Evolutionary Dynamics of Oncosuppression Under Selection Pressure
Abstract
1. Introduction
2. Materials and Methods
2.1. Relative Evolutionary Rate
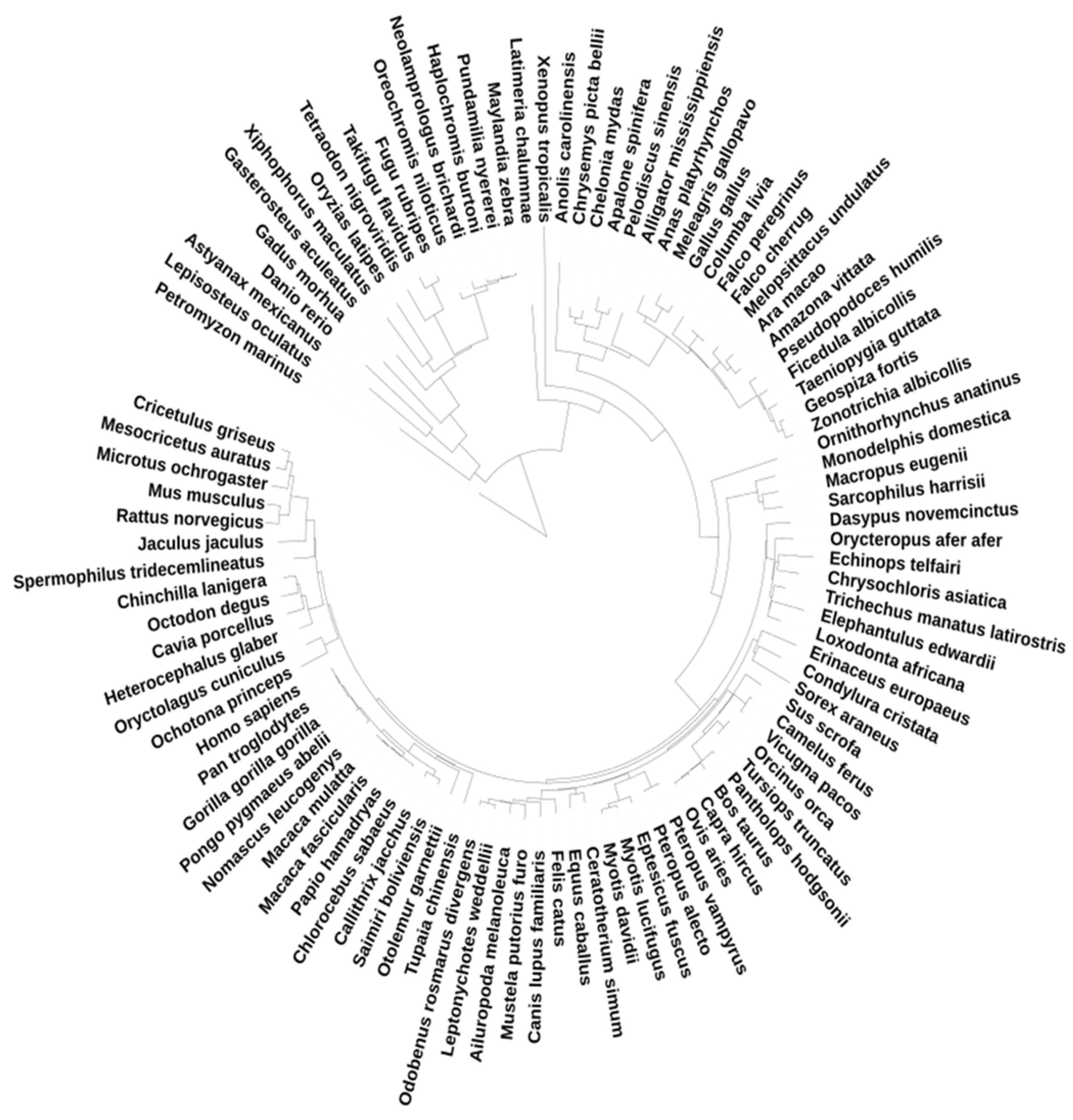
2.2. Genes and Species Datasets
2.3. Trees Construction
2.4. Traits
2.5. RER Calculation
2.6. Trait Analysis
2.7. Enrichment Analysis
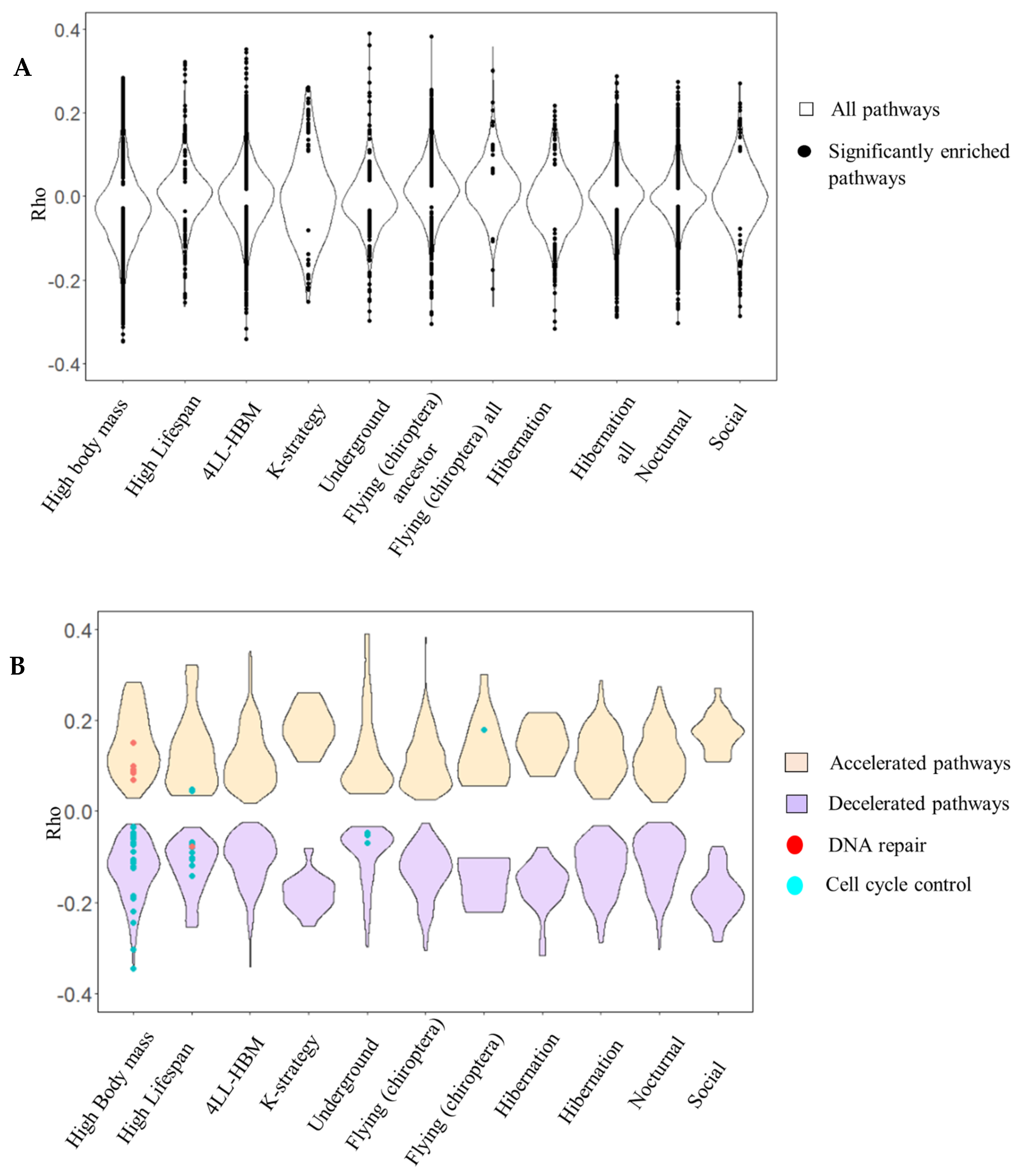
3. Results
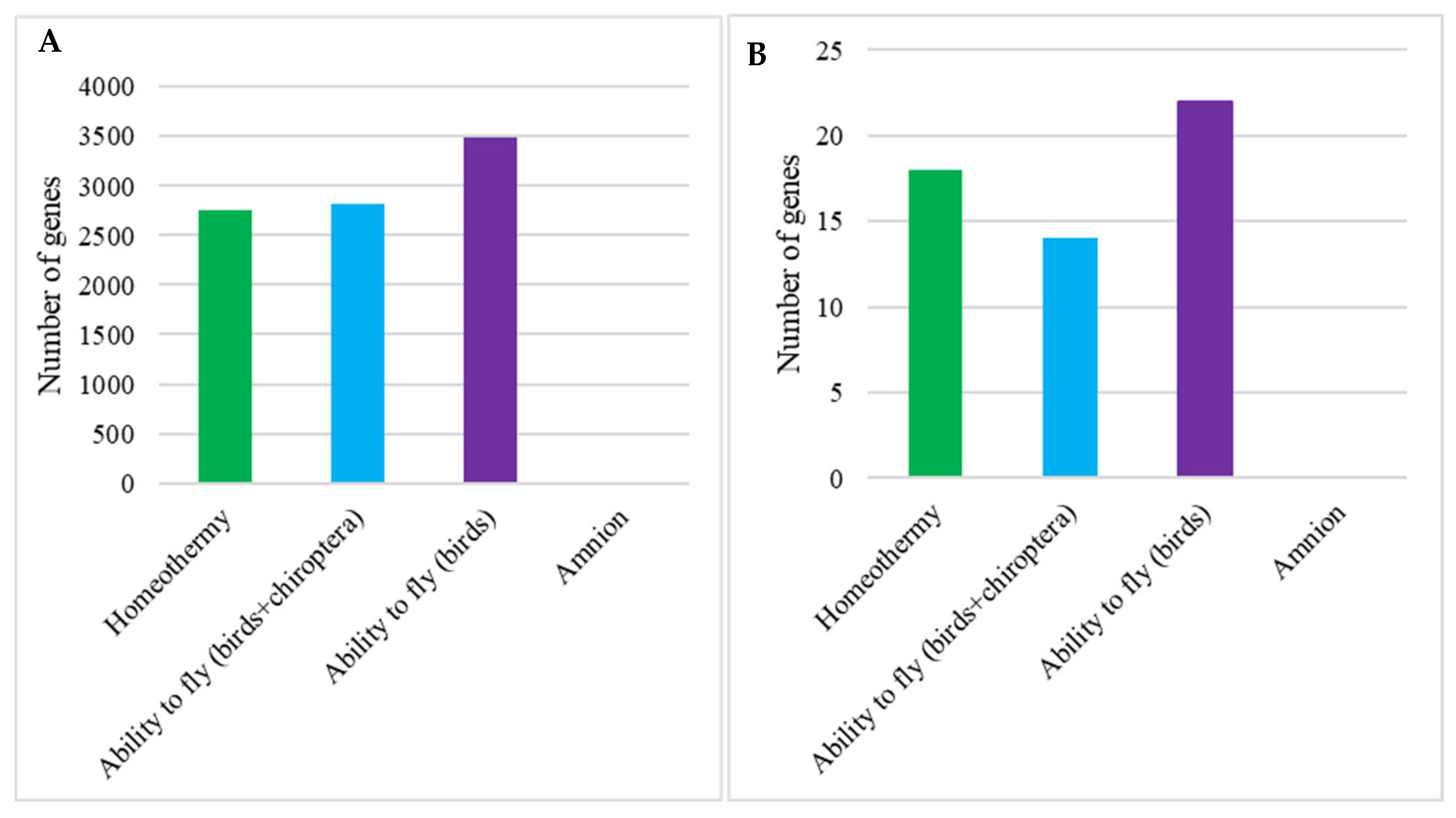
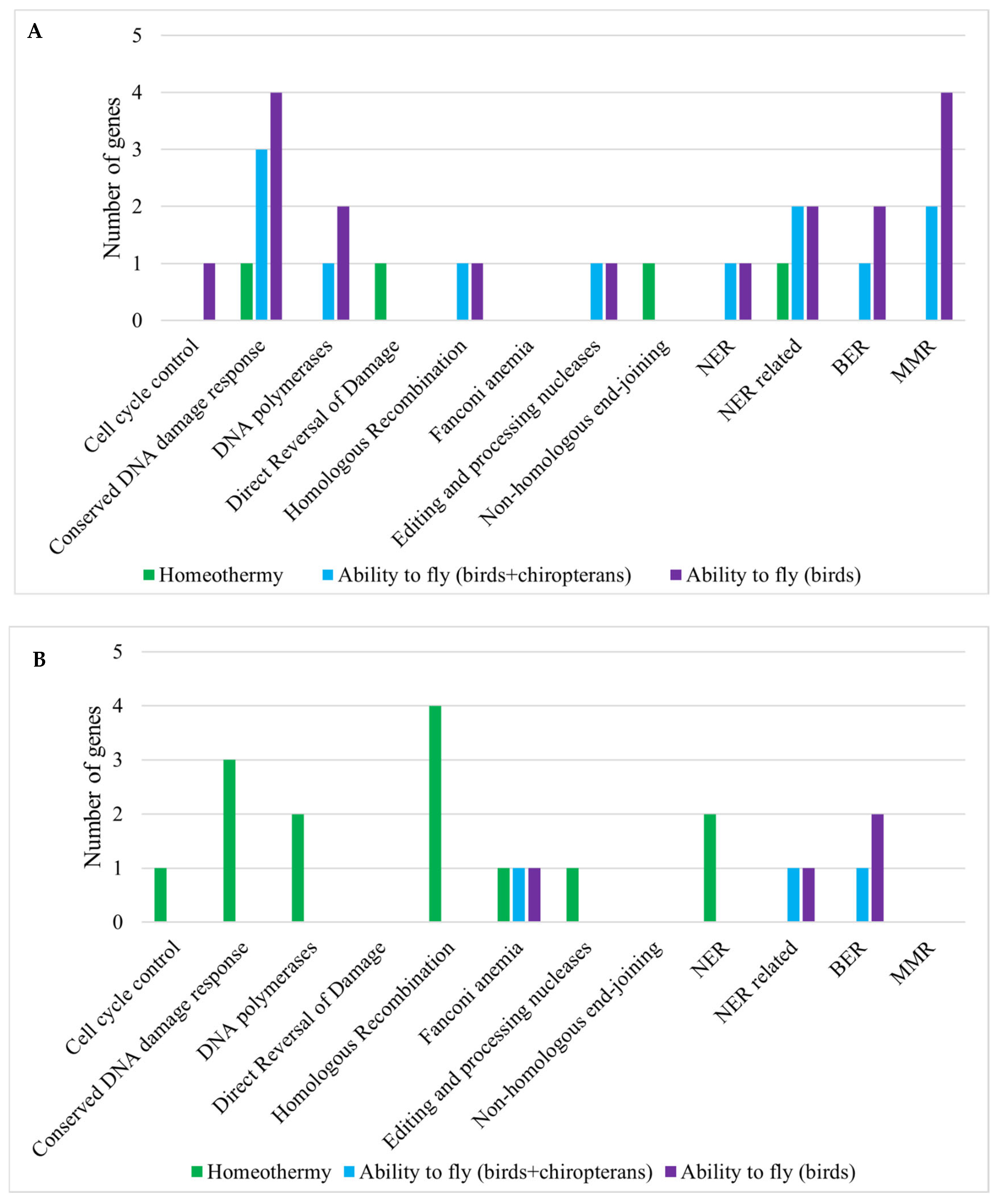
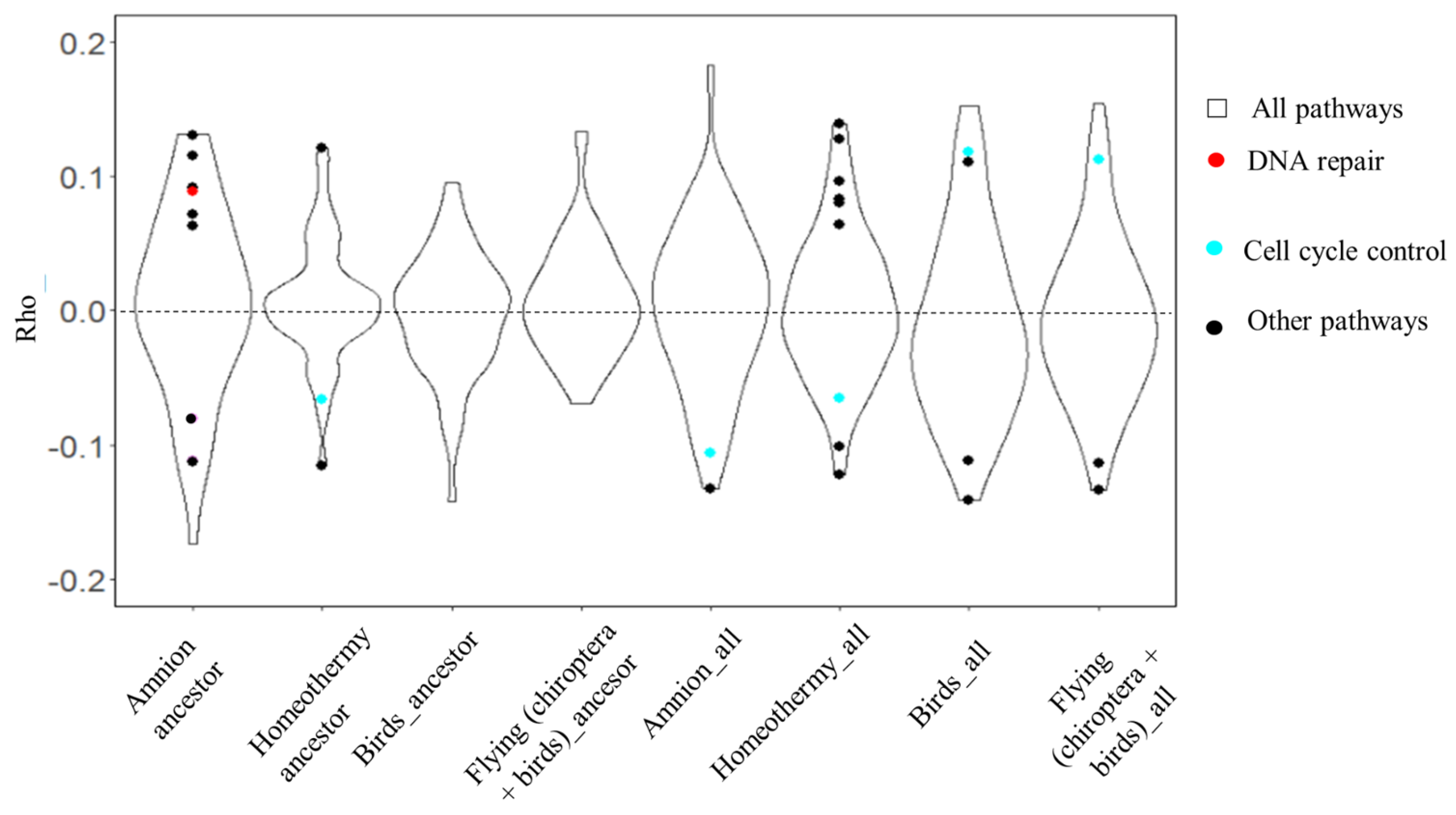
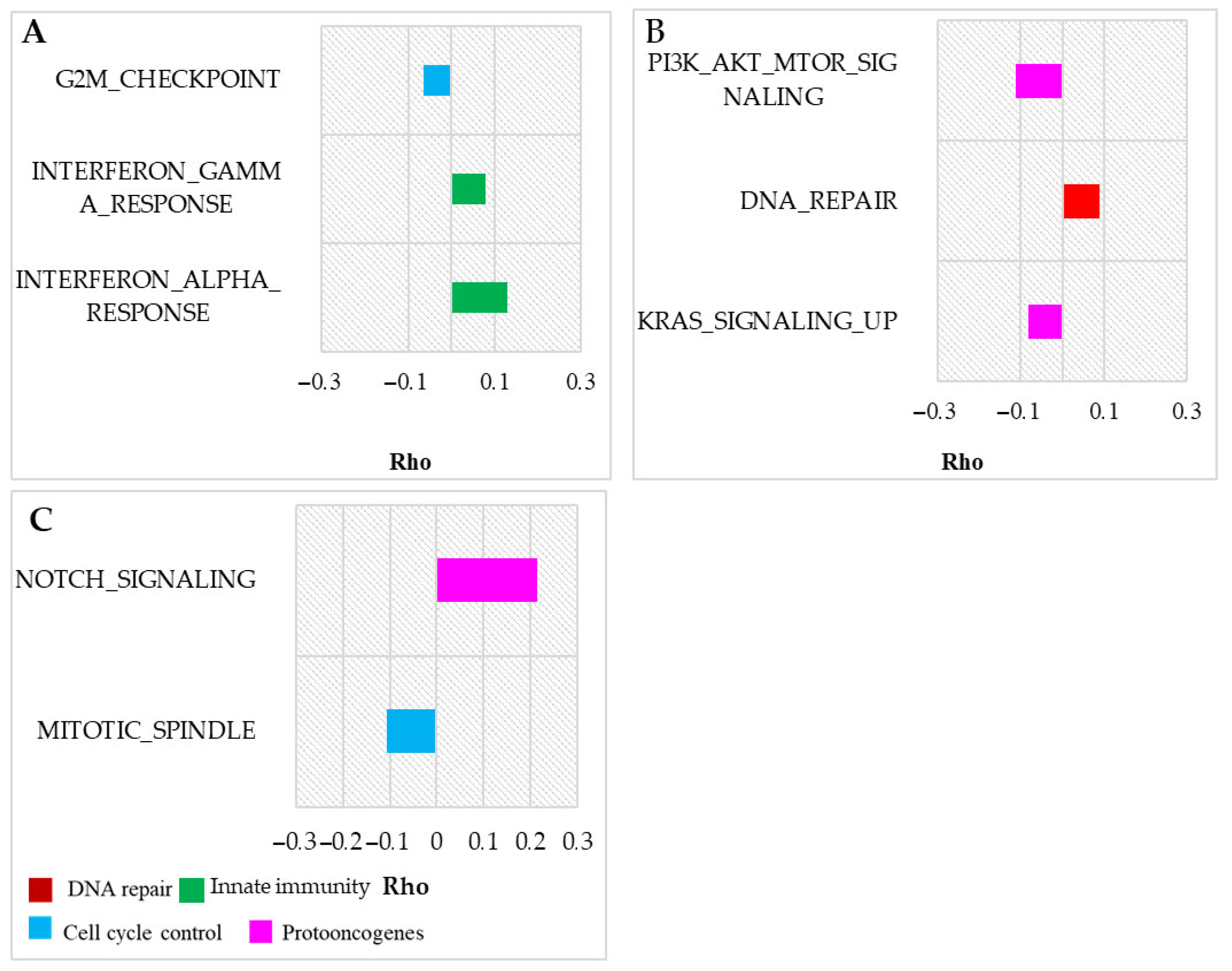
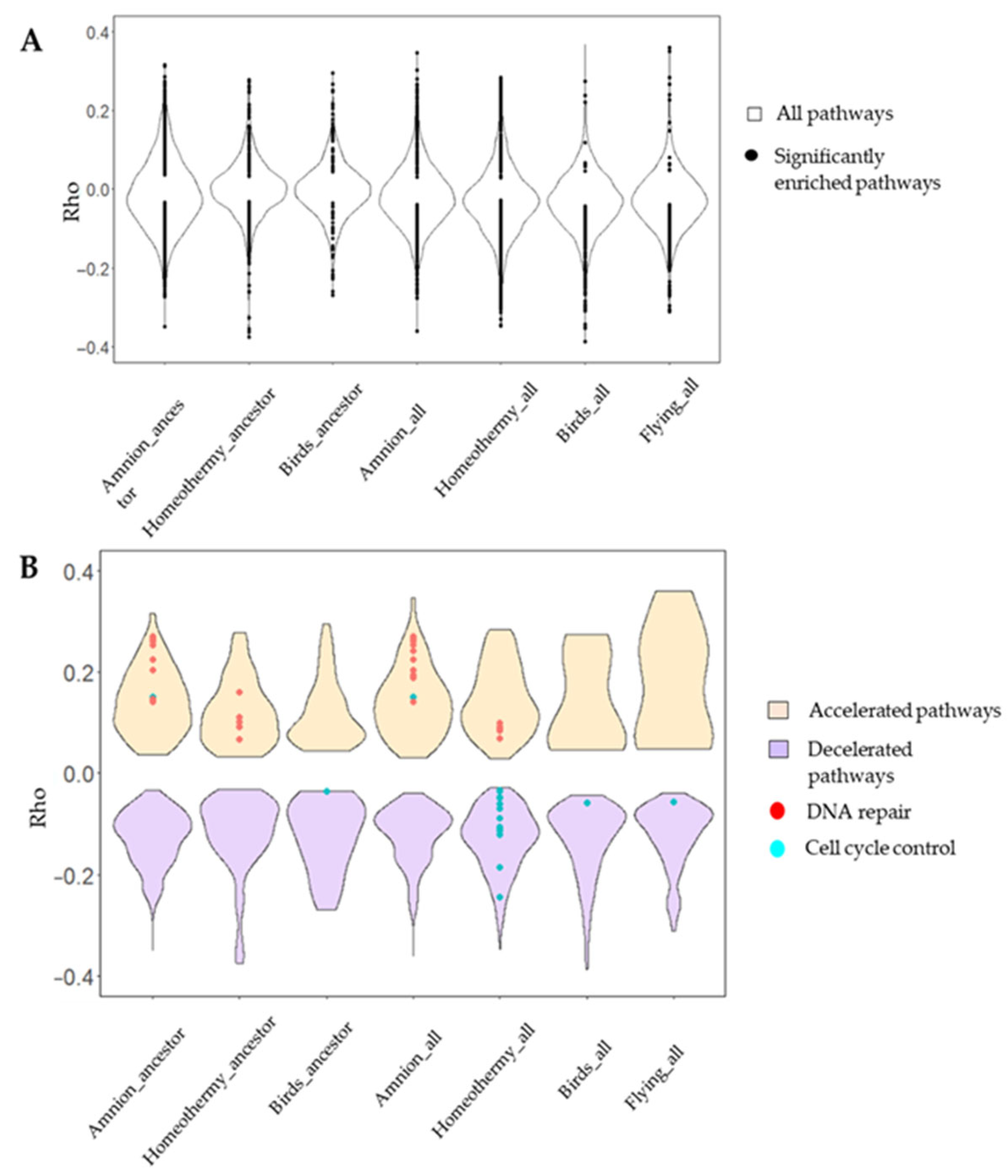
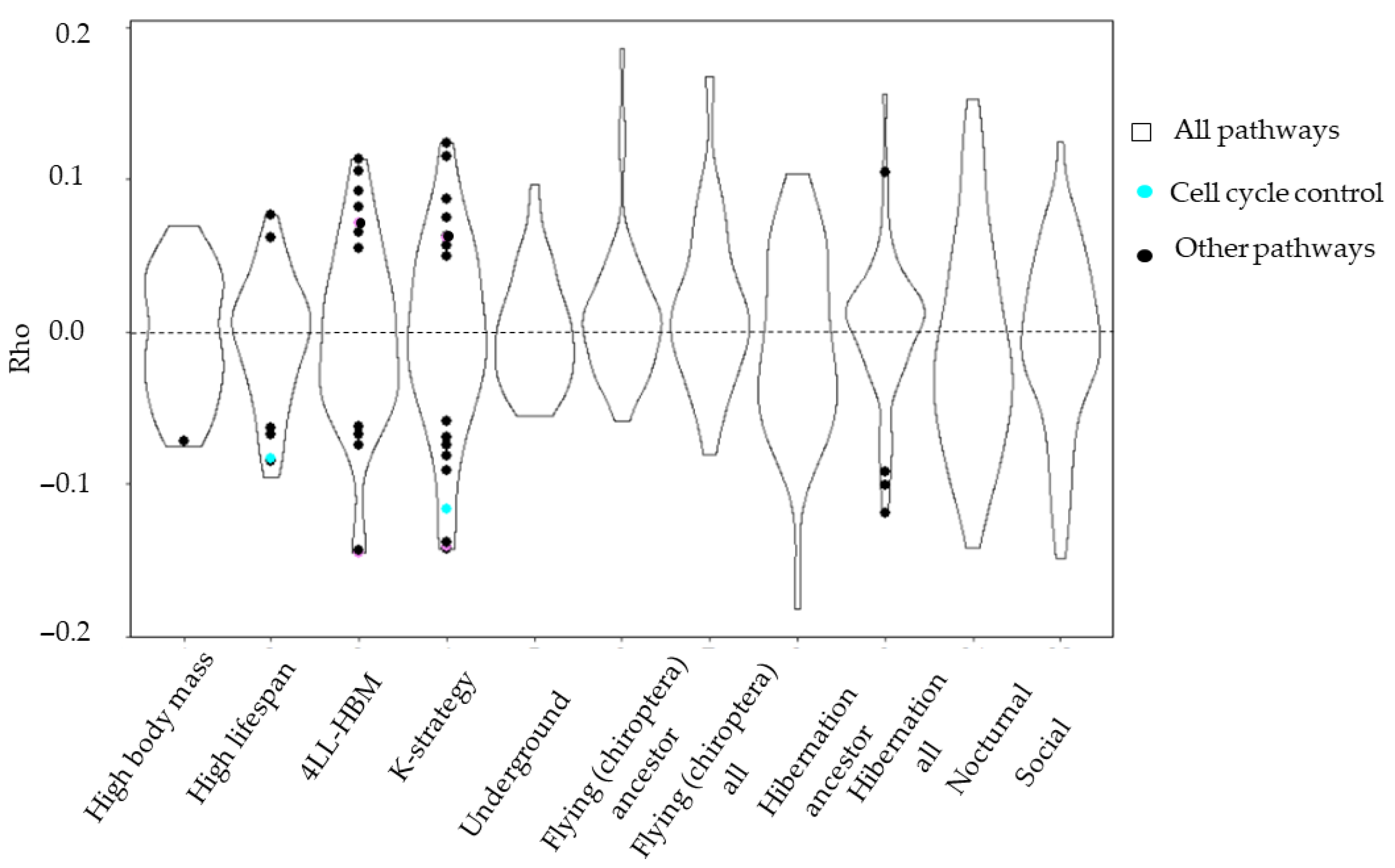
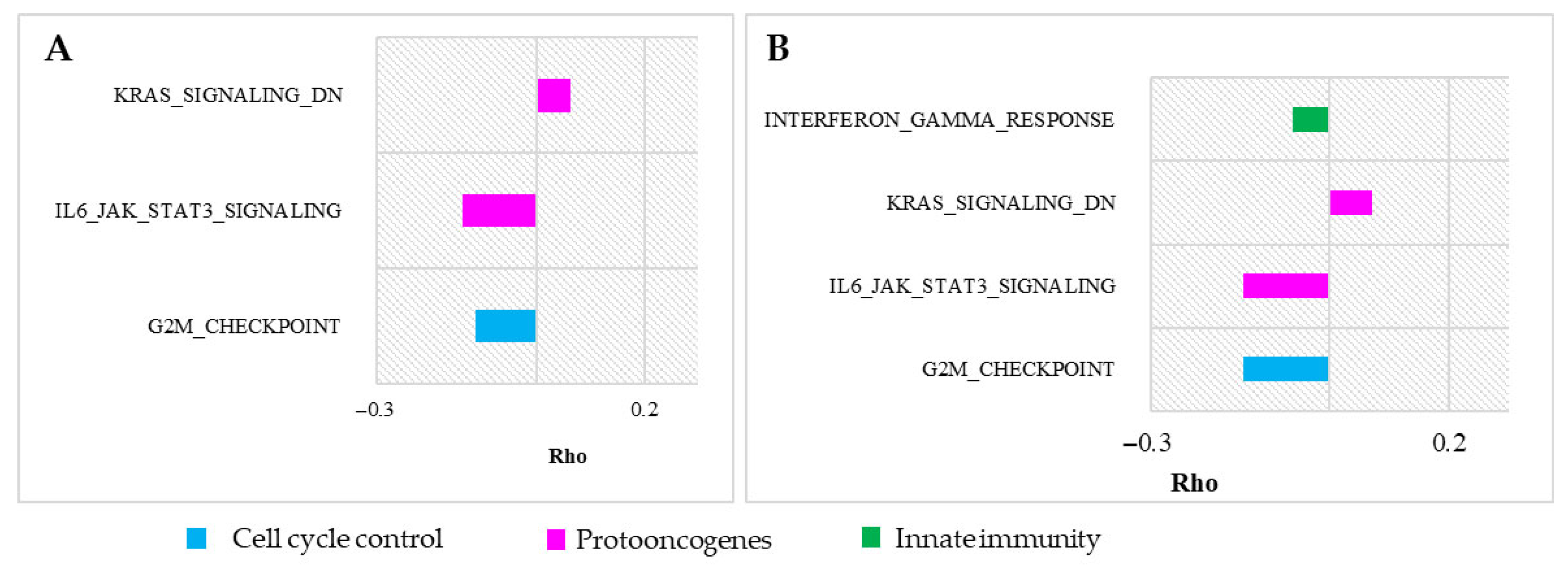
3.1. Vertebrate Trait Analysis
3.2. Mammal Trait Analysis
4. Discussion
4.1. Evolutionary Patterns in Oncosuppressor Genes
4.2. Insights into Mammalian-Specific Adaptations
4.3. Study Novelty and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DNA | Deoxyribonucleic Acid |
| RER | Relative Evolutionary Rate |
| UCSC | University of California Santa Cruz |
References
- World Health Organization. World Health Statistics 2024: Monitoring Health for the SDGs, Sustainable Development Goals; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Black, J.R.M.; McGranahan, N. Genetic and non-genetic clonal diversity in cancer evolution. Nat. Rev. Cancer 2021, 21, 379–392. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Garcês, A.; Pires, I.; Garcês, S. Ancient Diseases in Vertebrates: Tumours through the Ages. Animals 2024, 14, 1474. [Google Scholar] [CrossRef]
- Athena Aktipis, C.; Boddy, A.M.; Jansen, G.; Hibner, U.; Hochberg, M.E.; Maley, C.C.; Wilkinson, G.S. Cancer across the tree of life: Cooperation and cheating in multicellularity. Philos. Trans. R Soc. B Biol. Sci. 2015, 370, 20140219. [Google Scholar] [CrossRef] [PubMed]
- Vendramin, R.; Litchfield, K.; Swanton, C. Cancer evolution: Darwin and beyond. EMBO J. 2021, 40, e108389. [Google Scholar] [CrossRef]
- Carter, A.M.; Hsieh, S.T.; Dodson, P.; Sallan, L. Early amphibians evolved distinct vertebrae for habitat invasions. PLoS ONE 2021, 16, e0251983. [Google Scholar] [CrossRef]
- Cooney, C.R.; Thomas, G.H. Heterogeneous relationships between rates of speciation and body size evolution across vertebrate clades. Nat. Ecol. Evol. 2020, 5, 101–110. [Google Scholar] [CrossRef]
- Klabukov, I.; Smirnova, A.; Yakimova, A.; Kabakov, A.E.; Atiakshin, D.; Petrenko, D.; Shestakova, V.A.; Sulina, Y.; Yatsenko, E.; Stepanenko, V.N.; et al. Oncomatrix: Molecular Composition and Biomechanical Properties of the Extracellular Matrix in Human Tumors. J. Mol. Pathol. 2024, 5, 437–453. [Google Scholar] [CrossRef]
- Clarke, A.; Rothery, P.; Isaac, N.J.B. Scaling of basal metabolic rate with body mass and temperature in mammals. J. Anim. Ecol. 2010, 79, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Milholland, B.; Dong, X.; Zhang, L.; Hao, X.; Suh, Y.; Vijg, J. Differences between germline and somatic mutation rates in humans and mice. Nat. Commun. 2017, 8, 15183. [Google Scholar] [CrossRef]
- MacDonald, N.; Raven, N.; Diep, W.; Evans, S.; Pannipitiya, S.; Bramwell, G.; Vanbeek, C.; Thomas, F.; Russell, T.; Dujon, A.M.; et al. The molecular evolution of cancer associated genes in mammals. Sci. Rep. 2024, 14, 11650. [Google Scholar] [CrossRef]
- Vazquez, J.M.; Pena, M.T.; Muhammad, B.; Kraft, M.; Adams, L.B.; Lynch, V.J. Parallel evolution of reduced cancer risk and tumor suppressor duplications in Xenarthra. eLife 2022, 11, e82558. [Google Scholar] [CrossRef]
- Vincze, O.; Colchero, F.; Lemaître, J.F.; Conde, D.A.; Pavard, S.; Bieuville, M.; Urrutia, A.O.; Ujvari, B.; Boddy, A.M.; Maley, C.C.; et al. Cancer risk across mammals. Nature 2021, 601, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Boutry, J.; Dujon, A.M.; Gerard, A.L.; Tissot, S.; Macdonald, N.; Schultz, A.; Biro, P.A.; Beckmann, C.; Hamede, R.; Hamilton, D.G.; et al. Ecological and Evolutionary Consequences of Anticancer Adaptations. iScience 2020, 23, 101716. [Google Scholar] [CrossRef]
- Stephan, T.; Burgess, S.M.; Cheng, H.; Danko, C.G.; Gill, C.A.; Jarvis, E.D.; Koepfli, K.P.; Koltes, J.E.; Lyons, E.; Ronald, P.; et al. Darwinian genomics and diversity in the tree of life. Proc. Natl. Acad. Sci. USA 2022, 119, e2115644119. [Google Scholar] [CrossRef]
- O’Connell, M.J. Selection and the cell cycle: Positive darwinian selection in a well-Known DNA damage response pathway. J. Mol. Evol. 2010, 71, 444–457. [Google Scholar] [CrossRef]
- Dang, C.V. Links between metabolism and cancer. Genes Dev. 2012, 26, 877–890. [Google Scholar] [CrossRef] [PubMed]
- Tollis, M.; Schiffman, J.D.; Boddy, A.M. Evolution of cancer suppression as revealed by mammalian comparative genomics. Curr. Opin. Genet. Dev. 2017, 42, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Rozhok, A.I.; DeGregori, J. The Evolution of Lifespan and Age-Dependent Cancer Risk. Trends Cancer 2016, 2, 552–560. [Google Scholar] [CrossRef]
- Sarver, A.L.; Makielski, K.M.; DePauw, T.A.; Schulte, A.J.; Modiano, J.F.; Jaime Modiano, C.F. Increased risk of cancer in dogs and humans: A consequence of recent extension of lifespan beyond evolutionarily determined limitations? Aging Cancer 2022, 3, 3–19. [Google Scholar] [CrossRef]
- Gluckman, P.; Beedle, A.; Buklijas, T.; Low, F.H.M. Principles of Evolutionary Medicine, 2nd ed.; Oxford University Press: Oxford, UK, 2016. [Google Scholar]
- Greaves, M.; Maley, C.C. Clonal evolution in cancer. Nature 2012, 481, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, A.; Partha, R.; Clark, N.L.; Chikina, M. Pan-mammalian analysis of molecular constraints underlying extended lifespan. eLife 2020, 9, e51089. [Google Scholar] [CrossRef]
- Partha, R.; Kowalczyk, A.; Clark, N.L.; Chikina, M. Robust Method for Detecting Convergent Shifts in Evolutionary Rates. Mol. Biol. Evol. 2019, 36, 1817–1830. [Google Scholar] [CrossRef]
- Chikina, M.; Robinson, J.D.; Clark, N.L. Hundreds of Genes Experienced Convergent Shifts in Selective Pressure in Marine Mammals. Mol. Biol. Evol. 2016, 33, 2182–2192. [Google Scholar] [CrossRef]
- Benet-Pagès, A.; Rosenbloom, K.R.; Nassar, L.R.; Lee, C.M.; Raney, B.J.; Clawson, H.; Schmelter, D.; Casper, J.; Gonzalez, J.N.; Perez, G.; et al. Variant interpretation: UCSC Genome Browser Recommended Track Sets. Hum. Mutat. 2022, 43, 998–1011. [Google Scholar] [CrossRef]
- Rosenbloom, K.R.; Sloan, C.A.; Malladi, V.S.; Dreszer, T.R.; Learned, K.; Kirkup, V.M.; Wong, M.C.; Maddren, M.; Fang, R.; Heitner, S.G.; et al. ENCODE Data in the UCSC Genome Browser: Year 5 update. Nucleic Acids Res. 2013, 41, 56–63. [Google Scholar] [CrossRef]
- Forbes, S.A.; Beare, D.; Boutselakis, H.; Bamford, S.; Bindal, N.; Tate, J.; Cole, C.G.; Ward, S.; Dawson, E.; Ponting, L.; et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017, 45, D777–D783. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed]
- Li, L.Y.; Guan, Y.D.i.; Chen, X.S.; Yang, J.M.; Cheng, Y. DNA Repair Pathways in Cancer Therapy and Resistance. Front. Pharmacol. 2021, 11, 629266. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Pérez, F.; Granger, B.E. IPython: A System for. IEEE J. Mag. 2007, 9, 21–29. [Google Scholar]
- Talevich, E.; Invergo, B.M.; Cock, P.J.A.; Chapman, B.A. Bio.Phylo: A unified toolkit for processing, analyzing and visualizing phylogenetic trees in Biopython. BMC Bioinform. 2012, 13, 209. [Google Scholar] [CrossRef]
- Cherlin, V.A. Preadaptivity of Noncontractile Thermogenesis in the Evolution of Warm-Bloodedness in Vertebrates. Biol. Bull. Rev. 2023, 13, 647–664. [Google Scholar] [CrossRef]
- Jones, K.E.; Bielby, J.; Cardillo, M.; Fritz, S.A.; O’Dell, J.; Orme, C.D.L.; Safi, K.; Sechrest, W.; Boakes, E.H.; Carbone, C.; et al. PanTHERIA: A species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 2009, 90, 2648. [Google Scholar] [CrossRef]
- Lighten, J.; Incarnato, D.; Ward, B.J.; van Oosterhout, C.; Bradbury, I.; Hanson, M.; Bentzen, P. Adaptive phenotypic response to climate enabled by epigenetics in a K-strategy species, the fish Leucoraja ocellata (Rajidae). R. Soc. Open Sci. 2016, 3, 160299. [Google Scholar] [CrossRef]
- Thomas, F.; Asselin, K.; MacDonald, N.; Brazier, L.; Meliani, J.; Ujvari, B.; Dujon, A.M. Oncogenic processes: A neglected parameter in the evolutionary ecology of animals. Comptes Rendus. Biol. 2024, 347, 137–157. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Goodrich, J.M.; Dolinoy, D.C.; Ruden, D.M. Biological aging acceleration due to environmental exposures: An exciting new direction in toxicogenomics research. Genes 2023, 15, 16. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; He, J.; Hou, T.; Si, J.; Chen, S. Common pathogenetic mechanisms underlying aging and tumor and means of interventions. Aging Dis. 2022, 13, 1063. [Google Scholar] [CrossRef]
- Azpurua, J.; Seluanov, A. Long-lived cancer-resistant rodents as new model species for cancer research. Front. Genet. 2013, 3, 319. [Google Scholar] [CrossRef]
- Seluanov, A.; Ribeiro, A.A.C.M. That Differ in Size and Lifespan. Aging 2009, 7, 813–823. [Google Scholar] [CrossRef]
- Tian, X.; Azpurua, J.; Hine, C.; Vaidya, A.; Myakishev-Rempel, M.; Ablaeva, J.; Mao, Z.; Nevo, E.; Gorbunova, V.; Seluanov, A. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 2013, 499, 346–349. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Haxhinasto, S.; Mathis, D.; Benoist, C. The AKT–mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J. Exp. Med. 2008, 205, 565–574. [Google Scholar] [CrossRef]
- Barth, R.J.; Mule, J.J.; Spiess, P.J.; Rosenberg, S.A. Interferon gamma and tumor necrosis factor have a role in tumor regressions mediated by murine CD8+ tumor-infiltrating lymphocytes. J. Exp. Med. 1991, 173, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Castro, F.; Cardoso, A.P.; Gonçalves, R.M.; Serre, K.; Oliveira, M.J. Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front. Immunol. 2018, 9, 847. [Google Scholar] [CrossRef]
- De Araujo-Souza, P.S.; Hanschke, S.C.H.; Viola, J.P.B. Epigenetic control of interferon-gamma expression in CD8 T cells. J. Immunol. Res. 2015, 2015, 849573. [Google Scholar] [CrossRef]
- Uprety, D.; Adjei, A.A. KRAS: From undruggable to a druggable Cancer Target. Cancer Treat. Rev. 2020, 89, 102070. [Google Scholar] [CrossRef]
- Lane, P.J.; McConnell, F.M.; Anderson, G.; Nawaf, M.G.; Gaspal, F.M.; Withers, D.R. Evolving strategies for cancer and autoimmunity: Back to the future. Front. Immunol. 2014, 5, 154. [Google Scholar] [CrossRef]
- Porto, A.; Schmelter, R.; Vandeberg, J.L.; Marroig, G.; Cheverud, J.M. Evolution of the genotype-to-phenotype map and the cost of pleiotropy in mammals. Genetics 2016, 204, 1601–1612. [Google Scholar] [CrossRef] [PubMed]
- Wellenreuther, M.; Hansson, B. Detecting Polygenic Evolution: Problems, Pitfalls, and Promises. Trends Genet. 2016, 32, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Joshi, C.J.; Ke, W.; Drangowska-Way, A.; O’Rourke, E.J.; Lewis, N.E. What are housekeeping genes? PLOS Comput. Biol. 2022, 18, e1010295. [Google Scholar] [CrossRef]
- Chiari, Y.; Glaberman, S.; Lynch, V.J. Insights on cancer resistance in vertebrates: Reptiles as a parallel system to mammals. Nat. Rev. Cancer 2018, 18, 525. [Google Scholar] [CrossRef] [PubMed]
- White, C.R.; Phillips, N.F.; Seymour, R.S. The scaling and temperature dependence of vertebrate metabolism. Biol. Lett. 2006, 2, 125–127. [Google Scholar] [CrossRef]
- Stenvinkel, P.; Painer, J.; Johnson, R.J.; Natterson-Horowitz, B. Biomimetics—Nature’s roadmap to insights and solutions for burden of lifestyle diseases. J. Intern. Med. 2020, 287, 238–251. [Google Scholar] [CrossRef]
- Noble, R.; Kaltz, O.; Hochberg, M.E. Peto’s paradox and human cancers. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20150104. [Google Scholar] [CrossRef]
- Davies, K.T.J.; Bennett, N.C.; Faulkes, C.G.; Rossiter, S.J. Limited evidence for parallel molecular adaptations associated with the subterranean niche in mammals: A comparative study of three superorders. Mol. Biol. Evol. 2018, 35, 2544–2559. [Google Scholar] [CrossRef]
- Jiang, M.; Shi, L.; Li, X.; Dong, Q.; Sun, H.; Du, Y.; Zhang, Y.; Shao, T.; Cheng, H.; Chen, W.; et al. Genome-wide adaptive evolution to underground stresses in subterranean mammals: Hypoxia adaption, immunity promotion, and sensory specialization. Ecol. Evol. 2020, 10, 7377–7388. [Google Scholar] [CrossRef]
- Storey, K.B. Out Cold: Biochemical Regulation of Mammalian Hibernation—A Mini-Review. Gerontology 2010, 56, 220–230. [Google Scholar] [CrossRef]
- Clutton-Brock, T. Social evolution in mammals. Science 2021, 373, eabc9699. [Google Scholar] [CrossRef] [PubMed]
- Weinert, D. Ontogenetic Development of the Mammalian Circadian System. Chronobiol. Int. 2005, 22, 179–205. [Google Scholar] [CrossRef] [PubMed]
- Quintana-Murci, L.; Clark, A.G. Population genetic tools for dissecting innate immunity in humans. Nat. Rev. Immunol. 2013, 13, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Kerner, G.; Patin, E.; Quintana-Murci, L. New insights into human immunity from ancient genomics. Curr. Opin. Immunol. 2021, 72, 116–125. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Potievskiy, M.; Shatalov, P.A.; Klabukov, I.; Atiakshin, D.; Yakimova, A.; Baranovskii, D.; Shegai, P.V.; Kaprin, A.D. Evolutionary Dynamics of Oncosuppression Under Selection Pressure. Life 2025, 15, 1556. https://doi.org/10.3390/life15101556
Potievskiy M, Shatalov PA, Klabukov I, Atiakshin D, Yakimova A, Baranovskii D, Shegai PV, Kaprin AD. Evolutionary Dynamics of Oncosuppression Under Selection Pressure. Life. 2025; 15(10):1556. https://doi.org/10.3390/life15101556
Chicago/Turabian StylePotievskiy, Mikhail, Peter A. Shatalov, Ilya Klabukov, Dmitrii Atiakshin, Anna Yakimova, Denis Baranovskii, Peter V. Shegai, and Andrey D. Kaprin. 2025. "Evolutionary Dynamics of Oncosuppression Under Selection Pressure" Life 15, no. 10: 1556. https://doi.org/10.3390/life15101556
APA StylePotievskiy, M., Shatalov, P. A., Klabukov, I., Atiakshin, D., Yakimova, A., Baranovskii, D., Shegai, P. V., & Kaprin, A. D. (2025). Evolutionary Dynamics of Oncosuppression Under Selection Pressure. Life, 15(10), 1556. https://doi.org/10.3390/life15101556







