The Brazilian Caatinga Biome as a Hotspot for the Isolation of Antibiotic-Producing Actinomycetota
Abstract
1. Introduction
2. Material and Methods
2.1. Collection of Soil Samples and Isolation of Actinomycetota
2.2. Antimicrobial Activity Screening
2.3. Anti-Tubercular Potential of Actinomycetota Extracts
2.4. Microbial Identification Using Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry (MALDI-TOF MS)
2.5. LC-MS/MS Analyses and Molecular Networking
3. Results and Discussion
3.1. Recovery of Actinomycetota from the Caatinga Biome
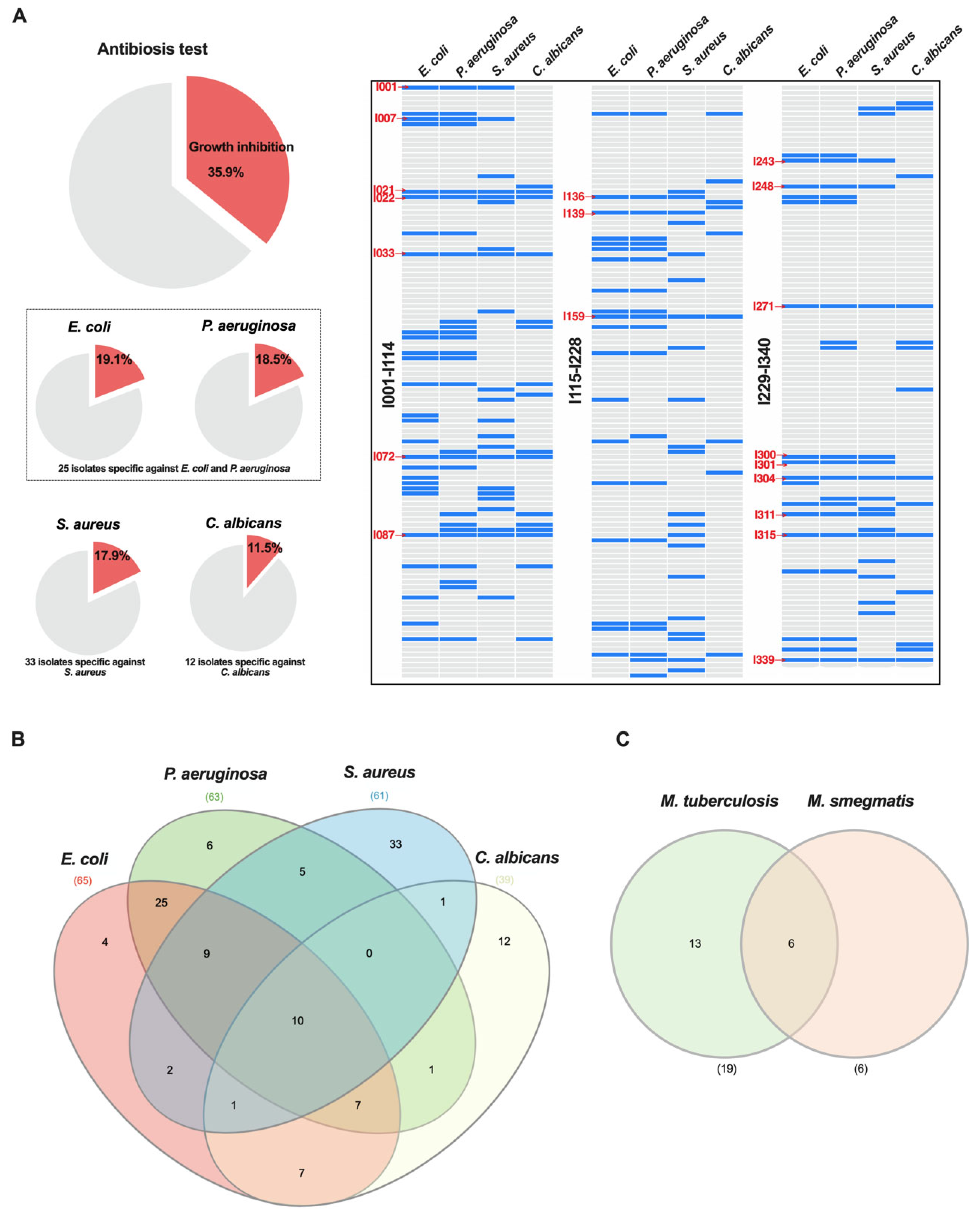
3.2. Antimicrobial Activity Spectrum of Actinomycetota Isolates from Caatinga Biome
3.3. Anti-Tubercular Activity of Actinomycetota Isolates from Caatinga Biome
| Actinomycetota Isolate ID | Biotyper Identification, Score and Best Actinomycetota Matched Pattern | Gram (−) | Gram (+) | Yeast | Mycobacteria | GNPS Dereplication | ||
|---|---|---|---|---|---|---|---|---|
| EC | PA | SA | CA | Mtb | Msmeg | |||
| I001 | NIP */<1.69/S. violaceoruber | - | - | - | - | |||
| I007 | S. nogalater/2.24/S. nogalater/ | - | - | - | - | |||
| I021 | NIP/<1.69/S. chartreusis | - | - | - | ||||
| I022 | NIP/<1.69/Streptomyces sp. HKI D58 HKJ | - | - | - | ||||
| I033 | NA | - | - | - | ||||
| I052 | NIP/<1.69/S. lavendulae | - | - | - | - | - | - | |
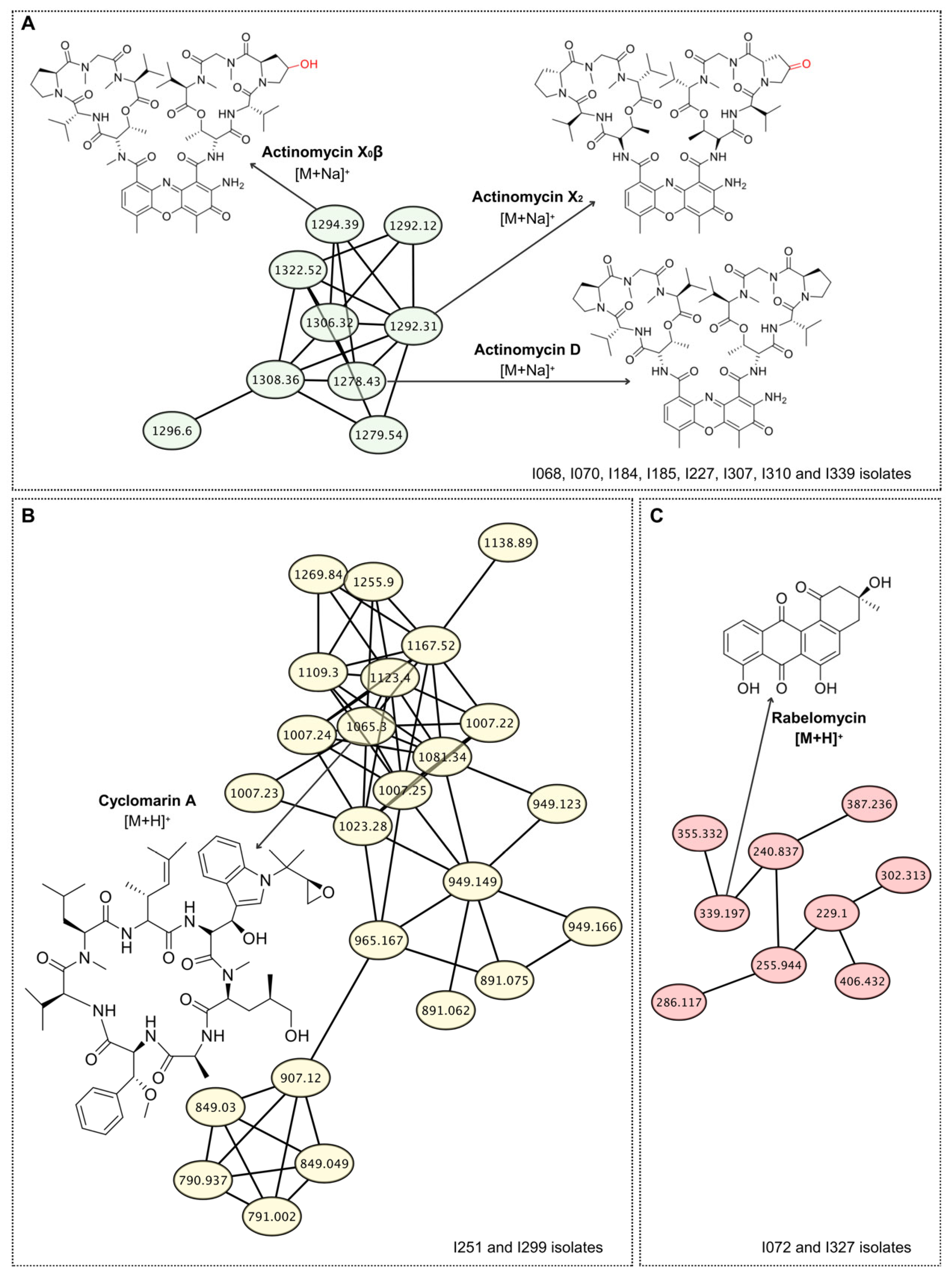
| I068 | NIP/<1.69/S. phaeochromogenes | - | - | - | - | Actinomycins | ||
| I070 | NIP/<1.69/S. nogalater | - | - | - | - | Actinomycins | ||
| I072 | Streptomyces sp./1.70/S. nogalater | - | - | Rabelomycin | ||||
| I082 | NA | - | - | - | - | - | - | |
| I087 | S. nogalater/2.32/S. nogalater | - | - | - | ||||
| I136 | NIP/<1.69/S. violaceoruber | - | - | |||||
| I139 | NIP/<1.69/S. violaceoruber | - | - | - | ||||
| I144 | Streptomyces sp./1.76/S. nogalater | - | - | - | - | - | - | |
| I152 | NIP/<1.69/S. nogalater | - | - | - | - | - | - | |
| I159 | NA | - | - | - | ||||
| I184 | NIP/<1.69/S. hirsutus | - | - | - | - | - | Actinomycins | |
| I185 | NIP/<1.69/S. chartreusis | - | - | - | - | - | Actinomycins | |
| I217 | NIP/<1.69/S. badius | - | - | - | - | - | - | |
| I227 | NIP/<1.69/S. chartreusis | - | - | - | - | - | Actinomycins | |
| I243 | NIP/<1.69/S. badius | - | - | - | - | |||
| I248 | NIP/<1.69/S. violaceoruber | - | - | - | ||||
| I251 | NIP/<1.69/S. violaceoruber | - | - | - | - | - | Cyclomarin A | |
| I271 | Streptomyces sp./1.74/S. violaceoruber | - | - | - | ||||
| I299 | NIP/<1.69/S. violaceoruber | - | - | - | - | - | Cyclomarin A | |
| I300 | Streptomyces sp./1.86/S. violaceoruber | - | - | - | - | |||
| I301 | NA | - | - | - | - | |||
| I304 | Streptomyces sp./1.86/S. nogalater | - | - | - | ||||
| I307 | NIP/<1.69/S. chartreusis | - | - | - | - | Actinomycins | ||
| I310 | NIP/<1.69/S. hirsutus | - | - | - | - | Actinomycins | ||
| I311 | S. nogalater/2.02/S. nogalater | - | - | - | - | |||
| I315 | Streptomyces sp./1.94/S. violaceoruber | - | - | - | - | |||
| I327 | S. violaceoruber/2.06/S. violaceoruber | - | Rabelomycin | |||||
| I339 | NIP/<1.69/S. chartreusis | Actinomycins | ||||||
3.4. MALDI-TOF MS-Based Characterization of Actinomycetota Isolates
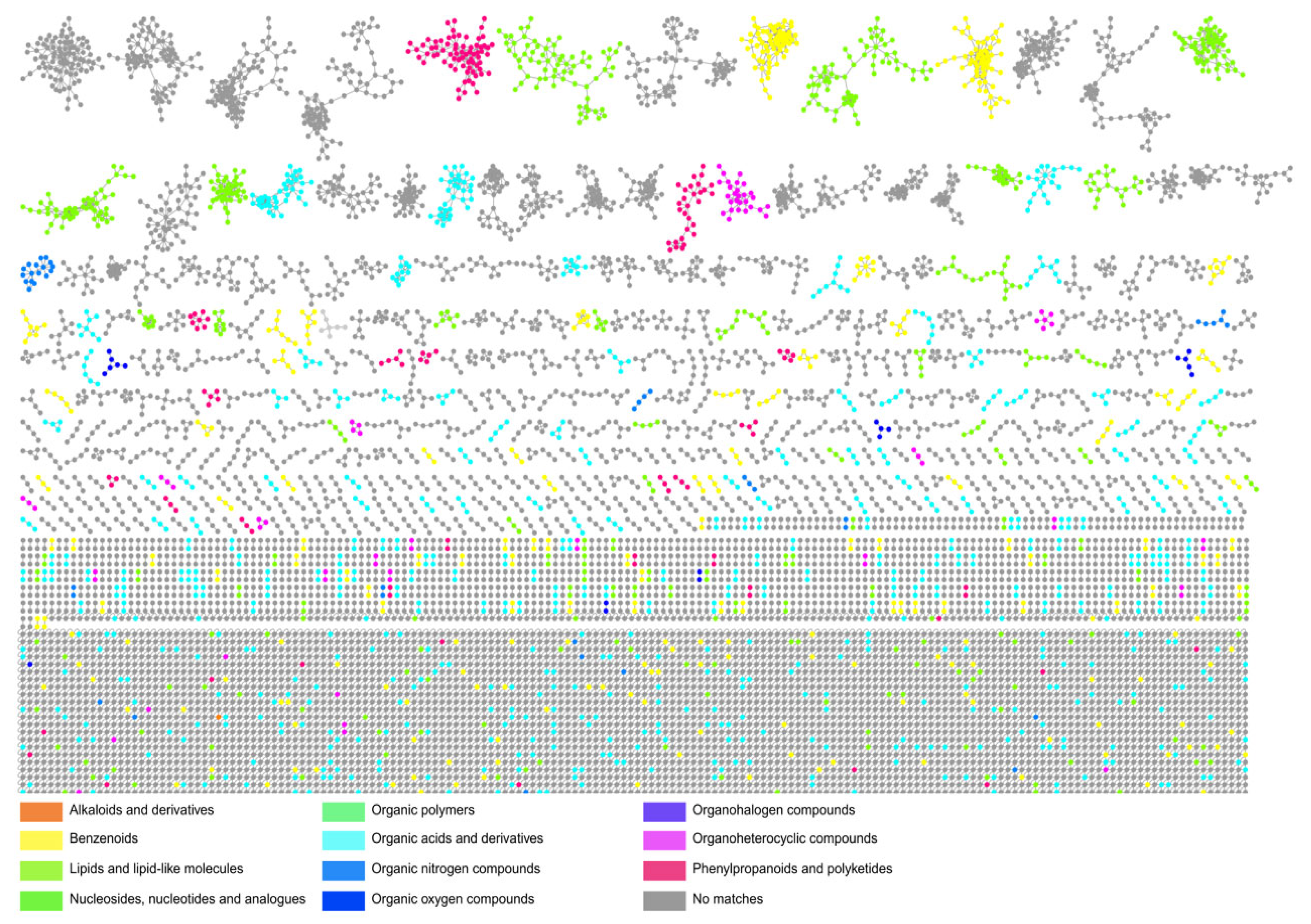
3.5. Metabolomic Fingerprint of Antibiotic-Producing Actinomycetota from Caatinga
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, B.; Prinzi, A.M.; Hill, B.K.; Tibbetts, R.; Salazar, H.; McAdams, D.; Rivera-Acosta, A.M.; Wungwattana, M.; Silbert, S.; Niles, D.T. Uniting Disciplines against Antimicrobial Resistance (AMR): Highlights from a Multidisciplinary Inaugural AMR Summit. Antimicrob. Steward. Healthc. Epidemiol. 2025, 5, e149. [Google Scholar] [CrossRef] [PubMed]
- van Bergeijk, D.A.; Terlouw, B.R.; Medema, M.H.; van Wezel, G.P. Ecology and Genomics of Actinobacteria: New Concepts for Natural Product Discovery. Nat. Rev. Microbiol. 2020, 18, 546–558. [Google Scholar] [CrossRef]
- Bérdy, J. Thoughts and Facts about Antibiotics: Where We Are Now and Where We Are Heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef]
- Hug, J.; Bader, C.; Remškar, M.; Cirnski, K.; Müller, R. Concepts and Methods to Access Novel Antibiotics from Actinomycetes. Antibiotics 2018, 7, 44. [Google Scholar] [CrossRef]
- Pylro, V.S.; Roesch, L.F.W.; Ortega, J.M.; do Amaral, A.M.; Tótola, M.R.; Hirsch, P.R.; Rosado, A.S.; Góes-Neto, A.; da Costa da Silva, A.L.; Rosa, C.A.; et al. Brazilian Microbiome Project: Revealing the Unexplored Microbial Diversity—Challenges and Prospects. Microb. Ecol. 2014, 67, 237–241. [Google Scholar] [CrossRef]
- Waksman, S. The Actinomycetes; The Williams & Wilkins Company: Baltimore, MD, USA, 1961; Volume II. [Google Scholar]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in Vitro Evaluating Antimicrobial Activity: A Review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Sidrônio, M.G.S.; Castelo Branco, A.P.O.T.; Abbadi, B.L.; Macchi, F.; Silveira, M.D.; Lock, G.d.A.; Costa, T.D.; de Araújo, D.M.; Cibulski, S.; Bizarro, C.V.; et al. Effects of Tafenoquine against Active, Dormant and Resistant Mycobacterium Tuberculosis. Tuberculosis 2021, 128, 102089. [Google Scholar] [CrossRef]
- Muradás, T.C.; Abbadi, B.L.; Villela, A.D.; Macchi, F.S.; Bergo, P.F.; de Freitas, T.F.; Sperotto, N.D.M.; Timmers, L.F.S.M.; Norberto de Souza, O.; Picada, J.N.; et al. Pre-Clinical Evaluation of Quinoxaline-Derived Chalcones in Tuberculosis. PLoS ONE 2018, 13, e0202568. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Ernst, M.; Kang, K.B.; Caraballo-Rodríguez, A.M.; Nothias, L.-F.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; et al. MolNetEnhancer: Enhanced Molecular Networks by Integrating Metabolome Mining and Annotation Tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef] [PubMed]
- Caatinga; Silva, J.M.C.d., Leal, I.R., Tabarelli, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; ISBN 978-3-319-68338-6. [Google Scholar]
- Naylor, D.; DeGraaf, S.; Purdom, E.; Coleman-Derr, D. Drought and Host Selection Influence Bacterial Community Dynamics in the Grass Root Microbiome. ISME J. 2017, 11, 2691–2704. [Google Scholar] [CrossRef] [PubMed]
- Marasco, R.; Fusi, M.; Rolli, E.; Ettoumi, B.; Tambone, F.; Borin, S.; Ouzari, H.; Boudabous, A.; Sorlini, C.; Cherif, A.; et al. Aridity Modulates Belowground Bacterial Community Dynamics in Olive Tree. Environ. Microbiol. 2021, 23, 6275–6291. [Google Scholar] [CrossRef] [PubMed]
- Rasuk, M.C.; Ferrer, G.M.; Kurth, D.; Portero, L.R.; Farías, M.E.; Albarracín, V.H. UV-Resistant Actinobacteria from High-Altitude Andean Lakes: Isolation, Characterization and Antagonistic Activities. Photochem. Photobiol. 2017, 93, 865–880. [Google Scholar] [CrossRef]
- Xie, F.; Pathom-aree, W. Actinobacteria from Desert: Diversity and Biotechnological Applications. Front. Microbiol. 2021, 12, 765531. [Google Scholar] [CrossRef]
- Mohammadipanah, F.; Wink, J. Actinobacteria from Arid and Desert Habitats: Diversity and Biological Activity. Front. Microbiol. 2015, 6, 1541. [Google Scholar] [CrossRef]
- Rateb, M.E.; Ebel, R.; Jaspars, M. Natural Product Diversity of Actinobacteria in the Atacama Desert. Antonie Leeuwenhoek 2018, 111, 1467–1477. [Google Scholar] [CrossRef]
- Vásquez-Dean, J.; Maza, F.; Morel, I.; Pulgar, R.; González, M. Microbial Communities from Arid Environments on a Global Scale. A Systematic Review. Biol. Res. 2020, 53, 29. [Google Scholar] [CrossRef]
- Bouskill, N.J.; Lim, H.C.; Borglin, S.; Salve, R.; Wood, T.E.; Silver, W.L.; Brodie, E.L. Pre-Exposure to Drought Increases the Resistance of Tropical Forest Soil Bacterial Communities to Extended Drought. ISME J. 2013, 7, 384–394. [Google Scholar] [CrossRef]
- Sati, H.; Carrara, E.; Savoldi, A.; Hansen, P.; Garlasco, J.; Campagnaro, E.; Boccia, S.; Castillo-Polo, J.A.; Magrini, E.; Garcia-Vello, P.; et al. The WHO Bacterial Priority Pathogens List 2024: A Prioritisation Study to Guide Research, Development, and Public Health Strategies against Antimicrobial Resistance. Lancet Infect. Dis. 2025, 25, 1033–1043. [Google Scholar] [CrossRef]
- Mishra, S.K.; Yasir, M.; Willcox, M. Candida auris: An Emerging Antimicrobial-Resistant Organism with the Highest Level of Concern. Lancet Microbe 2023, 4, e482–e483. [Google Scholar] [CrossRef]
- de Almeida, L.L.C.; Fernandes, S.P.; de Oliveira, G.D.; da Silveira Silva, M.; de Souza, T.A.; Rodrigues-Junior, V.S.; Cibulski, S.P. Harnessing Actinobacteria Secondary Metabolites for Tuberculosis Drug Discovery: Historical Trends, Current Status and Future Outlooks. Nat. Prod. Bioprospect. 2025, 15, 52. [Google Scholar] [CrossRef] [PubMed]
- Lelovic, N.; Mitachi, K.; Yang, J.; Lemieux, M.R.; Ji, Y.; Kurosu, M. Application of Mycobacterium Smegmatis as a Surrogate to Evaluate Drug Leads against Mycobacterium Tuberculosis. J. Antibiot. 2020, 73, 780–789. [Google Scholar] [CrossRef] [PubMed]
- Boone, D.R.; Castenholz, R.W.; Garrity, G.M. (Eds.) Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Springer: New York, NY, USA, 2001; Volume 1, ISBN 0387987711. [Google Scholar]
- Tsuchida, S.; Umemura, H.; Nakayama, T. Current Status of Matrix-Assisted Laser Desorption/Ionization–Time-of-Flight Mass Spectrometry (MALDI-TOF MS) in Clinical Diagnostic Microbiology. Molecules 2020, 25, 4775. [Google Scholar] [CrossRef] [PubMed]
- Ashfaq, M.Y.; Da’na, D.A.; Al-Ghouti, M.A. Application of MALDI-TOF MS for Identification of Environmental Bacteria: A Review. J. Environ. Manag. 2022, 305, 114359. [Google Scholar] [CrossRef]
- Zuffa, S.; Schmid, R.; Bauermeister, A.; Gomes, P.W.P.; Caraballo-Rodriguez, A.M.; El Abiead, Y.; Aron, A.T.; Gentry, E.C.; Zemlin, J.; Meehan, M.J.; et al. MicrobeMASST: A Taxonomically Informed Mass Spectrometry Search Tool for Microbial Metabolomics Data. Nat. Microbiol. 2024, 9, 336–345. [Google Scholar] [CrossRef]
- Gaudry, A.; Marcourt, L.; Kaiser, M.; Flückiger, J.; David, B.; Grondin, A.; Ioset, J.-R.; Mäser, P.; Queiroz, E.F.; Allard, P.-M.; et al. Efficient Constitution of a Library of Rotenoid Analogs Active against Trypanosoma cruzi from a Digitalized Plant Extract Collection. RSC Adv. 2025, 15, 15240–15251. [Google Scholar] [CrossRef]
- Gaudry, A.; Pagni, M.; Mehl, F.; Moretti, S.; Quiros-Guerrero, L.-M.; Cappelletti, L.; Rutz, A.; Kaiser, M.; Marcourt, L.; Queiroz, E.F.; et al. A Sample-Centric and Knowledge-Driven Computational Framework for Natural Products Drug Discovery. ACS Cent. Sci. 2024, 10, 494–510. [Google Scholar] [CrossRef]
- Liu, M.; Jia, Y.; Xie, Y.; Zhang, C.; Ma, J.; Sun, C.; Ju, J. Identification of the Actinomycin D Biosynthetic Pathway from Marine-Derived Streptomyces costaricanus SCSIO ZS0073. Mar. Drugs 2019, 17, 240. [Google Scholar] [CrossRef]
- Lo, Y.-S.; Tseng, W.-H.; Chuang, C.-Y.; Hou, M.-H. The Structural Basis of Actinomycin D–Binding Induces Nucleotide Flipping out, a Sharp Bend and a Left-Handed Twist in CGG Triplet Repeats. Nucleic Acids Res. 2013, 41, 4284–4294. [Google Scholar] [CrossRef]
- Wang, D.; Wang, C.; Gui, P.; Liu, H.; Khalaf, S.M.H.; Elsayed, E.A.; Wadaan, M.A.M.; Hozzein, W.N.; Zhu, W. Identification, Bioactivity, and Productivity of Actinomycins from the Marine-Derived Streptomyces heliomycini. Front. Microbiol. 2017, 8, 1147. [Google Scholar] [CrossRef]
- Schultz, A.W.; Oh, D.-C.; Carney, J.R.; Williamson, R.T.; Udwary, D.W.; Jensen, P.R.; Gould, S.J.; Fenical, W.; Moore, B.S. Biosynthesis and Structures of Cyclomarins and Cyclomarazines, Prenylated Cyclic Peptides of Marine Actinobacterial Origin. J. Am. Chem. Soc. 2008, 130, 4507–4516. [Google Scholar] [CrossRef]
- Schmitt, E.K.; Riwanto, M.; Sambandamurthy, V.; Roggo, S.; Miault, C.; Zwingelstein, C.; Krastel, P.; Noble, C.; Beer, D.; Rao, S.P.S.; et al. The Natural Product Cyclomarin Kills Mycobacterium Tuberculosis by Targeting the ClpC1 Subunit of the Caseinolytic Protease. Angew. Chem. Int. Ed. 2011, 50, 5889–5891. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, D.; Rao, S.P.S.; Noble, C.G. Structural Basis of Mycobacterial Inhibition by Cyclomarin A. J. Biol. Chem. 2013, 288, 30883–30891. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-C.; Parker, W.L.; Slusarchyk, D.S.; Greenwood, G.L.; Graham, S.F.; Meyers, E. Isolation, characterization, and structure of rabelomycin, a new antibiotic. J. Antibiot. 1970, 23, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Kharel, M.K.; Pahari, P.; Shepherd, M.D.; Tibrewal, N.; Nybo, S.E.; Shaaban, K.A.; Rohr, J. Angucyclines: Biosynthesis, Mode-of-Action, New Natural Products, and Synthesis. Nat. Prod. Rep. 2012, 29, 264–325. [Google Scholar] [CrossRef]
- Rohr, J.; Thiericke, R. Angucycline Group Antibiotics. Nat. Prod. Rep. 1992, 9, 103. [Google Scholar] [CrossRef]
- Liu, H.-S.; Chen, H.-R.; Huang, S.-S.; Li, Z.-H.; Wang, C.-Y.; Zhang, H. Bioactive Angucyclines/Angucyclinones Discovered from 1965 to 2023. Mar. Drugs 2025, 23, 25. [Google Scholar] [CrossRef]
- Ribeiro, I.; Girão, M.; Alexandrino, D.A.M.; Ribeiro, T.; Santos, C.; Pereira, F.; Mucha, A.P.; Urbatzka, R.; Leão, P.N.; Carvalho, M.F. Diversity and Bioactive Potential of Actinobacteria Isolated from a Coastal Marine Sediment in Northern Portugal. Microorganisms 2020, 8, 1691. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, S.P.; de Almeida, L.L.C.; de Andrade, A.S.A.; Abreu, L.S.; Nascimento, Y.M.; de Souza, T.A.; da Silva, E.F.; Volpato, F.C.Z.; Barth, A.L.; Tavares, J.F.; et al. The Brazilian Caatinga Biome as a Hotspot for the Isolation of Antibiotic-Producing Actinomycetota. Life 2025, 15, 1494. https://doi.org/10.3390/life15101494
Fernandes SP, de Almeida LLC, de Andrade ASA, Abreu LS, Nascimento YM, de Souza TA, da Silva EF, Volpato FCZ, Barth AL, Tavares JF, et al. The Brazilian Caatinga Biome as a Hotspot for the Isolation of Antibiotic-Producing Actinomycetota. Life. 2025; 15(10):1494. https://doi.org/10.3390/life15101494
Chicago/Turabian StyleFernandes, Sayoane Pessoa, Luana Layse Câmara de Almeida, Adrielly Silva Albuquerque de Andrade, Lucas Silva Abreu, Yuri Mangueira Nascimento, Thalisson Amorim de Souza, Evandro Ferreira da Silva, Fabiana Caroline Zempulski Volpato, Afonso Luis Barth, Josean Fechine Tavares, and et al. 2025. "The Brazilian Caatinga Biome as a Hotspot for the Isolation of Antibiotic-Producing Actinomycetota" Life 15, no. 10: 1494. https://doi.org/10.3390/life15101494
APA StyleFernandes, S. P., de Almeida, L. L. C., de Andrade, A. S. A., Abreu, L. S., Nascimento, Y. M., de Souza, T. A., da Silva, E. F., Volpato, F. C. Z., Barth, A. L., Tavares, J. F., de Araújo, D. A. M., Rodrigues-Junior, V. d. S., & Cibulski, S. P. (2025). The Brazilian Caatinga Biome as a Hotspot for the Isolation of Antibiotic-Producing Actinomycetota. Life, 15(10), 1494. https://doi.org/10.3390/life15101494








