The First Complete Mitogenome Characterization of Brown Alga Dictyota coriacea (Phaeophyceae, Heterokontophyta) and Its Phylogenetic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, DNA Extraction, and Sequencing
2.2. Sequence Assembly, Annotation and Analysis
2.3. Phylogenetic Analysis
3. Results and Discussion
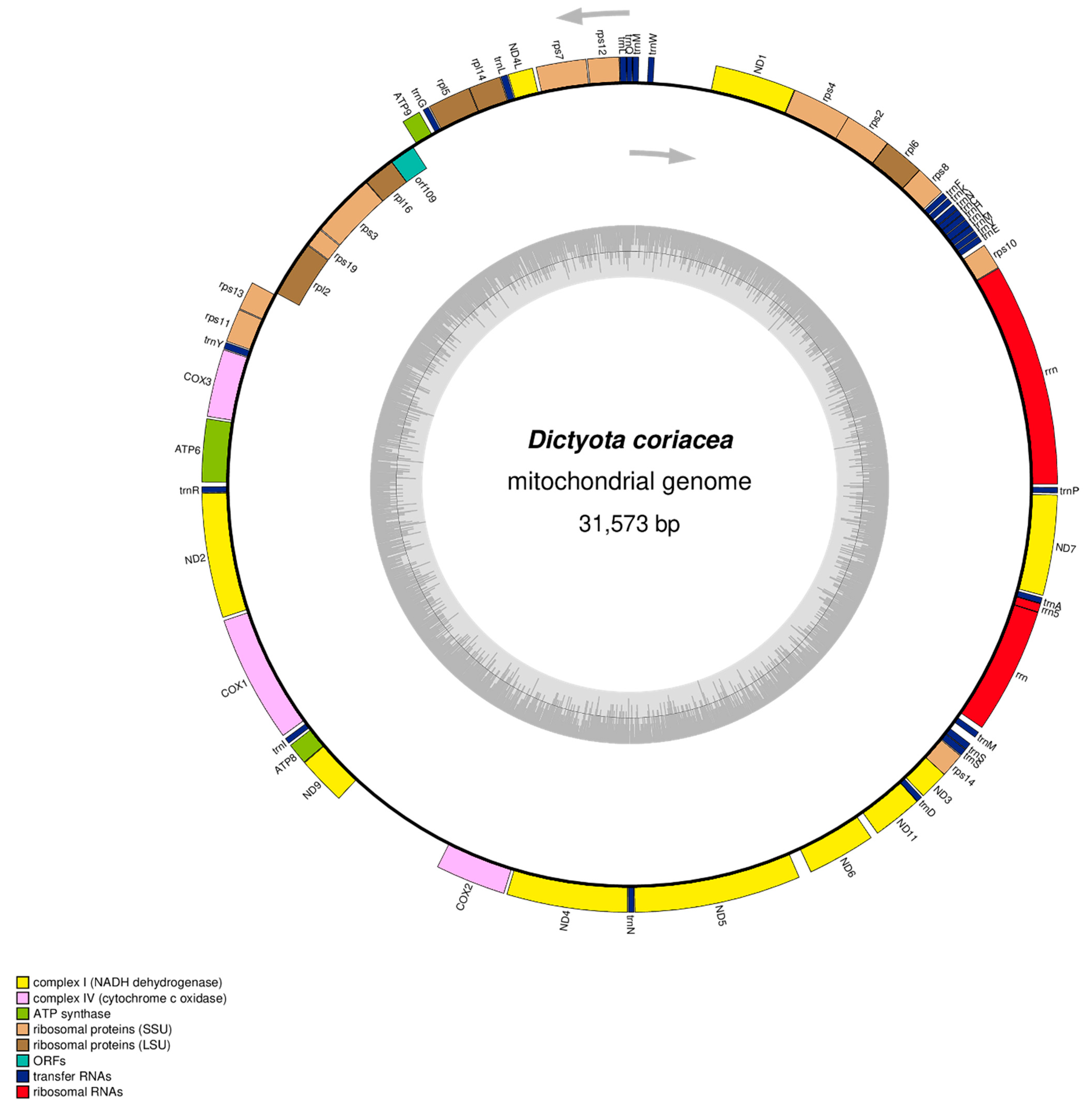
3.1. Mitogenome Structure and Nucleotide Composition
3.2. Protein-Coding Gene Features
3.3. Ribosomal and Transfer RNA Genes
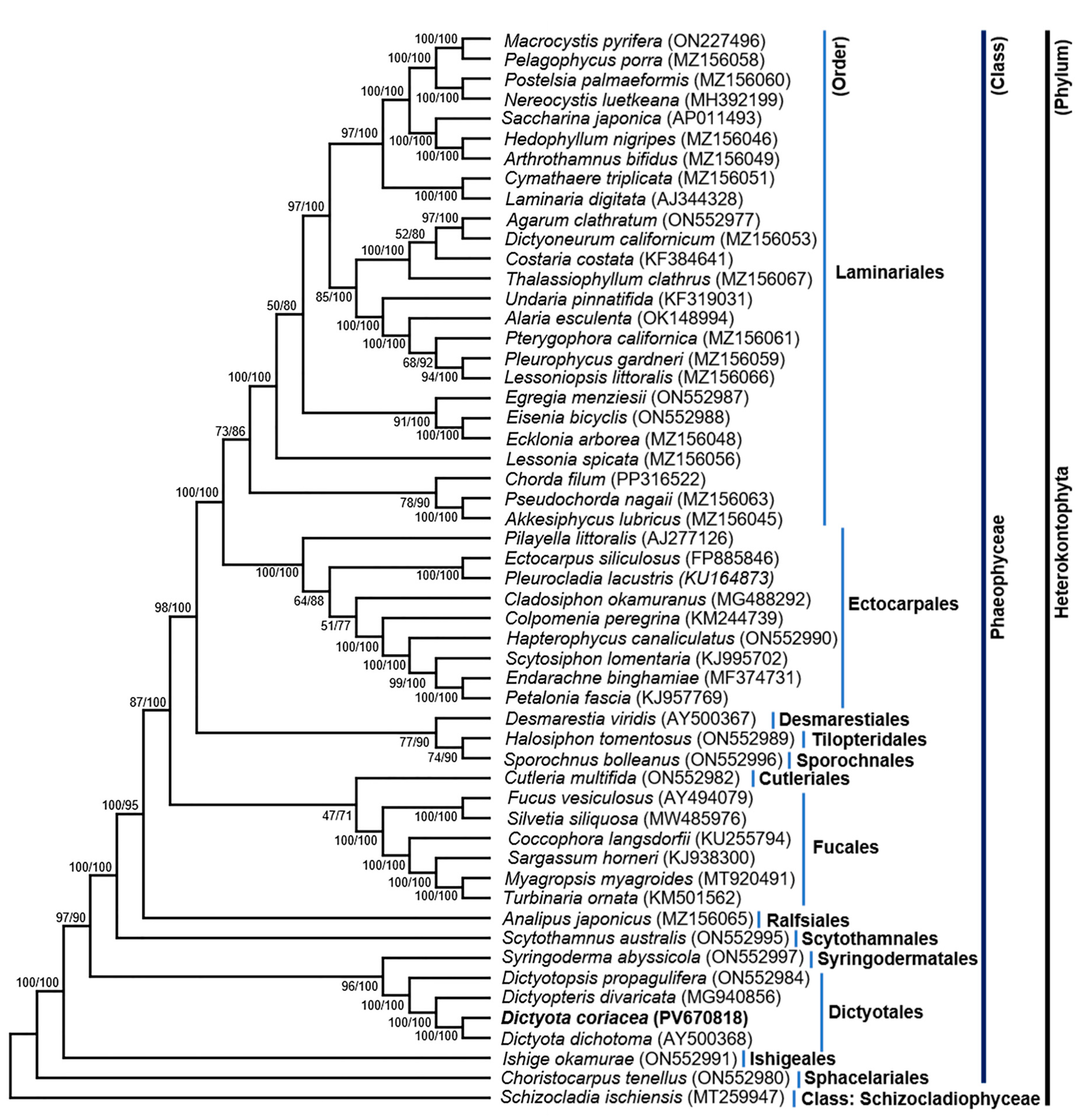
3.4. Phylogenetic Relationship Within Phaeophyceae
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fong, P.; Paul, V.J. Coral Reef Algae. In Coral Reefs: An Ecosystem in Transition; Dubinsky, Z., Stambler, N., Eds.; Springer: Dordrecht, The Netherlands, 2011. [Google Scholar] [CrossRef]
- Mejia, A.Y.; Puncher, G.N.; Engelen, A.H. Macroalgae in Tropical Marine Coastal Systems. In Seaweed Biology: Novel Insights into Ecophysiology, Ecology and Utilization; Wiencke, C., Bischof, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 219. [Google Scholar] [CrossRef]
- Rushdi, M.I.; Abdel-Rahman, I.A.M.; Attia, E.Z.; Saber, H.; Saber, A.A.; Bringmann, G.; Abdelmohsen, U.R. The biodiversity of the genus Dictyota: Phytochemical and pharmacological natural products prospectives. Molecules 2022, 27, 672. [Google Scholar] [CrossRef]
- Georgii, A.D.N.P.; Teixeira, V.L. Dictyota and Canistrocarpus Brazilian Brown Algae and Their Bioactive Diterpenes—A Review. Mar. Drugs 2023, 21, 484. [Google Scholar] [CrossRef] [PubMed]
- Piao, M.J.; Kang, K.A.; Herath, H.M.U.L.; Fernando, P.D.S.M.; Lee, N.H.; Hyun, J.W. Extract of seaweed Dictyota coriacea scavenges superoxide anion and hydroxyl radical. Food Suppl. Biomater. Health 2023, 3, e23. [Google Scholar] [CrossRef]
- Hwang, I.K.; Kim, H.S.; Lee, W.J. Evidence for taxonomic status of Pachydictyon coriaceum (Holmes) Okamura (Dictyotales, Phaeophyceae) based on morphology and plastid protein coding rbcL, psaA, and psbA gene sequences. Algae 2004, 19, 175–190. [Google Scholar] [CrossRef]
- Gissi, C.; Iannelli, F.; Pesole, G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity 2008, 101, 301–320. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Pang, S.; Li, X.; Li, J. Complete mitochondrial genome of the brown alga Sargassum horneri (Sargassaceae, Phaeophyceae): Genome organization and phylogenetic analyses. J. Appl. Phycol. 2015, 27, 469–478. [Google Scholar] [CrossRef]
- Starko, S.; Bringloe, T.T.; Gomez, M.S.; Darby, H.; Graham, S.W.; Martone, P.T. Genomic rearrangements and sequence evolution across brown algal organelles. Genome Biol. Evol. 2021, 13, evab124. [Google Scholar] [CrossRef]
- Graf, L.; Kim, Y.J.; Cho, G.Y.; Miller, K.A.; Yoon, H.S. Plastid and mitochondrial genomes of Coccophora langsdorfii (Fucales, Phaeophyceae) and the utility of molecular markers. PLoS ONE 2017, 12, e0187104. [Google Scholar] [CrossRef]
- Engel, C.R.; Billard, M.V.E.; Viard, F. Conservation and polymorphism of mitochondrial intergenic sequences in brown algae (Phaeophyceae). Eur. J. Phycol. 2008, 43, 195–205. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, Y.; Bi, Y.; Chen, W.; Moejes, F.W. Understanding the evolution of mitochondrial genomes in Phaeophyceae inferred from mitogenomes of Ishige okamurae (Ishigeales) and Dictyopteris divaricata (Dictyotales). J. Mol. Evol. 2019, 87, 16–26. [Google Scholar] [CrossRef]
- Liu, Y.J.; Zhang, T.Y.; Wang, Q.Q.; Draisma, S.G.; Hu, Z.M. Comparative structure and evolution of the organellar genomes of Padina usoehtunii (Dictyotales) with the brown algal crown radiation clade. BMC Genom. 2024, 25, 747. [Google Scholar] [CrossRef] [PubMed]
- Bringloe, T.T.; Starko, S.; Wade, R.M.; Vieira, C.; Kawai, H.; De Clerck, O.; Cock, J.M.; Coelho, S.M.; Destombe, C.; Valero, M.; et al. Phylogeny and evolution of the brown algae. Crit. Rev. Plant Sci. 2020, 39, 281–321. [Google Scholar] [CrossRef]
- Denoeud, F.; Godfroy, O.; Cruaud, C.; Heesch, S.; Nehr, Z.; Tadrent, N.; Couloux, A.; Brillet-Guéguen, L.; Delage, L.; Mckeown, D.; et al. Evolutionary genomics of the emergence of brown algae as key components of coastal ecosystems. Cell 2024, 187, 6943–6965. [Google Scholar] [CrossRef]
- Rana, S.; Valentin, K.; Riehl, J.; Blanfuné, A.; Reynes, L.; Thibaut, T.; Bartsch, I.; Eichinger, L.; Glöckner, G. Analysis of organellar genomes in brown algae reveals an independent introduction of similar foreign sequences into the mitochondrial genome. Genomics 2021, 113, 646–654. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Beck, N.; Lang, B. MFannot, Organelle Genome Annotation Webserver; Université de Montréal: Montreal, QC, Canada, 2010; Available online: https://megasun.bch.umontreal.ca/cgi-bin/dev_mfa/mfannotInterface.pl (accessed on 12 October 2025).
- Gish, W.; States, D.J. Identification of protein coding regions by database similarity search. Nat. Genet. 1993, 3, 266–272. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. In Gene Prediction: Methods and Protocols; Springer: New York, NY, USA, 2019; Volume 1962, pp. 1–14. [Google Scholar] [CrossRef]
- Lang, B.F.; Laforest, M.J.; Burger, G. Mitochondrial introns: A critical view. Trends Genet. 2007, 23, 119–125. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Secq, M.P.O.L.; Goër, S.L.D.; Stam, W.T.; Olsen, J.L. Complete mitochondrial genomes of the three brown algae (Heterokonta: Phaeophyceae) Dictyota dichotoma, Fucus vesiculosus and Desmarestia viridis. Curr. Genet. 2006, 49, 47–58. [Google Scholar] [CrossRef]
- West, J.A.; Motomura, T.; Choi, S.; De-Goër, S.L.; Miller, K.A.; Karsten, U.; Yoon, H.S.; Lim, P.E.; Nagasato, C. Light and electron microscopy, molecular phylogeny and mannitol content of Dictyotopsis propagulifera (Dictyotales, Heterokontophyta) isolated from mangrove mud in Perak, Malaysia. Phycol. Res. 2025, 73, 210–221. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, J.; Jia, S.; Wang, G.; Liu, T.; Wang, X. The complete mitogenome of three continuous generations of ‘Rongfu’: A crucial Saccharina cultivation variety. Mitochondrial DNA Part B 2019, 4, 744–745. [Google Scholar] [CrossRef]
- Xia, Y.; Guo, Z.; Zhang, X.; Zhang, J. The complete mitogenome of ‘Dongfang No. 3’, an important Saccharina cultivar in China. Mitochondrial DNA Part B 2021, 6, 1098–1100. [Google Scholar] [CrossRef] [PubMed]
- Moeckel, C.; Zaravinos, A.; Georgakopoulos-Soares, I. Strand asymmetries across genomic processes. Comput. Struct. Biotechnol. J. 2023, 21, 2036–2047. [Google Scholar] [CrossRef] [PubMed]
- Patil, M.P.; Kim, J.O.; Kim, Y.R.; Nirmal, N.; Kim, G.D.; Kim, K. Characterization of the complete mitochondrial genome of the red alga Ahnfeltiopsis flabelliformis (Rhodophyta, Gigartinales, Phyllophoraceae) and its phylogenetic analysis. Biology 2025, 14, 638. [Google Scholar] [CrossRef]
- Butenko, A.; Lukeš, J.; Speijer, D.; Wideman, J.G. Mitochondrial genomes revisited: Why do different lineages retain different genes? BMC Biol. 2024, 22, 15. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M.; Li, W.H. The codon adaptation index—A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, L. Ribosome structural changes dynamically affect ribosome function. Int. J. Mol. Sci. 2024, 25, 11186. [Google Scholar] [CrossRef]
- Parks, M.M.; Kurylo, C.M.; Batchelder, J.E.; Vincent, C.T.; Blanchard, S.C. Implications of sequence variation on the evolution of rRNA. Chromosome Res. 2019, 27, 89–93. [Google Scholar] [CrossRef]
- Silberfeld, T.; Rousseau, F.; de Reviers, B. An updated classification of brown algae (Ochrophyta, Phaeophyceae). Cryptogam. Algol. 2014, 35, 117–156. [Google Scholar] [CrossRef]
| Gene | Position | Size (bp) | Coding Strand | IN | Codon | Anti-Codon | Amino Acids | ||
|---|---|---|---|---|---|---|---|---|---|
| Start | End | Start | Stop | ||||||
| rnl | 1 | 2668 | 2668 | H | 32 | - | - | - | - |
| rps10 | 2673 | 2987 | 315 | H | 4 | ATG | TAG | - | 104 |
| trnE | 3037 | 3108 | 72 | H | 49 | - | - | TTC | - |
| trnV | 3115 | 3188 | 74 | H | 6 | - | - | TAC | - |
| trnM | 3196 | 3268 | 73 | H | 7 | - | - | CAT | - |
| trnL | 3271 | 3354 | 84 | H | 2 | - | - | TAA | - |
| trnH | 3360 | 3433 | 74 | H | 5 | - | - | GTG | - |
| trnC | 3436 | 3508 | 73 | H | 2 | - | - | GCA | - |
| trnN | 3512 | 3584 | 73 | H | 3 | - | - | GTT | - |
| trnK | 3608 | 3680 | 73 | H | 23 | - | - | TTT | - |
| trnF | 3694 | 3766 | 73 | H | 13 | - | - | GAA | - |
| rps8 | 3783 | 4154 | 372 | H | 16 | ATG | TGA | - | 123 |
| rpl6 | 4151 | 4642 | 492 | H | −4 | ATG | TAA | - | 163 |
| rps2 | 4642 | 5199 | 558 | H | −1 | ATG | TAA | - | 185 |
| rps4 | 5189 | 5902 | 714 | H | −11 | ATG | TAG | - | 237 |
| nad1 | 5904 | 6872 | 969 | H | 1 | ATG | TAA | - | 322 |
| tatC | 7586 | 6861 | 726 | L | −12 | ATG | TAA | - | 241 |
| trnW | 7600 | 7672 | 73 | H | 13 | - | - | CCA | - |
| trnM | 7784 | 7856 | 73 | H | 111 | - | - | CAT | - |
| trnQ | 7858 | 7929 | 72 | H | 1 | - | - | TTG | - |
| trnL | 7930 | 8009 | 80 | H | 0 | - | - | TAG | - |
| rps12 | 8013 | 8393 | 381 | H | 3 | ATG | TAG | - | 126 |
| rps7 | 8404 | 9003 | 600 | H | 10 | ATG | TAA | - | 199 |
| nad4L | 9042 | 9344 | 303 | H | 38 | ATG | TAA | - | 100 |
| trnL | 9352 | 9433 | 82 | H | 7 | - | - | CAA | - |
| rpl14 | 9439 | 9825 | 387 | H | 5 | ATG | TGA | - | 128 |
| rpl5 | 9828 | 10,343 | 516 | H | 2 | ATG | TAG | - | 171 |
| trnG | 10,354 | 10,426 | 73 | H | 10 | - | - | GCC | - |
| atp9 | 10,474 | 10,701 | 228 | H | 47 | ATG | TAA | - | 75 |
| orf109 | 11,086 | 10,757 | 330 | L | 55 | ATG | TAA | - | 109 |
| rpl16 | 11,490 | 11,083 | 408 | L | −4 | ATG | TGA | - | 135 |
| rps3 | 12,314 | 11,493 | 822 | L | 2 | ATG | TAA | - | 273 |
| rps19 | 12,565 | 12,323 | 243 | L | 8 | ATG | TAA | - | 80 |
| rpl2 | 13,291 | 12,572 | 720 | L | 6 | ATG | TAA | - | 239 |
| rps13 | 13,316 | 13,660 | 345 | H | 24 | ATG | TAA | - | 114 |
| rps11 | 13,667 | 14,071 | 405 | H | 6 | ATG | TAA | - | 134 |
| trnY | 14,083 | 14,164 | 82 | H | 11 | - | - | GTA | - |
| cox3 | 14,172 | 14,984 | 813 | H | 7 | ATG | TAA | - | 270 |
| atp6 | 15,008 | 15,760 | 753 | H | 23 | ATG | TAA | - | 250 |
| trnR | 15,811 | 15,883 | 73 | H | 50 | - | - | TCT | - |
| nad2 | 15,888 | 17,375 | 1488 | H | 4 | ATG | TAA | - | 495 |
| cox1 | 17,410 | 18,939 | 1530 | H | 34 | ATG | TAA | - | 509 |
| trnI | 18,980 | 19,051 | 72 | H | 40 | - | - | GAT | - |
| atp8 | 19,078 | 19,338 | 261 | H | 26 | ATG | TGA | - | 86 |
| nad9 | 19,341 | 19,928 | 588 | H | 2 | ATG | TAA | - | 195 |
| cob | 19,943 | 21,097 | 1155 | H | 14 | ATG | TGA | - | 384 |
| cox2 | 21,338 | 22,183 | 846 | H | 240 | ATG | TAA | - | 281 |
| nad4 | 22,215 | 23,657 | 1443 | H | 31 | ATG | TAA | - | 480 |
| trnN | 23,664 | 23,735 | 72 | H | 6 | - | - | ATT | - |
| nad5 | 23,739 | 25,730 | 1992 | H | 3 | ATG | TAA | - | 663 |
| nad6 | 25,876 | 26,697 | 822 | H | 145 | ATG | TAA | - | 273 |
| nad11 | 26,782 | 27,384 | 603 | H | 84 | ATG | TGA | - | 200 |
| trnD | 27,383 | 27,455 | 73 | H | −2 | - | - | GTC | - |
| nad3 | 27,476 | 27,835 | 360 | H | 20 | ATG | TAA | - | 119 |
| rps14 | 27,829 | 28,125 | 297 | H | −7 | GTG | TAA | - | 98 |
| trnS | 28,132 | 28,217 | 86 | H | 6 | - | - | GCT | - |
| trnS | 28,221 | 28,305 | 85 | H | 3 | - | - | TGA | - |
| trnM | 28,397 | 28,472 | 76 | H | 91 | - | - | CAT | - |
| rns | 28,537 | 30,061 | 1525 | H | 64 | - | - | - | - |
| rrn5 | 30,051 | 30,163 | 113 | H | −11 | - | - | - | - |
| trnA | 30,158 | 30,230 | 73 | H | −6 | - | - | TGC | - |
| nad7 | 30,254 | 31,450 | 1197 | H | 23 | ATG | TAA | - | 399 |
| trnP | 31,470 | 31,541 | 72 | H | 19 | - | - | TGG | - |
| Species | Size (bp) | Nucleotide Composition (%) | AT-Skew | GC-Skew | Number of Genes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | T | G | C | A+T | G+C | PCG | tRNA | rRNA | ORF * | ||||
| Dictyota coriacea | 31,573 | 25.4 | 35.8 | 22.4 | 16.5 | 61.2 | 38.8 | −0.170 | 0.153 | 34 | 25 | 3 | 1 |
| Dictyota dichotoma | 31,617 | 25.9 | 37.5 | 21.6 | 14.9 | 63.5 | 36.5 | −0.183 | 0.182 | 35 | 25 | 3 | 3 |
| Dictyopteris divaricata | 32,021 | 25.5 | 36.2 | 21.8 | 16.5 | 61.7 | 38.3 | −0.174 | 0.139 | 35 | 24 | 3 | 3 |
| Dictyotopsis propagulifera | 30,995 | 32.4 | 40.7 | 15.3 | 11.5 | 73.2 | 26.8 | −0.113 | 0.140 | 35 | 23 | 3 | 0 |
| PCG | Length (bp) | Nucleotide Composition (%) | AT-Skewness | GC-Skewness | |||||
|---|---|---|---|---|---|---|---|---|---|
| A | T | G | C | A+T | G+C | ||||
| rps10 | 315 | 29.8 | 35.6 | 19.7 | 14.9 | 65.4 | 34.6 | −0.087 | 0.138 |
| rps8 | 372 | 23.7 | 34.9 | 26.3 | 15.1 | 58.6 | 41.4 | −0.193 | 0.273 |
| rpl6 | 492 | 28.9 | 32.7 | 23.4 | 15.0 | 61.6 | 38.4 | −0.063 | 0.217 |
| rps2 | 558 | 29.2 | 35.1 | 20.8 | 14.9 | 64.3 | 35.7 | −0.092 | 0.166 |
| rps4 | 714 | 30.1 | 33.1 | 19.7 | 17.1 | 63.2 | 36.8 | −0.047 | 0.072 |
| nad1 | 969 | 21.3 | 39.5 | 23.7 | 15.5 | 60.8 | 39.2 | −0.301 | 0.211 |
| tatC | 726 | 33.3 | 34.7 | 12.7 | 19.3 | 68.0 | 32.0 | −0.020 | −0.207 |
| rps12 | 381 | 31.2 | 27.6 | 23.6 | 17.6 | 58.8 | 41.2 | 0.063 | 0.146 |
| rps7 | 600 | 30.8 | 33.3 | 19.0 | 16.8 | 64.2 | 35.8 | −0.039 | 0.060 |
| nad4L | 303 | 24.1 | 43.6 | 19.5 | 12.9 | 67.7 | 32.3 | −0.288 | 0.204 |
| rpl14 | 387 | 28.4 | 29.2 | 25.3 | 17.1 | 57.6 | 42.4 | −0.013 | 0.195 |
| rpl5 | 516 | 27.7 | 39.0 | 20.0 | 13.4 | 66.7 | 33.3 | −0.169 | 0.198 |
| atp9 | 228 | 19.7 | 37.7 | 27.6 | 14.9 | 57.5 | 42.5 | −0.313 | 0.299 |
| orf109 | 330 | 40.9 | 25.5 | 16.1 | 17.6 | 66.4 | 33.6 | 0.233 | −0.045 |
| rpl16 | 408 | 37.7 | 21.6 | 21.1 | 19.6 | 59.3 | 40.7 | 0.273 | 0.036 |
| rps3 | 822 | 43.3 | 23.5 | 16.1 | 17.2 | 66.8 | 33.2 | 0.297 | −0.033 |
| rps19 | 243 | 37.9 | 24.7 | 15.6 | 21.8 | 62.6 | 37.4 | 0.211 | −0.165 |
| rpl2 | 720 | 36.1 | 22.4 | 22.4 | 19.2 | 58.5 | 41.5 | 0.235 | 0.077 |
| rps13 | 345 | 28.4 | 33.9 | 21.4 | 16.2 | 62.3 | 37.7 | −0.088 | 0.138 |
| rps11 | 405 | 32.3 | 29.4 | 22.2 | 16.0 | 61.7 | 38.3 | 0.048 | 0.161 |
| cox3 | 813 | 19.7 | 38.1 | 26.0 | 16.2 | 57.8 | 42.2 | −0.319 | 0.230 |
| atp6 | 753 | 20.3 | 40.9 | 23.0 | 15.8 | 61.2 | 38.8 | −0.336 | 0.185 |
| nad2 | 1488 | 21.0 | 43.9 | 19.6 | 15.5 | 64.9 | 35.1 | −0.353 | 0.117 |
| cox1 | 1530 | 20.6 | 37.5 | 22.9 | 19.1 | 58.0 | 42.0 | −0.291 | 0.090 |
| atp8 | 261 | 26.4 | 42.9 | 20.3 | 10.3 | 69.3 | 30.7 | −0.238 | 0.325 |
| nad9 | 588 | 25.3 | 33.7 | 24.0 | 17.0 | 59.0 | 41.0 | −0.141 | 0.170 |
| cob | 1155 | 22.0 | 38.8 | 22.6 | 16.6 | 60.8 | 39.2 | −0.276 | 0.152 |
| cox2 | 846 | 24.7 | 35.8 | 23.3 | 16.2 | 60.5 | 39.5 | −0.184 | 0.180 |
| nad4 | 1443 | 21.8 | 40.3 | 21.6 | 16.4 | 62.0 | 38.0 | −0.298 | 0.139 |
| nad5 | 1992 | 21.6 | 39.1 | 22.4 | 16.9 | 60.7 | 39.3 | −0.289 | 0.139 |
| nad6 | 822 | 22.6 | 44.8 | 18.0 | 14.6 | 67.4 | 32.6 | −0.329 | 0.104 |
| nad11 | 603 | 25.9 | 34.0 | 26.0 | 14.1 | 59.9 | 40.1 | −0.136 | 0.298 |
| nad3 | 360 | 22.5 | 42.8 | 20.0 | 14.7 | 65.3 | 34.7 | −0.311 | 0.152 |
| rps14 | 297 | 33.7 | 30.6 | 20.9 | 14.8 | 64.3 | 35.7 | 0.047 | 0.170 |
| nad7 | 1197 | 26.3 | 30.5 | 25.4 | 17.8 | 56.8 | 43.2 | −0.074 | 0.176 |
| Total | 23,982 | 26.1 | 35.8 | 21.7 | 16.5 | 61.8 | 38.2 | - | - |
| PCG | Length (bp) | Amino Acids | Start Codon | Stop Codon | Strand |
|---|---|---|---|---|---|
| rps10 | 315 a,b/330 c/291 d | 104 a,b/109 c/96 d | ATG | TAG a/TAA b,d/TGA c | H |
| rps8 | 372 | 123 | ATG | TGA | H |
| rpl6 | 492 | 163 | ATG | TAA | H |
| rps2 | 558 a/567 b,d/570 c | 185 a/188 b,d/189 c | ATG | TAA | H |
| rps4 | 714 a,b/711 c/705 d | 237 a,b/236 c/234 d | ATG | TAG a,b,c/TAA d | H |
| nad1 | 969 a,b/975 c,d | 322 a,b/324 c,d | ATG | TAA | H |
| tatC | 726 a/720 b,d/768 c | 241 a/239 b,d/255 c | ATG | TAA a,c/TAG b,d | L |
| orf * | 114 b/117 c | 37 b/38 c | TTG b/CTG c | TAA b,c | H b,c |
| rps12 | 381 a,b/384 c/375 d | 126 a,b/127 c/124 d | ATG | TAG a,b/TGA c/TAA d | H |
| rps7 | 600 a/594 b/597 c/531 d | 199 a/197 b/198 c/176 d | ATG | TAA a,b,d/TGA c | H |
| nad4L | 303 | 100 | ATG | TAA a,d/TGA b,c | H |
| rpl14 | 387 a,b,c/381 d | 128 a,b,c/126 d | ATG | TGA a/TAA b,c,d | H |
| rpl5 | 516 a,b/519 c/531 d | 171 a,b/172 c/176 d | ATG | TAG a/TAA b,c,d | H |
| atp9 | 228 | 75 | ATG | TAA | H |
| orf * | 330 a/336 b/366 c | 109 a/111 b/121 c | ATG a,b,c | TAA a,b,c | L a,b,c |
| rpl16 | 408 | 135 | ATG | TGA a,b/TAA c,d | L |
| rps3 | 822 a/834 b/837 c/777 d | 273 a/277 b/278 c/258 d | ATG | TAA | L |
| rps19 | 243 a,b/258 c/255 d | 80 a,b/85 c/84 d | ATG | TAA a,b,c/TGA d | L |
| rpl2 | 720 a,b/732 c/690 d | 239 a,b/243 c/229 d | ATG | TAA | L |
| rps13 | 345 a/348 b,c/351 d | 114 a/115 b,c/116 d | ATG | TAA | H |
| rps11 | 405 a/417 b/402 c/396 d | 134 a/138 b/133 c/131 d | ATG | TAA a,c,d/TAG b | H |
| cox3 | 813 a/807 b,c/801 d | 270 a/268 b,c/266 d | ATG | TAA a,d/TGA b,c | H |
| atp6 | 753 | 250 | ATG | TAA a,b,d/TGA c | H |
| nad2 | 1488 a,b,c/1491 d | 495 a,b,c/496 d | ATG | TAA | H |
| cox1 | 1530 a,b/1539 c/1485 d | 509 a,b/512 c/494 d | ATG | TAA a,b,c/TAG d | H |
| atp8 | 261 | 86 | ATG | TGA a,b/TAA c,d | H |
| nad9 | 588 a/579 b/573 c/582 d | 195 a/192 b/190 c/193 d | ATG | TAA a,c,d/TGA b | H |
| cob | 1155 a,b/1152 c/1158 d | 384 a,b/383 c/385 d | ATG | TGA a,b,c/TAA d | H |
| rpl31 | 201 b/231 c/207 d | 66 b/76 c/68 d | ATG b,c,d | TAG b/TGA c/TAA d | H b,c,d |
| cox2 | 846 a/885 b/873 c/888 d | 281 a/294 b/290 c/295 d | ATG | TAA a,c,d/ TAG b | H |
| nad4 | 1443 a,b/1437 c,d | 480 a,b/478 c,d | ATG | TAA a,b,d/TAG c | H |
| nad5 | 1992 a/1989 b,c/1998 d | 663 a/662 b,c/665 d | ATG | TAA | H |
| orf * | 165 b/156 c | 54 b/51 c | ATG b,c | TGA b/TAG c | H b,c |
| nad6 | 822 a,b/834 c/792 d | 273 a,b/277 c/262 d | ATG | TAA | H |
| nad11 | 603 | 200 | ATG | TGA | H |
| nad3 | 360 a,b,d/363 c | 119 a,b,d/200 c | ATG | TAA a,b,d/TGA c | H |
| rps14 | 297 | 98 | GTG a,b/ATG c,d | TAA | H |
| nad7 | 1197 a,b,c/1200 d | 398 a,b,c/399 d | ATG | TAA | H |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patil, M.P.; Woo, H.-E.; Jeon, Y.J.; Kitamura, S.-I.; Kim, Y.-R.; Kim, J.-O.; Kim, K. The First Complete Mitogenome Characterization of Brown Alga Dictyota coriacea (Phaeophyceae, Heterokontophyta) and Its Phylogenetic Analysis. Life 2025, 15, 1605. https://doi.org/10.3390/life15101605
Patil MP, Woo H-E, Jeon YJ, Kitamura S-I, Kim Y-R, Kim J-O, Kim K. The First Complete Mitogenome Characterization of Brown Alga Dictyota coriacea (Phaeophyceae, Heterokontophyta) and Its Phylogenetic Analysis. Life. 2025; 15(10):1605. https://doi.org/10.3390/life15101605
Chicago/Turabian StylePatil, Maheshkumar Prakash, Hee-Eun Woo, Young Jae Jeon, Shin-Ichi Kitamura, Young-Ryun Kim, Jong-Oh Kim, and Kyunghoi Kim. 2025. "The First Complete Mitogenome Characterization of Brown Alga Dictyota coriacea (Phaeophyceae, Heterokontophyta) and Its Phylogenetic Analysis" Life 15, no. 10: 1605. https://doi.org/10.3390/life15101605
APA StylePatil, M. P., Woo, H.-E., Jeon, Y. J., Kitamura, S.-I., Kim, Y.-R., Kim, J.-O., & Kim, K. (2025). The First Complete Mitogenome Characterization of Brown Alga Dictyota coriacea (Phaeophyceae, Heterokontophyta) and Its Phylogenetic Analysis. Life, 15(10), 1605. https://doi.org/10.3390/life15101605







