Computational–Experimental Identification of Palindromic Motifs Bound by Bacterial XRE Family Transcriptional Regulators
Abstract
1. Introduction
2. Materials and Methods
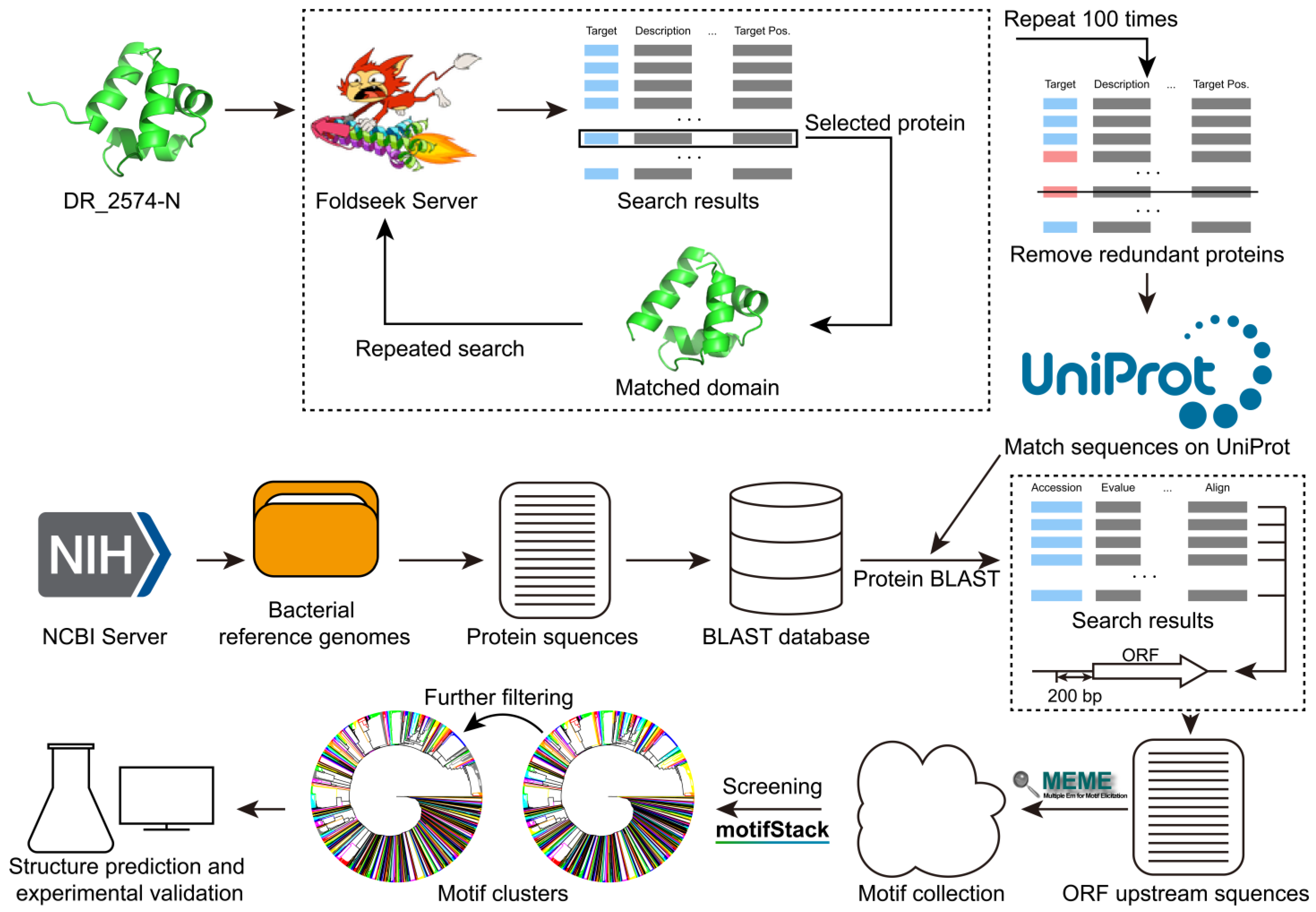
2.1. The Collection of XRE Family Proteins
2.2. The Construction of the BLAST Database
2.3. Motif Discovery and Clustering
2.4. AlphaFold Interaction Structure Prediction
2.5. Protein Induction Expression
2.6. Protein Purification
2.7. EMSA
2.8. Genomic Motif Searching
3. Results
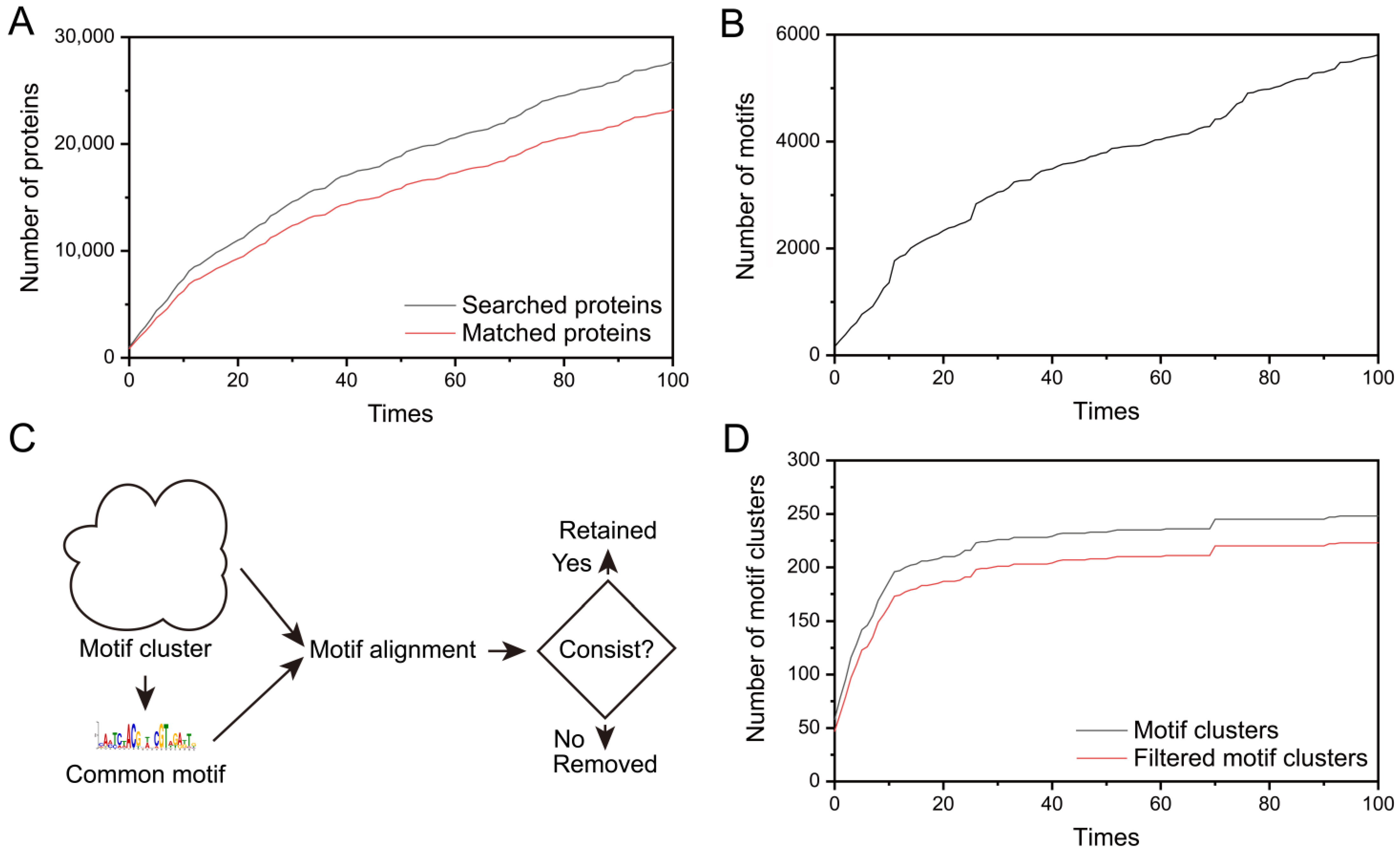
3.1. The Workflow and Result of Motif Discovery
3.2. Motif Analysis and Clustering
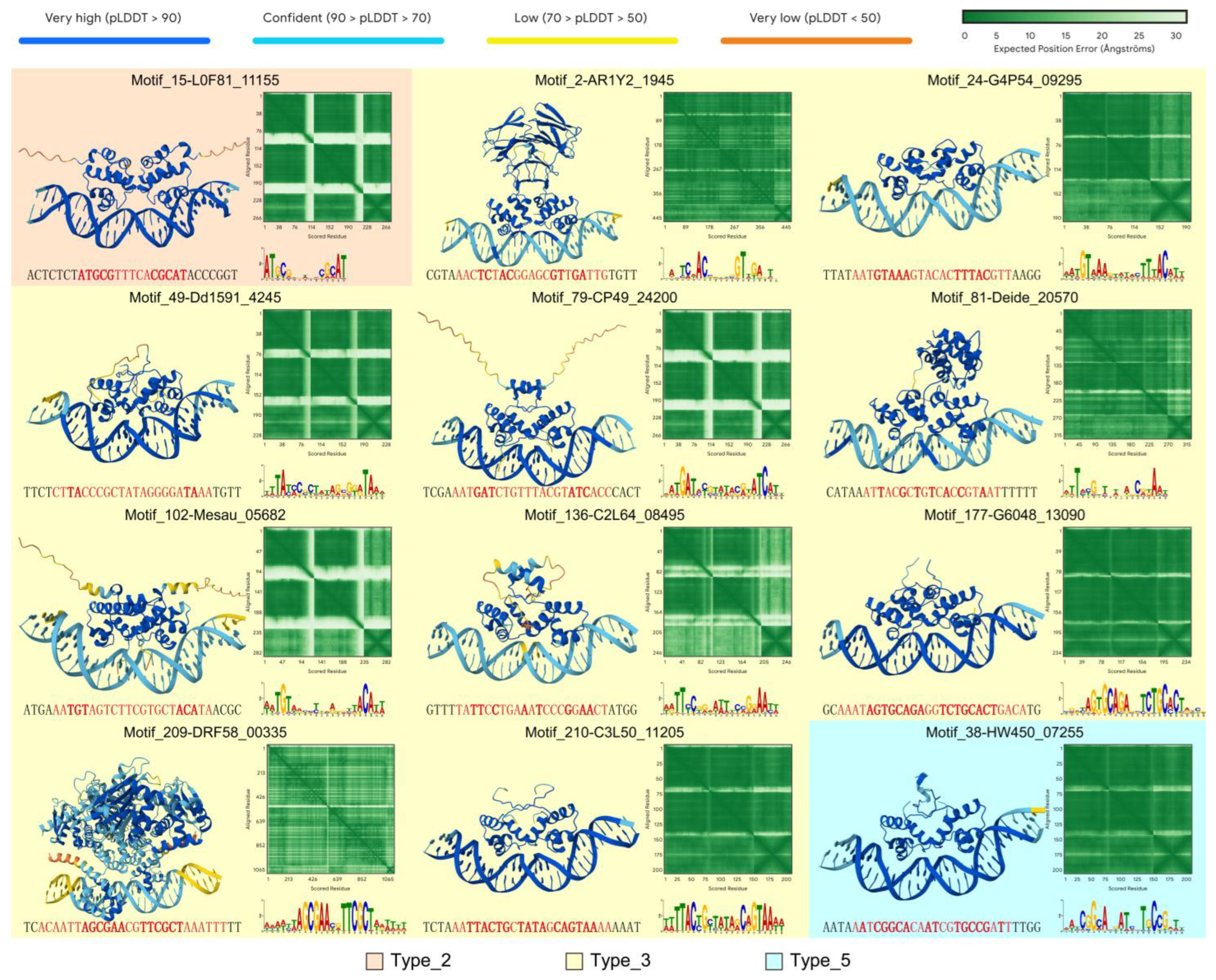
3.3. Structure Prediction of Protein–DNA Motif Complex
3.4. Experimental Validation of Protein–DNA Interactions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maddocks, S.E.; Oyston, P. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 2008, 154, 3609–3623. [Google Scholar] [CrossRef]
- Ramos, J.L.; Martinez-Bueno, M.; Molina-Henares, A.J.; Teran, W.; Watanabe, K.; Zhang, X.; Gallegos, M.T.; Brennan, R.; Tobes, R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005, 69, 326–356. [Google Scholar] [CrossRef]
- Cortes-Avalos, D.; Martinez-Perez, N.; Ortiz-Moncada, M.A.; Juarez-Gonzalez, A.; Banos-Vargas, A.A.; Estrada, L.S.P.; Perez-Rueda, E.; Ibarra, J.A. An update of the unceasingly growing and diverse AraC/XylS family of transcriptional activators. Fems Microbiol. Rev. 2021, 45, fuab020. [Google Scholar] [CrossRef]
- Tulin, G.; Figueroa, N.R.; Checa, S.K.; Soncini, F.C. The multifarious MerR family of transcriptional regulators. Mol. Microbiol. 2024, 121, 230–242. [Google Scholar] [CrossRef]
- Brown, N.L.; Stoyanov, J.V.; Kidd, S.P.; Hobman, J.L. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 2003, 27, 145–163. [Google Scholar] [CrossRef]
- Liu, G.F.; Wang, X.X.; Su, H.Z.; Lu, G.T. Progress on the GntR family transcription regulators in bacteria. Yi Chuan 2021, 43, 66–73. [Google Scholar] [CrossRef]
- Jain, D. Allosteric control of transcription in GntR family of transcription regulators: A structural overview. IUBMB Life 2015, 67, 556–563. [Google Scholar] [CrossRef]
- Murarka, P.; Srivastava, P. An improved method for the isolation and identification of unknown proteins that bind to known DNA sequences by affinity capture and mass spectrometry. PLoS ONE 2018, 13, e0202602. [Google Scholar] [CrossRef]
- Wang, J.; Dong, X.; Shao, Y.; Guo, H.; Pan, L.; Hui, W.; Kwok, L.Y.; Zhang, H.; Zhang, W. Genome adaptive evolution of Lactobacillus casei under long-term antibiotic selection pressures. BMC Genom. 2017, 18, 320. [Google Scholar] [CrossRef]
- Wang, R.; Lin, X.; Zha, G.; Wang, J.; Huang, W.; Wang, J.; Hou, Y.; Mou, H.; Zhang, T.; Zhu, H.; et al. Mechanism of enrofloxacin-induced multidrug resistance in the pathogenic Vibrio harveyi from diseased abalones. Sci. Total Environ. 2022, 830, 154738. [Google Scholar] [CrossRef]
- Eckstein, S.; Brehm, J.; Seidel, M.; Lechtenfeld, M.; Heermann, R. Two novel XRE-like transcriptional regulators control phenotypic heterogeneity in Photorhabdus luminescens cell populations. BMC Microbiol. 2021, 21, 63. [Google Scholar] [CrossRef]
- Lu, H.; Wang, L.; Li, S.; Pan, C.; Cheng, K.; Luo, Y.; Xu, H.; Tian, B.; Zhao, Y.; Hua, Y. Structure and DNA damage-dependent derepression mechanism for the XRE family member DG-DdrO. Nucleic Acids Res. 2019, 47, 9925–9933. [Google Scholar] [CrossRef]
- Si, M.; Chen, C.; Zhong, J.; Li, X.; Liu, Y.; Su, T.; Yang, G. MsrR is a thiol-based oxidation-sensing regulator of the XRE family that modulates C. glutamicum oxidative stress resistance. Microb. Cell Fact. 2020, 19, 189. [Google Scholar] [CrossRef]
- Qiu, H.; Li, Y.; Yuan, M.; Chen, H.; Dandekar, A.A.; Dai, W. Uncovering a hidden functional role of the XRE-cupin protein PsdR as a novel quorum-sensing regulator in Pseudomonas aeruginosa. PLOS Pathog. 2024, 20, e1012078. [Google Scholar] [CrossRef]
- Trouillon, J.; Ragno, M.; Simon, V.; Attree, I.; Elsen, S. Transcription Inhibitors with XRE DNA-Binding and Cupin Signal-Sensing Domains Drive Metabolic Diversification in Pseudomonas. Msystems 2021, 6, 00753-20. [Google Scholar] [CrossRef]
- Zhu, Y.; Lu, T.; Zhang, J.; Zhang, P.; Tao, M.; Pang, X. A novel XRE family regulator that controls antibiotic production and development in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 2020, 104, 10075–10089. [Google Scholar] [CrossRef]
- Tian, J.; Ye, L.; Yang, Y.; Zhang, Y.; Hu, C.; Liao, G. Transposon-based screen identifies a XRE family regulator crucial for candicidin biosynthesis in Streptomyces albus J1074. Sci. China Life Sci. 2020, 63, 1421–1424. [Google Scholar] [CrossRef]
- Jung, J.; Glatter, T.; Herfurth, M.; Sogaard-Andersen, L. DdiA, an XRE family transcriptional regulator, is a co-regulator of the DNA damage response in Myxococcus xanthus. J. Bacteriol. 2025, 207, e0018425. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, S.; Pan, Z.; Yu, Y.; Yao, H.; Liu, Y.; Liu, G. XRE family transcriptional regulator XtrSs modulates Streptococcus suis fitness under hydrogen peroxide stress. Arch. Microbiol. 2022, 204, 244. [Google Scholar] [CrossRef]
- Hu, Y.; Hu, Q.; Wei, R.; Li, R.; Zhao, D.; Ge, M.; Yao, Q.; Yu, X. The XRE Family Transcriptional Regulator SrtR in Streptococcus suis Is Involved in Oxidant Tolerance and Virulence. Front. Cell Infect. Microbiol. 2018, 8, 452. [Google Scholar] [CrossRef]
- Mclaughlin, M.; Fiebig, A.; Crosson, S. XRE transcription factors conserved in Caulobacter and phiCbK modulate adhesin development and phage production. PLoS Genet. 2023, 19, e1011048. [Google Scholar] [CrossRef]
- Bose, B.; Auchtung, J.M.; Lee, C.A.; Grossman, A.D. A conserved anti-repressor controls horizontal gene transfer by proteolysis. Mol. Microbiol. 2008, 70, 570–582. [Google Scholar] [CrossRef]
- Auchtung, J.M.; Lee, C.A.; Garrison, K.L.; Grossman, A.D. Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis. Mol. Microbiol. 2007, 64, 1515–1528. [Google Scholar] [CrossRef]
- Masiewicz, P.; Brzostek, A.; Wolanski, M.; Dziadek, J.; Zakrzewska-Czerwinska, J. A novel role of the PrpR as a transcription factor involved in the regulation of methylcitrate pathway in Mycobacterium tuberculosis. PLoS ONE 2012, 7, e43651. [Google Scholar] [CrossRef]
- Caliandro, R.; de Diego, I.; Gomis-Ruth, F.X. Crystal structure report of the ImmR transcriptional regulator DNA-binding domain of the Bacillus subtilis ICEBs1 transposon. Sci. Rep. 2022, 12, 5258. [Google Scholar] [CrossRef]
- Xu, Q.; van Wezel, G.P.; Chiu, H.J.; Jaroszewski, L.; Klock, H.E.; Knuth, M.W.; Miller, M.D.; Lesley, S.A.; Godzik, A.; Elsliger, M.A.; et al. Structure of an MmyB-like regulator from C. aurantiacus, member of a new transcription factor family linked to antibiotic metabolism in actinomycetes. PLoS ONE 2012, 7, e41359. [Google Scholar] [CrossRef]
- Wang, L.; Lu, H.; Wang, Y.; Yang, S.; Xu, H.; Cheng, K.; Zhao, Y.; Tian, B.; Hua, Y. An Improved Method for Identifying Specific DNA-Protein-Binding Sites In Vitro. Mol. Biotechnol. 2017, 59, 59–65. [Google Scholar] [CrossRef]
- Makarova, K.S.; Omelchenko, M.V.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A.; Zhai, M.; Lapidus, A.; Copeland, A.; Kim, E.; Land, M.; et al. Deinococcus geothermalis: The pool of extreme radiation resistance genes shrinks. PLoS ONE 2007, 2, e955. [Google Scholar] [CrossRef]
- Novichkov, P.S.; Laikova, O.N.; Novichkova, E.S.; Gelfand, M.S.; Arkin, A.P.; Dubchak, I.; Rodionov, D.A. RegPrecise: A database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 2010, 38, D111–D118. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- van Kempen, M.; Kim, S.S.; Tumescheit, C.; Mirdita, M.; Lee, J.; Gilchrist, C.; Soding, J.; Steinegger, M. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 2024, 42, 243–246. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Ludanyi, M.; Blanchard, L.; Dulermo, R.; Brandelet, G.; Bellanger, L.; Pignol, D.; Lemaire, D.; de Groot, A. Radiation response in Deinococcus deserti: IrrE is a metalloprotease that cleaves repressor protein DdrO. Mol. Microbiol. 2014, 94, 434–449. [Google Scholar] [CrossRef]
- Wang, L.; Zhong, S.; Song, S.; Wang, L.; Hua, Y.; Lu, H. Bacterial PprI-DdrO system evolved from ImmA-ImmR system in the prophage. Protein Sci. 2025, 34, e70239. [Google Scholar] [CrossRef]
- Haag, A.F.; Podkowik, M.; Ibarra-Chavez, R.; Gallego, D.S.F.; Ram, G.; Chen, J.; Marina, A.; Novick, R.P.; Penades, J.R. A regulatory cascade controls Staphylococcus aureus pathogenicity island activation. Nat. Microbiol. 2021, 6, 1300–1308. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Zhong, S.; Wang, L.; Lu, H.; Hua, Y. Computational–Experimental Identification of Palindromic Motifs Bound by Bacterial XRE Family Transcriptional Regulators. Life 2025, 15, 1577. https://doi.org/10.3390/life15101577
Wang L, Zhong S, Wang L, Lu H, Hua Y. Computational–Experimental Identification of Palindromic Motifs Bound by Bacterial XRE Family Transcriptional Regulators. Life. 2025; 15(10):1577. https://doi.org/10.3390/life15101577
Chicago/Turabian StyleWang, Linjia, Shitong Zhong, Liangyan Wang, Huizhi Lu, and Yuejin Hua. 2025. "Computational–Experimental Identification of Palindromic Motifs Bound by Bacterial XRE Family Transcriptional Regulators" Life 15, no. 10: 1577. https://doi.org/10.3390/life15101577
APA StyleWang, L., Zhong, S., Wang, L., Lu, H., & Hua, Y. (2025). Computational–Experimental Identification of Palindromic Motifs Bound by Bacterial XRE Family Transcriptional Regulators. Life, 15(10), 1577. https://doi.org/10.3390/life15101577






