9. [Google Scholar] [CrossRef]
- Mueller, N.T.; Bakacs, E.; Combellick, J.; Grigoryan, Z.; Dominguez-Bello, M.G. The infant microbiome development: Mom matters. Trends Mol. Med. 2015, 21, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Bennett, P.R.; Brown, R.G.; MacIntyre, D.A. Vaginal Microbiome in Preterm Rupture of Membranes. Obstet. Gynecol. Clin. N. Am. 2020, 47, 503–521. [Google Scholar] [CrossRef]
- Huang, C.; Gin, C.; Fettweis, J.; Foxman, B.; Gelaye, B.; MacIntyre, D.A.; Subramaniam, A.; Fraser, W.; Tabatabaei, N.; Callahan, B. Meta-analysis reveals the vaginal microbiome is a better predictor of earlier than later preterm birth. BMC Biol. 2023, 21, 199. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 20 March 2024).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 2011, 17, 3. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- France, M.T.; Ma, B.; Gajer, P.; Brown, S.; Humphrys, M.S.; Holm, J.B.; Waetjen, L.E.; Brotman, R.M.; Ravel, J. VALENCIA: A nearest centroid classification method for vaginal microbial communities based on composition. Microbiome 2020, 8, 166. [Google Scholar] [CrossRef]
- Usyk, M.; Schlecht, N.F.; Pickering, S.; Williams, L.; Sollecito, C.C.; Gradissimo, A.; Porras, C.; Safaeian, M.; Pinto, L.; Herrero, R.; et al. molBV reveals immune landscape of bacterial vaginosis and predicts human papillomavirus infection natural history. Nat. Commun. 2022, 13, 233. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan Community Ecology Package; R Core Team: Vienna, Austria, 2024; Available online: https://vegandevs.github.io/vegan/ (accessed on 14 August 2025).
- Peschel, S.; Muller, C.L.; von Mutius, E.; Boulesteix, A.L.; Depner, M. NetCoMi: Network construction and comparison for microbiome data in R. Brief Bioinform. 2021, 22, bbaa290. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The igraph software. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Pedersen, T.L.; Pedersen, M.; LazyData, T.; Rcpp, I.; Rcpp, L. Package ‘ggraph’. Retrieved January 2018. Available online: https://github.com/thomasp85/ggraph/ (accessed on 14 August 2025).
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, NY, USA, 2016; pp. 189–201. [Google Scholar]
- Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. 2023. Available online: https://rpkgs.datanovia.com/ggpubr/ (accessed on 14 August 2025).
- Amabebe, E.; Anumba, D.O.C. The Vaginal Microenvironment: The Physiologic Role of Lactobacilli. Front. Med. 2018, 5, 181. [Google Scholar] [CrossRef] [PubMed]
- Donders, G.G.; Van Calsteren, K.; Bellen, G.; Reybrouck, R.; Van den Bosch, T.; Riphagen, I.; Van Lierde, S. Predictive value for preterm birth of abnormal vaginal flora, bacterial vaginosis and aerobic vaginitis during the first trimester of pregnancy. BJOG 2009, 116, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- Gajer, P.; Brotman, R.M.; Bai, G.; Sakamoto, J.; Schutte, U.M.; Zhong, X.; Koenig, S.S.; Fu, L.; Ma, Z.S.; Zhou, X.; et al. Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med. 2012, 4, 132ra152. [Google Scholar] [CrossRef]
- Rampersaud, R.; Planet, P.J.; Randis, T.M.; Kulkarni, R.; Aguilar, J.L.; Lehrer, R.I.; Ratner, A.J. Inerolysin, a cholesterol-dependent cytolysin produced by Lactobacillus iners. J. Bacteriol. 2011, 193, 1034–1041. [Google Scholar] [CrossRef]
- Briselden, A.M.; Moncla, B.J.; Stevens, C.E.; Hillier, S.L. Sialidases (neuraminidases) in bacterial vaginosis and bacterial vaginosis-associated microflora. J. Clin. Microbiol. 1992, 30, 663–666. [Google Scholar] [CrossRef]
- Menon, R.; Fortunato, S.J. Infection and the role of inflammation in preterm premature rupture of the membranes. Best Pract. Res. Clin. Obstet. Gynaecol. 2007, 21, 467–478. [Google Scholar] [CrossRef]
- Feduniw, S.; Zeber-Lubecka, N.; Pruc, M.; Gaca, Z.; Szarpak, L.; Ciebiera, M. Does the Vaginal Microbiota Influence the Incidence of the Preterm Premature Rupture of Membranes? J. Clin. Med. 2025, 14, 6577. [Google Scholar] [CrossRef]
- Zheng, N.; Guo, R.; Wang, J.; Zhou, W.; Ling, Z. Contribution of Lactobacillus iners to Vaginal Health and Diseases: A Systematic Review. Front. Cell Infect. Microbiol. 2021, 11, 792787. [Google Scholar] [CrossRef]
- Holm, J.B.; Carter, K.A.; Ravel, J.; Brotman, R.M. Lactobacillus iners and genital health: Molecular clues to an enigmatic vaginal species. Curr. Infect. Dis. Rep. 2023, 25, 67–75. [Google Scholar] [CrossRef]
- Nelson, D.B.; Hanlon, A.; Hassan, S.; Britto, J.; Geifman-Holtzman, O.; Haggerty, C.; Fredricks, D.N. Preterm labor and bacterial vaginosis-associated bacteria among urban women. J. Perinat. Med. 2009, 37, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liang, T.; Liang, Z.; Jiang, T.; Chen, Y.; Chen, T.; Dong, B.; Wu, Q.; Gao, Y. Increased vaginal Gardnerella vaginalis abundance and reduced D-galactose metabolism are associated with preterm birth in older mothers with columnar ectopy in South China. mSystems 2025, 10, e0082525. [Google Scholar] [CrossRef] [PubMed]
- Nelson, T.M.; Borgogna, J.L.; Brotman, R.M.; Ravel, J.; Walk, S.T.; Yeoman, C.J. Vaginal biogenic amines: Biomarkers of bacterial vaginosis or precursors to vaginal dysbiosis? Front. Physiol. 2015, 6, 253. [Google Scholar] [CrossRef] [PubMed]
- Borgogna, J.C.; Shardell, M.D.; Grace, S.G.; Santori, E.K.; Americus, B.; Li, Z.; Ulanov, A.; Forney, L.; Nelson, T.M.; Brotman, R.M.; et al. Biogenic Amines Increase the Odds of Bacterial Vaginosis and Affect the Growth of and Lactic Acid Production by Vaginal Lactobacillus spp. Appl. Environ. Microbiol. 2021, 87, e03068-20. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, R.; Kavyani, B.; Nabizadeh, E.; Kadkhoda, H.; Asghari Ozma, M.; Abdi, M. Microbiota metabolites in the female reproductive system: Focused on the short-chain fatty acids. Heliyon 2023, 9, e14562. [Google Scholar] [CrossRef] [PubMed]
- Vidmar Simic, M.; Maver, A.; Zimani, A.N.; Hocevar, K.; Peterlin, B.; Kovanda, A.; Premru-Srsen, T. Oral microbiome and preterm birth. Front. Med. 2023, 10, 1177990. [Google Scholar] [CrossRef] [PubMed]

| PPROM (n = 8) | HC (n = 7) | p-Value | |
|---|---|---|---|
| Maternal age, years (mean ± SD) | 36.75 ± 1.75 | 34.57 ± 4.04 | 0.378 |
| Pre-pregnancy BMI (kg/m2) | 36.16 ± 6.34 | 32.50 ± 3.35 | 0.444 |
| Gestational age at sampling, weeks (mean ± SD) | 34.39 ± 2.52 | 36.56 ± 0.87 | 0.022 |
| Gestational age at delivery, weeks (mean ± SD) | 34.10 ± 2.38 | 38.44 ± 1.00 | 3.11 × 10−4 |
| Cervical length, cm (mean ± SD) | 2.45 ± 0.79 | 3.34 ± 0.52 | 0.066 |
| 1 min Apgar score (median (Q1–Q3)) | 6.5 (5–9) | 9 (8–9) | 0.114 |
| Moderate (4–6), n (%) | 4 (50%) | 1 (14%) | |
| Vigorous (7–10), n (%) | 4 (50%) | 6 (86%) | |
| 5 min Apgar score (median (Q1–Q3)) | 8.5 (6.5–10) | 10 (10–10) | 0.093 |
| Moderate (4–6), n (%) | 2 (25%) | 0 (0%) | |
| Vigorous (7–10), n (%) | 6 (75%) | 7 (100%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nam, S.; Hong, S.; Park, I.Y.; Shin, S. Vaginal Microbiome and Functional Pathway Alterations in Preterm Premature Rupture of Membranes Revealed by 16S rRNA Sequencing. Life 2025, 15, 1604. https://doi.org/10.3390/life15101604
Nam S, Hong S, Park IY, Shin S. Vaginal Microbiome and Functional Pathway Alterations in Preterm Premature Rupture of Membranes Revealed by 16S rRNA Sequencing. Life. 2025; 15(10):1604. https://doi.org/10.3390/life15101604
Chicago/Turabian StyleNam, Sangho, Subeen Hong, In Yang Park, and Sun Shin. 2025. "Vaginal Microbiome and Functional Pathway Alterations in Preterm Premature Rupture of Membranes Revealed by 16S rRNA Sequencing" Life 15, no. 10: 1604. https://doi.org/10.3390/life15101604
APA StyleNam, S., Hong, S., Park, I. Y., & Shin, S. (2025). Vaginal Microbiome and Functional Pathway Alterations in Preterm Premature Rupture of Membranes Revealed by 16S rRNA Sequencing. Life, 15(10), 1604. https://doi.org/10.3390/life15101604






