Scent of COVID-19: Whole-Genome Sequencing Analysis Reveals the Role of ACE2, IFI44, and NDUFAF4 in Long-Lasting Olfactory Dysfunction
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
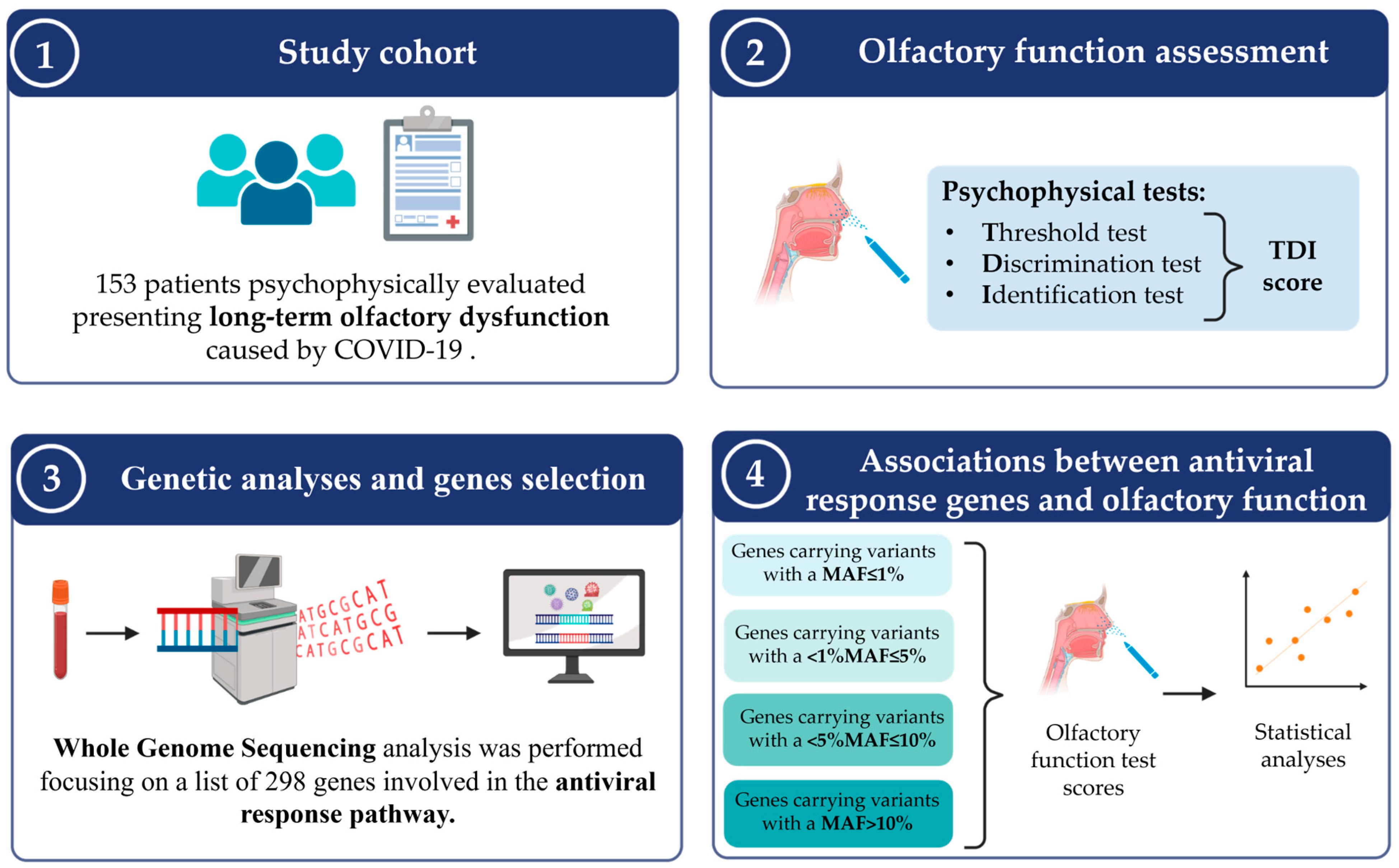
2.2. Study Cohort and Clinical Evaluations
2.3. Psychophysical Assessment of Olfactory Function
2.4. Biological Sample Collection and WGS Analysis
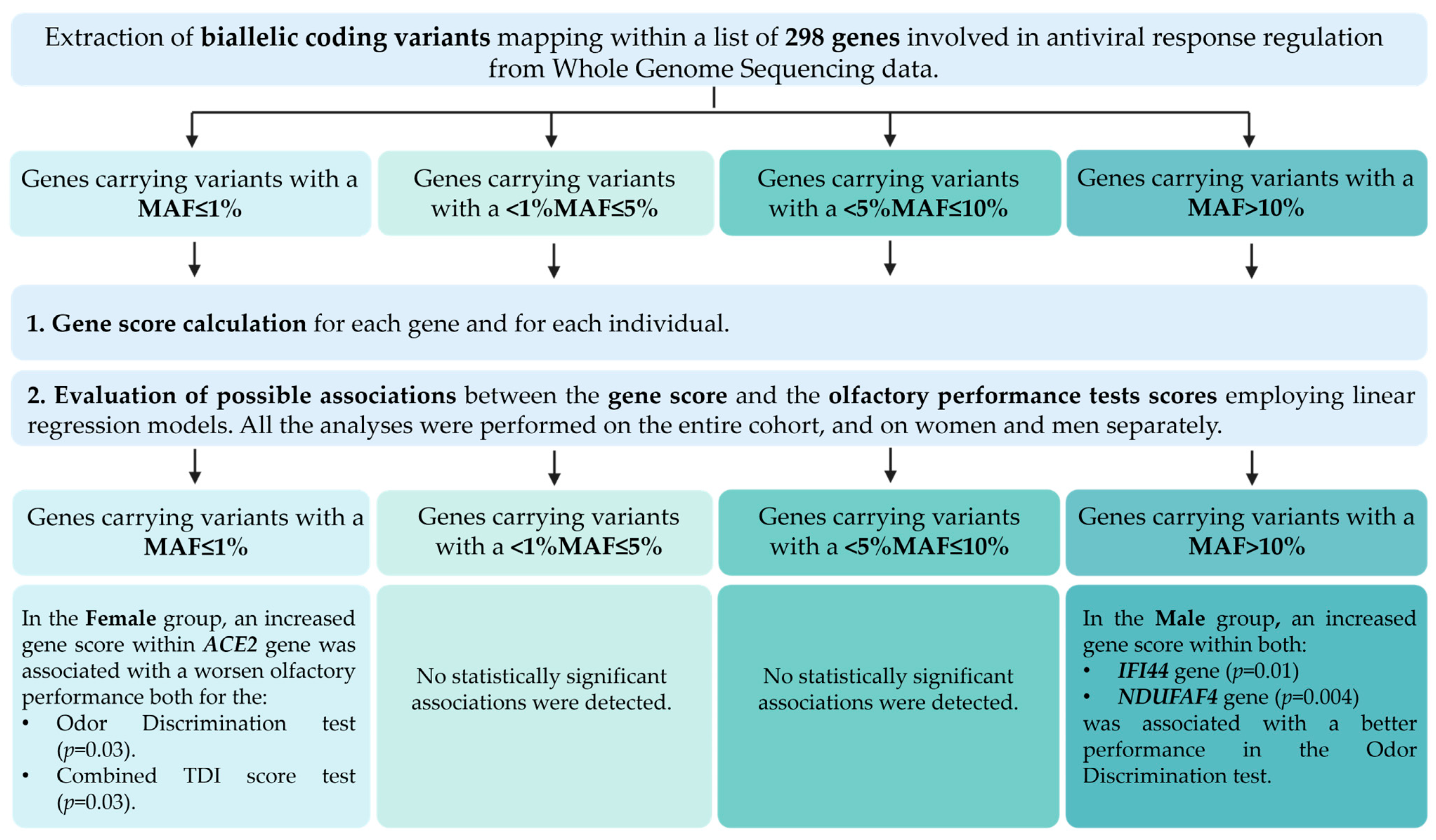
2.5. Gene Selection
2.6. Statistical Analysis
3. Results
3.1. Demographic and Clinical Data
3.2. Association Analyses Between Rare Variants and Olfactory Performance
3.3. Association Analyses Between Common Variants and Olfactory Performance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boesveldt, S.; Parma, V. The importance of the olfactory system in human well-being, through nutrition and social behavior. Cell Tissue Res. 2021, 383, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Boesveldt, S.; Postma, E.M.; Boak, D.; Welge-Luessen, A.; Schöpf, V.; Mainland, J.D.; Martens, J.; Ngai, J.; Duffy, V.B. Anosmia—A Clinical Review. Chem. Senses 2017, 42, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Croy, I.; Nordin, S.; Hummel, T. Olfactory Disorders and Quality of Life—An Updated Review. Chem. Senses 2014, 39, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Pinto, J.M. The Epidemiology of Olfactory Disorders. Curr. Otorhinolaryngol. Rep. 2016, 4, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Landis, B.N.; Hummel, T.; Hugentobler, M.; Giger, R.; Lacroix, J.S. Ratings of Overall Olfactory Function. Chem. Senses 2003, 28, 691–694. [Google Scholar] [CrossRef]
- Whitcroft, K.L.; Altundag, A.; Balungwe, P.; Boscolo-Rizzo, P.; Douglas, R.; Enecilla, M.L.B.; Fjaeldstad, A.W.; Fornazieri, M.A.; Frasnelli, J.; Gane, S.; et al. Position paper on olfactory dysfunction: 2023 Executive Summary. Rhinology 2023, 61, 1–108. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Y.; Vaira, L.A.; Boscolo-Rizzo, P.; Walker, A.; Hopkins, C. Post-viral olfactory loss and parosmia. BMJ Med. 2023, 2, e000382. [Google Scholar] [CrossRef] [PubMed]
- Spinato, G.; Fabbris, C.; Polesel, J.; Cazzador, D.; Borsetto, D.; Hopkins, C.; Boscolo-Rizzo, P. Alterations in Smell or Taste in Mildly Symptomatic Outpatients With SARS-CoV-2 Infection. JAMA 2020, 323, 2089–2090. [Google Scholar] [CrossRef]
- Boscolo-Rizzo, P.; Hummel, T.; Invitto, S.; Spinato, G.; Tomasoni, M.; Emanuelli, E.; Tofanelli, M.; Cavicchia, A.; Grill, V.; Vaira, L.A.; et al. Psychophysical assessment of olfactory and gustatory function in post-mild COVID-19 patients: A matched case-control study with 2-year follow-up. Int. Forum. Allergy Rhinol. 2023, 13, 1864–1875. [Google Scholar] [CrossRef] [PubMed]
- Saniasiaya, J.; Islam, M.A.; Abdullah, B. Prevalence of Olfactory Dysfunction in Coronavirus Disease 2019 (COVID-19): A Meta-analysis of 27,492 Patients. Laryngoscope 2021, 131, 865–878. [Google Scholar] [CrossRef]
- Krishnakumar, H.N.; Momtaz, D.A.; Sherwani, A.; Mhapankar, A.; Gonuguntla, R.K.; Maleki, A.; Abbas, A.; Ghali, A.N.; Al Afif, A. Pathogenesis and progression of anosmia and dysgeusia during the COVID-19 pandemic. Eur. Arch. Oto.-Rhino.-Laryngol. 2023, 280, 505–509. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Clavijo, A.; Gonzalez-Velandia, K.Y.; Rangaswamy, U.; Guarneri, G.; Boscolo-Rizzo, P.; Tofanelli, M.; Gardenal, N.; Sanges, R.; Dibattista, M.; Tirelli, G.; et al. Supporting Cells of the Human Olfactory Epithelium Co-Express the Lipid Scramblase TMEM16F and ACE2 and May Cause Smell Loss by SARS-CoV-2 Spike-Induced Syncytia. Cell. Physiol. Biochem. 2022, 56, 254–269. [Google Scholar] [CrossRef] [PubMed]
- Krishna, B.; Wills, M.; Sithole, N. Long COVID: What is known and what gaps need to be addressed. Br. Med. Bull. 2023, 147, 6–19. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.W.; Toong, P.J.; Guarnera, E.; Berezovsky, I.N. Disrupted chromatin architecture in olfactory sensory neurons: Looking for the link from COVID-19 infection to anosmia. Sci. Rep. 2023, 13, 5906. [Google Scholar] [CrossRef] [PubMed]
- Shelton, J.F.; Shastri, A.J.; Fletez-Brant, K.; Aslibekyan, S.; Auton, A. The UGT2A1/UGT2A2 locus is associated with COVID-19-related loss of smell or taste. Nat. Genet. 2022, 54, 121–124. [Google Scholar] [CrossRef]
- Neiers, F.; Jarriault, D.; Menetrier, F.; Faure, P.; Briand, L.; Heydel, J.-M. The odorant metabolizing enzyme UGT2A1: Immunolocalization and impact of the modulation of its activity on the olfactory response. PLoS ONE 2021, 16, e0249029. [Google Scholar] [CrossRef]
- Ren, J.; Zhang, Y.; Guo, W.; Feng, K.; Yuan, Y.; Huang, T.; Cai, Y.-D. Identification of Genes Associated with the Impairment of Olfactory and Gustatory Functions in COVID-19 via Machine-Learning Methods. Life 2023, 13, 798. [Google Scholar] [CrossRef] [PubMed]
- Matuozzo, D.; Talouarn, E.; Marchal, A.; Zhang, P.; Manry, J.; Seeleuthner, Y.; Zhang, Y.; Bolze, A.; Chaldebas, M.; Milisavljevic, B.; et al. Correction: Rare predicted loss-of-function variants of type I IFN immunity genes are associated with life-threatening COVID-19. Genome Med. 2024, 16, 6. [Google Scholar] [CrossRef]
- Khadzhieva, M.B.; Gracheva, A.S.; Belopolskaya, O.B.; Kolobkov, D.S.; Kashatnikova, D.A.; Redkin, I.V.; Kuzovlev, A.N.; Grechko, A.V.; Salnikova, L.E. COVID-19 severity: Does the genetic landscape of rare variants matter? Front. Genet. 2023, 14, 1152768. [Google Scholar] [CrossRef]
- Adimulam, T.; Arumugam, T.; Gokul, A.; Ramsuran, V. Genetic Variants within SARS-CoV-2 Human Receptor Genes May Contribute to Variable Disease Outcomes in Different Ethnicities. Multidiscip. Digit. Publ. Inst. 2023, 24, 8711. [Google Scholar] [CrossRef]
- Binny, R.N.; Priest, P.; French, N.P.; Parry, M.; Lustig, A.; Hendy, S.C.; Maclaren, O.J.; Ridings, K.M.; Steyn, N.; Vattiato, G.; et al. Sensitivity of Reverse Transcription Polymerase Chain Reaction Tests for Severe Acute Respiratory Syndrome Coronavirus 2 Through Time. J. Infect. Dis. 2023, 227, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Fang, Y.; Li, W.; Pan, C.; Qin, P.; Zhong, Y.; Liu, X.; Huang, M.; Liao, Y.; Li, S. CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19). Eur. Radiol. 2020, 30, 4407–4416. [Google Scholar] [CrossRef] [PubMed]
- Oleszkiewicz, A.; Schriever, V.A.; Croy, I.; Hähner, A.; Hummel, T. Updated Sniffin’ Sticks normative data based on an extended sample of 9139 subjects. Eur. Arch. Oto.-Rhino.-Laryngol. 2019, 276, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Boscolo-Rizzo, P.; Hummel, T.; Menini, A.; Maniaci, A.; Uderzo, F.; Bigolin, L.; Tirelli, G. Adherence to olfactory training improves orthonasal and retronasal olfaction in post-COVID-19 olfactory loss. Rhinol. J. 2024, 62, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Tarasov, A.; Vilella, A.J.; Cuppen, E.; Nijman, I.J.; Prins, P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics 2015, 31, 2032–2034. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Loh, P.-R.; Danecek, P.; Palamara, P.F.; Fuchsberger, C.; Reshef, Y.A.; Finucane, H.K.; Schoenherr, S.; Forer, L.; McCarthy, S.; Abecasis, C.F.G.R.; et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016, 48, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Smedley, D.; Haider, S.; Ballester, B.; Holland, R.; London, D.; Thorisson, G.; Kasprzyk, A. BioMart–biological queries made easy. BMC Genom. 2009, 10, 22. [Google Scholar] [CrossRef]
- Thaweethai, T.; Jolley, S.E.; Karlson, E.W.; Levitan, E.B.; Levy, B.; McComsey, G.A.; McCorkell, L.; Nadkarni, G.N.; Parthasarathy, S.; Singh, U.; et al. Development of a Definition of Postacute Sequelae of SARS-CoV-2 Infection. JAMA 2023, 329, 1934–1946. [Google Scholar] [CrossRef] [PubMed]
- Groff, D.; Sun, A.; Ssentongo, A.E.; Ba, D.M.; Parsons, N.; Poudel, G.R.; Lekoubou, A.; Oh, J.S.; Ericson, J.E.; Ssentongo, P.; et al. Short-term and Long-term Rates of Postacute Sequelae of SARS-CoV-2 Infection: A Systematic Review. JAMA Netw. Open 2021, 4, e2128568. [Google Scholar] [CrossRef]
- Garg, M.; Maralakunte, M.; Garg, S.; Dhooria, S.; Sehgal, I.; Bhalla, A.S.; Vijayvergiya, R.; Grover, S.; Bhatia, V.; Jagia, P.; et al. The conundrum of “long-covid-19”: A narrative review. Dove. Med. Press Ltd. 2021, 14, 2491–2506. [Google Scholar] [CrossRef]
- Park, J.W.; Wang, X.; Xu, R.-H. Revealing the mystery of persistent smell loss in Long COVID patients. Int. J. Biol. Sci. 2022, 18, 4795–4808. [Google Scholar] [CrossRef] [PubMed]
- Butowt, R.; Bilinska, K.; von Bartheld, C.S. Olfactory dysfunction in COVID-19: New insights into the underlying mechanisms. Trends Neurosci. 2023, 46, 75–90. [Google Scholar] [CrossRef] [PubMed]
- de Melo, G.D.; Lazarini, F.; Levallois, S.; Hautefort, C.; Michel, V.; Larrous, F.; Verillaud, B.; Aparicio, C.; Wagner, S.; Gheusi, G.; et al. COVID-19–related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci. Transl. Med. 2021, 13, eabf8396. [Google Scholar] [CrossRef]
- Rodriguez, S.; Cao, L.; Rickenbacher, G.T.; Benz, E.G.; Magdamo, C.; Gomez, L.R.; Holbrook, E.H.; Albers, A.D.; Gallagher, R.; Westover, M.B.; et al. Innate immune signaling in the olfactory epithelium reduces odorant receptor levels: Modeling transient smell loss in COVID-19 patients. medRxiv 2020. [Google Scholar] [CrossRef]
- Zazhytska, M.; Kodra, A.; Hoagland, D.A.; Frere, J.; Fullard, J.F.; Shayya, H.; McArthur, N.G.; Moeller, R.; Uhl, S.; Omer, A.D.; et al. Non-cell-autonomous disruption of nuclear architecture as a potential cause of COVID-19-induced anosmia. Cell 2022, 185, 1052–1064. [Google Scholar] [CrossRef]
- Lane, A.P.; Turner, J.; May, L.; Reed, R. A Genetic Model of Chronic Rhinosinusitis-Associated Olfactory Inflammation Reveals Reversible Functional Impairment and Dramatic Neuroepithelial Reorganization. J. Neurosci. 2010, 30, 2324. [Google Scholar] [CrossRef]
- Bomba, L.; Walter, K.; Soranzo, N. The impact of rare and low-frequency genetic variants in common disease. Genome Biol. 2017, 18, 77. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, G.R.; Rivas, M.A. Rare and common variant discovery in complex disease: The IBD case study. Hum. Mol. Genet. 2019, 28, R162–R169. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Di Gaetano, C.; Cugliari, G.; Matullo, G. Advances in the genetics of hypertension: The effect of rare variants. Int. J. Mol. Sci. 2018, 19, 688. [Google Scholar] [CrossRef] [PubMed]
- Urpa, L.; Kurki, M.I.; Rahikkala, E.; Hämäläinen, E.; Salomaa, V.; Suvisaari, J.; Keski-Filppula, R.; Rauhala, M.; Korpi-Heikkilä, S.; Komulainen-Ebrahim, J.; et al. Evidence for the additivity of rare and common variant burden throughout the spectrum of intellectual disability. Eur. J. Hum. Genet. 2024, 32, 576–583. [Google Scholar] [CrossRef]
- Turner, A.J. Chapter 25-ACE2 Cell Biology, Regulation, and Physiological Functions. In The Protective Arm of the Renin Angiotensin System (RAS); Unger, T., Steckelings, U.M., Santos, R.A.S.D., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 185–189. [Google Scholar] [CrossRef]
- Banu, N.; Panikar, S.S.; Leal, L.R.; Leal, A.R. Protective role of ACE2 and its downregulation in SARS-CoV-2 infection leading to Macrophage Activation Syndrome: Therapeutic implications. Life Sci. 2020, 256, 117905. [Google Scholar] [CrossRef]
- Oudit, G.Y.; Wang, K.; Viveiros, A.; Kellner, M.J.; Penninger, J.M. Angiotensin-converting enzyme 2—At the heart of the COVID-19 pandemic. Cell 2023, 186, 906–922. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, G.; Abdelgadir, Y.; Abdelghani, A.; Simpson, P.; Barbeau, J.; Basel, D.; Barrios, C.S.; A Smith, B.; Schilter, K.F.; Udani, R.; et al. Reduction in ACE2 expression in peripheral blood mononuclear cells during COVID-19–implications for post COVID-19 conditions. BMC Infect. Dis. 2024, 24, 663. [Google Scholar] [CrossRef]
- Ekholm, M.; Kahan, T.; Jörneskog, G.; Bröijersen, A.; Wallén, N.H. Angiotensin II infusion in man is proinflammatory but has no short-term effects on thrombin generation in vivo. Thromb. Res. 2009, 124, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Ferrario, C.M.; Trask, A.J.; Jessup, J.A. Advances in biochemical and functional roles of angiotensin-converting enzyme 2 and angiotensin-(1–7) in regulation of cardiovascular function. Am. J. Physiol.-Heart Circ. Physiol. 2005, 289, H2281–H2290. [Google Scholar] [CrossRef] [PubMed]
- Khazaal, S.; Harb, J.; Rima, M.; Annweiler, C.; Wu, Y.; Cao, Z.; Khattar, Z.A.; Legros, C.; Kovacic, H.; Fajloun, Z.; et al. The Pathophysiology of Long COVID throughout the Renin-Angiotensin System. Molecules 2022, 27, 2903. [Google Scholar] [CrossRef] [PubMed]
- Finlay, J.B.; Brann, D.H.; Hachem, R.A.; Jang, D.W.; Oliva, A.D.; Ko, T.; Gupta, R.; Wellford, S.A.; Moseman, E.A.; Jang, S.S.; et al. Persistent post-COVID-19 smell loss is associated with immune cell infiltration and altered gene expression in olfactory epithelium. Sci. Transl. Med. 2022, 14, eadd0484. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Inc. 2014, 32, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Dediego, M.L.; Nogales, A.; Martinez-Sobrido, L.; Topham, D.J. Interferon-induced protein 44 interacts with cellular fk506-binding protein 5, negatively regulates host antiviral responses, and supports virus replication. Am. Soc. Microbiol. 2019, 10, e01839-19. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Ramaswamy, S.; Harilal, D.; Uddin, M.; Loney, T.; Nowotny, N.; Alsuwaidi, H.; Varghese, R.; Deesi, Z.; Alkhajeh, A.; et al. Host transcriptomic profiling of COVID-19 patients with mild, moderate, and severe clinical outcomes. Comput. Struct. Biotechnol. J. 2021, 19, 153–160. [Google Scholar] [CrossRef]
- Zheng, Q.; Wang, D.; Lin, R.; Lv, Q.; Wang, W. IFI44 is an immune evasion biomarker for SARS-CoV-2 and Staphylococcus aureus infection in patients with RA. Front. Immunol. 2022, 13, 1013322. [Google Scholar] [CrossRef]
- Balsa, E.; Marco, R.; Perales-Clemente, E.; Szklarczyk, R.; Calvo, E.; Landázuri, M.O.; Enríquez, J.A. NDUFA4 Is a Subunit of Complex IV of the Mammalian Electron Transport Chain. Cell Metab. 2012, 16, 378–386. [Google Scholar] [CrossRef]
- Pan, W.; Shen, Z.; Wang, H.; He, H. The host cellular protein Ndufaf4 interacts with the vesicular stomatitis virus M protein and affects viral propagation. Virus Genes 2021, 57, 250–257. [Google Scholar] [CrossRef]
- West, A.P.; Shadel, G.S.; Ghosh, S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 2011, 11, 389–402. [Google Scholar] [CrossRef]
- Peppercorn, K.; Edgar, C.D.; Kleffmann, T.; Tate, W.P. A pilot study on the immune cell proteome of long COVID patients shows changes to physiological pathways similar to those in myalgic encephalomyelitis/chronic fatigue syndrome. Sci. Rep. 2023, 13, 22068. [Google Scholar] [CrossRef] [PubMed]
- Coelho, D.H.; Reiter, E.R.; Budd, S.G.; Shin, Y.; Kons, Z.A.; Costanzo, R.M. Quality of life and safety impact of COVID-19 associated smell and taste disturbances. Am. J. Otolaryngol. 2021, 42, 103001. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.V.; Reiter, E.R.; DiNardo, L.J.; Costanzo, R.M. Hazardous Events Associated with Impaired Olfactory Function. Arch. Otolaryngol. Head Neck Surg. 2004, 130, 317–319. [Google Scholar] [CrossRef] [PubMed]
- Blomkvist, A.; Hofer, M. Olfactory Impairment and Close Social Relationships. A Narrat. Review. Chem. Senses 2021, 46, bjab037. [Google Scholar] [CrossRef] [PubMed]
- Hummel, T.; Nordin, S. Olfactory disorders and their consequences for quality of life. Acta Otolaryngol. 2005, 125, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Imam, S.A.; Lao, W.P.; Reddy, P.; Nguyen, S.A.; Schlosser, R.J. Is SARS-CoV-2 (COVID-19) postviral olfactory dysfunction (PVOD) different from other PVOD? World J. Otorhinolaryngol. Head Neck Surg. 2020, 6, S26–S32. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, M.; Aiba, T.; Mori, J.; Nakai, Y. An Epidemiological Study of Postviral Olfactory Disorder. Acta Otolaryngol. 1998, 118, 191–196. [Google Scholar] [CrossRef]
- Gemmati, D.; Bramanti, B.; Serino, M.L.; Secchiero, P.; Zauli, G.; Tisato, V. COVID-19 and individual genetic susceptibility/receptivity: Role of ACE1/ACE2 genes, immunity, inflammation and coagulation. might the double x-chromosome in females be protective against SARS-COV-2 compared to the single x-chromosome in males? Int. J. Mol. Sci. 2020, 21, 3474. [Google Scholar] [CrossRef]
| Demographic Data and Comorbidities | Total, N = 153 | Females, N = 111 | Males, N = 42 |
|---|---|---|---|
| Age | |||
| Mean (SD) | 49.2 (14.3) | 51.1 (14.0) | 44.2 (14.2) |
| Range | 18.0, 90.0 | 18.0, 90.0 | 20.0, 76.0 |
| Hypertension | |||
| No | 131 (87.3%) | 95 (87.2%) | 36 (87.8%) |
| Yes | 19 (12.7%) | 14 (12.8%) | 5 (12.2%) |
| Not reported | 3 | 2 | 1 |
| Diabetes | |||
| No | 147 (97.4%) | 106 (96.4%) | 41 (100%) |
| Yes | 4 (2.6%) | 4 (3.6%) | 0 (0%) |
| Not reported | 2 | 1 | 1 |
| Cardiovascular disease | |||
| No | 143 (94.7%) | 104 (94.5%) | 39 (95.1%) |
| Yes | 8 (5.3%) | 6 (5.5%) | 2 (4.9%) |
| Not reported | 2 | 1 | 1 |
| Active cancer | |||
| No | 150 (99.3%) | 109 (99.1%) | 41 (100.0%) |
| Yes | 1 (0.7%) | 1 (0.9%) | 0 (0%) |
| Not reported | 2 | 1 | 1 |
| Chronic respiratory disease | |||
| No | 148 (98.0%) | 109 (99.1%) | 39 (95.1%) |
| Yes | 3 (2.0%) | 1 (0.9%) | 2 (4.9%) |
| Not reported | 2 | 1 | 1 |
| Olfactory Performance Data | Total, N = 153 | Females, N = 111 | Males, N = 42 |
|---|---|---|---|
| Odor Threshold Test | |||
| Mean (SD) | 4.5 (2.9) | 4.8 (2.8) | 3.9 (3.2) |
| Range | 0.0, 11.3 | 0.0, 11.3 | 0.0, 10.5 |
| Odor Discrimination Test | |||
| Mean (SD) | 9.4 (2.7) | 9.5 (2.6) | 9.1 (2.7) |
| Range | 2.0, 14.0 | 2.0, 14.0 | 4.0, 14.0 |
| Odor Identification Test | |||
| Mean (SD) | 9.4 (3.2) | 9.5 (3.3) | 9.0 (3.0) |
| Range | 1.0, 16.0 | 1.0, 16.0 | 3.0, 15.0 |
| Combined TDI Score | |||
| Mean (SD) | 23.3 (7.0) | 23.8 (6.9) | 22.0 (7.1) |
| Range | 4.0, 37.5 | 4.0, 35.5 | 7.0, 37.5 |
| Group | Olfactory Function Test | Gene | β Gene | Adjusted p-Value |
|---|---|---|---|---|
| Female | Odor Discrimination Test | ACE2 | −8.21 | 0.03 |
| Female | Combined TDI Score | ACE2 | −21.70 | 0.03 |
| Group | Olfactory Function Test | Gene | β Gene | Adjusted p-Value |
|---|---|---|---|---|
| Male | Odor Discrimination Test | IFI44 | 0.39 | 0.01 |
| Male | Odor Discrimination Test | NDUFAF4 | 0.26 | 0.004 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spedicati, B.; Pecori, A.; Concas, M.P.; Santin, A.; Ruberto, R.; Nardone, G.G.; D’Alessandro, A.; Tirelli, G.; Boscolo-Rizzo, P.; Girotto, G. Scent of COVID-19: Whole-Genome Sequencing Analysis Reveals the Role of ACE2, IFI44, and NDUFAF4 in Long-Lasting Olfactory Dysfunction. Life 2025, 15, 56. https://doi.org/10.3390/life15010056
Spedicati B, Pecori A, Concas MP, Santin A, Ruberto R, Nardone GG, D’Alessandro A, Tirelli G, Boscolo-Rizzo P, Girotto G. Scent of COVID-19: Whole-Genome Sequencing Analysis Reveals the Role of ACE2, IFI44, and NDUFAF4 in Long-Lasting Olfactory Dysfunction. Life. 2025; 15(1):56. https://doi.org/10.3390/life15010056
Chicago/Turabian StyleSpedicati, Beatrice, Alessandro Pecori, Maria Pina Concas, Aurora Santin, Romina Ruberto, Giuseppe Giovanni Nardone, Andrea D’Alessandro, Giancarlo Tirelli, Paolo Boscolo-Rizzo, and Giorgia Girotto. 2025. "Scent of COVID-19: Whole-Genome Sequencing Analysis Reveals the Role of ACE2, IFI44, and NDUFAF4 in Long-Lasting Olfactory Dysfunction" Life 15, no. 1: 56. https://doi.org/10.3390/life15010056
APA StyleSpedicati, B., Pecori, A., Concas, M. P., Santin, A., Ruberto, R., Nardone, G. G., D’Alessandro, A., Tirelli, G., Boscolo-Rizzo, P., & Girotto, G. (2025). Scent of COVID-19: Whole-Genome Sequencing Analysis Reveals the Role of ACE2, IFI44, and NDUFAF4 in Long-Lasting Olfactory Dysfunction. Life, 15(1), 56. https://doi.org/10.3390/life15010056







