Cell Immortality: In Vitro Effective Techniques to Achieve and Investigate Its Applications and Challenges
Abstract
1. Introduction
2. The Application of Immortalized Cell Lines
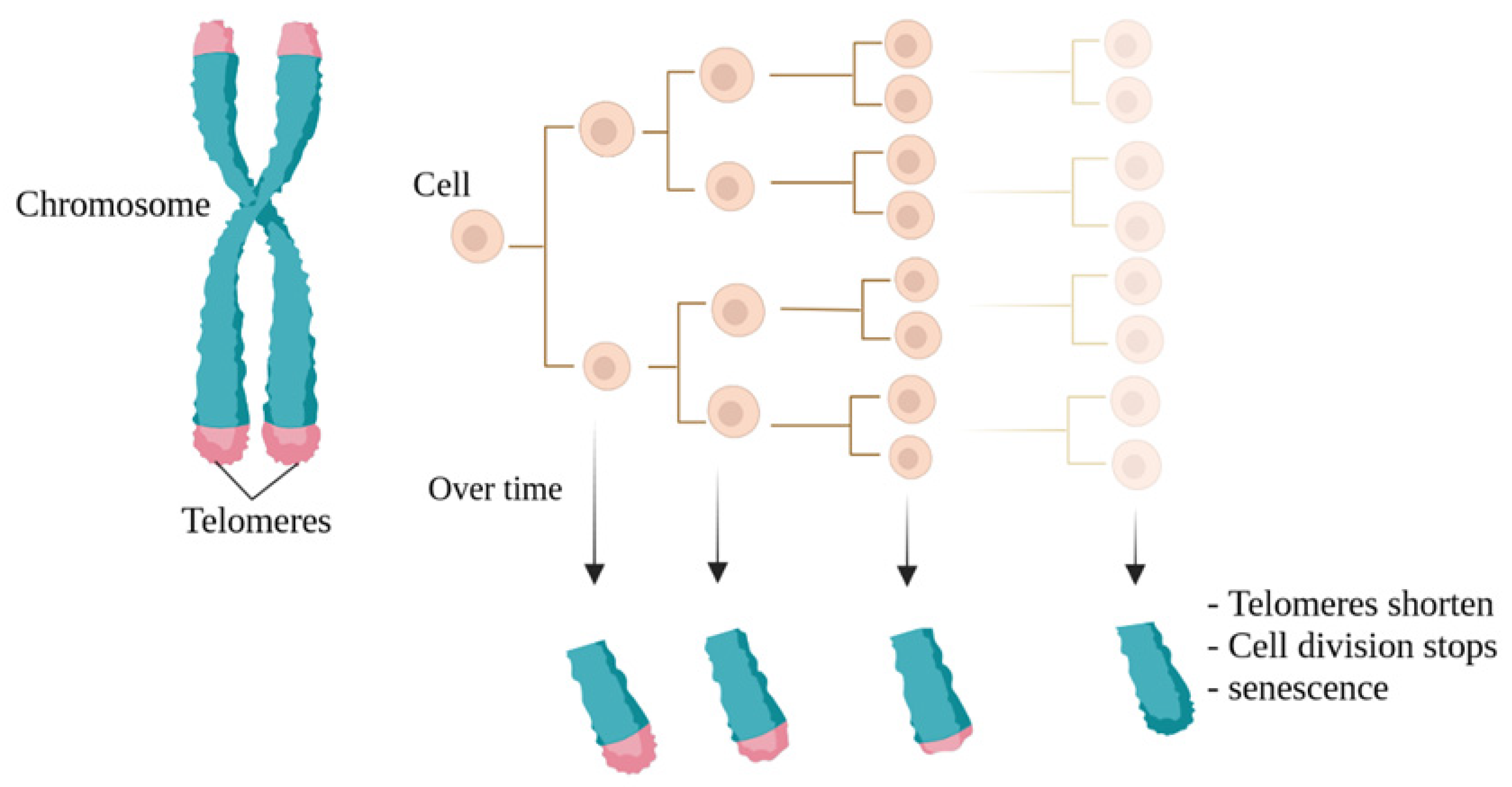
3. Telomeres
4. Immortality and a Review of Its Techniques
4.1. Viral Genes Can Control the Cell Cycle
4.1.1. Simian Virus 40
4.1.2. Human Papillomavirus (HPV)
4.1.3. Human T-Lymphotropic Virus (HTLV)
4.1.4. Adenoviruses
4.1.5. Epstein–Barr Virus (EBV)
4.2. The Overexpression of Specific Genes for Immortalization
4.2.1. The HOX Gene Family and Lhx2
4.2.2. c-Myc Gene Expression
4.2.3. CDK4 Gene Expression
4.2.4. TERT Gene Expression
4.3. Cancer Cells and Immortalization
4.4. Combining Methods
4.5. Chemical Components and Rays Role in Immortalization
4.6. Gene Manipulation and Immortalization
5. Challenges of Using Cell Lines
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hayflick, L. The Limited In Vitro Lifetime of Human Diploid Cell Strains. Exp. Cell Res. 1965, 37, 614–636. [Google Scholar] [CrossRef] [PubMed]
- Hayflick, L.; Moorhead, P.S. The Serial Cultivation of Human Diploid Cell strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Mancera, P.A.; Young, A.R.; Narita, M. Inside and out: The activities of senescence in cancer. Nat. Rev. Cancer 2014, 14, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, E.H. Telomeres: No end in sight. Cell 1994, 77, 621–623. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Funk, W.D.; Wang, S.-S.; Weinrich, S.L.; Avilion, A.A.; Chiu, C.-P.; Adams, R.R.; Chang, E.; Allsopp, R.C.; Yu, J. The RNA component of human telomerase. Science 1995, 269, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres shorten during ageing of human fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Shay, J.W.; Wright, W.E. Senescence and immortalization: Role of telomeres and telomerase. Carcinogenesis 2005, 26, 867–874. [Google Scholar] [CrossRef]
- Bajpai, V.K.; Andreadis, S.T. Stem cell sources for vascular tissue engineering and regeneration. Tissue Eng. Part. B Rev. 2012, 18, 405–425. [Google Scholar] [CrossRef]
- Addis, R.; Campesi, I.; Fois, M.; Capobianco, G.; Dessole, S.; Fenu, G.; Montella, A.; Cattaneo, M.G.; Vicentini, L.M.; Franconi, F. Human umbilical endothelial cells (HUVECs) have a sex: Characterisation of the phenotype of male and female cells. Biol. Sex. Differ. 2014, 5, 18. [Google Scholar] [CrossRef]
- Lorenz, M.; Blaschke, B.; Benn, A.; Hammer, E.; Witt, E.; Kirwan, J.; Fritsche-Guenther, R.; Gloaguen, Y.; Bartsch, C.; Vietzke, A. Sex-specific metabolic and functional differences in human umbilical vein endothelial cells from twin pairs. Atherosclerosis 2019, 291, 99–106. [Google Scholar] [CrossRef]
- Lorenz, M.; Koschate, J.; Kaufmann, K.; Kreye, C.; Mertens, M.; Kuebler, W.M.; Baumann, G.; Gossing, G.; Marki, A.; Zakrzewicz, A. Does cellular sex matter? Dimorphic transcriptional differences between female and male endothelial cells. Atherosclerosis 2015, 240, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lingappan, K. Differential sex-specific effects of oxygen toxicity in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 2017, 486, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Kulshreshtha, A.; Agrawal, A. Regulation by non-coding RNAs in respiratory disorders. In RNA-Based Regulation in Human Health and Disease; Elsevier: Amsterdam, The Netherlands, 2020; pp. 233–249. [Google Scholar]
- Gomez-Lechon, M.; Donato, M.; Castell, J.; Jover, R. Human hepatocytes as a tool for studying toxicity and drug metabolism. Curr. Drug Metab. 2003, 4, 292–312. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, C. Development of new cell lines for animal cell biotechnology. Crit. Rev. Biotechnol. 1990, 10, 155–178. [Google Scholar] [CrossRef]
- Schurr, M.J.; Foster, K.N.; Centanni, J.M.; Comer, A.R.; Wicks, A.; Gibson, A.L.; Thomas-Virnig, C.L.; Schlosser, S.J.; Faucher, L.D.; Lokuta, M.A. Phase I/II clinical evaluation of StrataGraft: A consistent, pathogen-free human skin substitute. J. Trauma 2009, 66, 866. [Google Scholar] [CrossRef] [PubMed]
- Masters, J.R. HeLa cells 50 years on: The good, the bad and the ugly. Nat. Rev. Cancer 2002, 2, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Svalastog, A.L.; Martinelli, L. Representing life as opposed to being: The bio-objectification process of the HeLa cells and its relation to personalized medicine. Croat. Med. J. 2013, 54, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Adey, A.; Burton, J.N.; Kitzman, J.O.; Hiatt, J.B.; Lewis, A.P.; Martin, B.K.; Qiu, R.; Lee, C.; Shendure, J. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature 2013, 500, 207–211. [Google Scholar] [CrossRef]
- Robin, T.; Bairoch, A.; Müller, M.; Lisacek, F.; Lane, L. Large-Scale Reanalysis of Publicly Available HeLa Cell Proteomics Data in the Context of the Human Proteome Project. J. Proteome Res. 2018, 17, 4160–4170. [Google Scholar] [CrossRef]
- Zhang, P.; Zhao, X.; Zhang, W.; He, A.; Lei, B.; Zhang, W.; Chen, Y. Leukemia-associated gene MLAA-34 reduces arsenic trioxide-induced apoptosis in HeLa cells via activation of the Wnt/β-catenin signaling pathway. PLoS ONE 2017, 12, e0186868. [Google Scholar] [CrossRef]
- Sağlam Metiner, P.; Can, H.; Ayyıldız Tamiş, D.; Karakavuk, M.; Kımız Geboloğlu, I.; Gülçe İz, S.; Atalay Şahar, E.; Değirmenci Döşkaya, A.; Gürüz, Y.; Deliloğlu Gürhan, S.; et al. The use of Toxoplasma gondii tachyzoites produced in HeLa cells adhered to Cytodex 1 microcarriers as antigen in serological assays: An application of microcarrier technology. Cytotechnology 2019, 71, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Olea-Flores, M.; Kan, J.; Carlson, A.; Syed, S.A.; McCann, C.; Mondal, V.; Szady, C.; Ricker, H.M.; McQueen, A.; Navea, J.G.; et al. ZIP11 Regulates Nuclear Zinc Homeostasis in HeLa Cells and Is Required for Proliferation and Establishment of the Carcinogenic Phenotype. Front. Cell Dev. Biol. 2022, 10, 895433. [Google Scholar] [CrossRef] [PubMed]
- Ivanković, M.; Cukusić, A.; Gotić, I.; Skrobot, N.; Matijasić, M.; Polancec, D.; Rubelj, I. Telomerase activity in HeLa cervical carcinoma cell line proliferation. Biogerontology 2007, 8, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Scherer, W.F.; Syverton, J.T.; Gey, G.O. Studies on the propagation in vitro of poliomyelitis viruses. IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J. Exp. Med. 1953, 97, 695–710. [Google Scholar] [CrossRef] [PubMed]
- Landry, J.J.; Pyl, P.T.; Rausch, T.; Zichner, T.; Tekkedil, M.M.; Stütz, A.M.; Jauch, A.; Aiyar, R.S.; Pau, G.; Delhomme, N.; et al. The genomic and transcriptomic landscape of a HeLa cell line. G3 2013, 3, 1213–1224. [Google Scholar] [CrossRef]
- Irfan Maqsood, M.; Matin, M.M.; Bahrami, A.R.; Ghasroldasht, M.M. Immortality of cell lines: Challenges and advantages of establishment. Cell Biol. Int. 2013, 37, 1038–1045. [Google Scholar] [CrossRef] [PubMed]
- Geller, H.M.; Quiñones-Jenab, V.; Poltorak, M.; Freed, W.J. Applications of immortalized cells in basic and clinical neurology. J. Cell Biochem. 1991, 45, 279–283. [Google Scholar] [CrossRef]
- Yoon, Y.; Kim, H.S.; Jeon, I.; Noh, J.-E.; Park, H.J.; Lee, S.; Park, I.-H.; Stevanato, L.; Hicks, C.; Corteling, R. Implantation of the clinical-grade human neural stem cell line, CTX0E03, rescues the behavioral and pathological deficits in the quinolinic acid-lesioned rodent model of Huntington’s disease. Stem Cells 2020, 38, 936–947. [Google Scholar] [CrossRef]
- Chen, X.; Lungova, V.; Zhang, H.; Mohanty, C.; Kendziorski, C.; Thibeault, S.L. Novel immortalized human vocal fold epithelial cell line: In vitro tool for mucosal biology. FASEB J. 2021, 35, e21243. [Google Scholar] [CrossRef]
- Hindul, N.L.; Jhita, A.; Oprea, D.G.; Hussain, T.A.; Gonchar, O.; Campillo, M.A.M.; O’Regan, L.; Kanemaki, M.T.; Fry, A.M.; Hirota, K. Construction of a human hTERT RPE-1 cell line with inducible Cre for editing of endogenous genes. Biol. Open 2022, 11, bio059056. [Google Scholar] [CrossRef]
- Hong, H.X.; Zhang, Y.M.; Xu, H.; Su, Z.Y.; Sun, P. Immortalization of swine umbilical vein endothelial cells with human telomerase reverse transcriptase. Mol. Cells 2007, 24, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Lathuiliere, A.; Vernet, R.; Charrier, E.; Urwyler, M.; Von Rohr, O.; Belkouch, M.-C.; Saingier, V.; Bouvarel, T.; Guillarme, D.; Engel, A. Immortalized human myoblast cell lines for the delivery of therapeutic proteins using encapsulated cell technology. Mol. Ther. Methods Clin. Dev. 2022, 26, 441–458. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Shay, J.W.; Minna, J.D. Immortalized normal human lung epithelial cell models for studying lung cancer biology. Respir. Investig. 2020, 58, 344–354. [Google Scholar] [CrossRef]
- Zhuang, Y.; Grainger, J.M.; Vedell, P.T.; Yu, J.; Moyer, A.M.; Gao, H.; Fan, X.Y.; Qin, S.; Liu, D.; Kalari, K.R.; et al. Establishment and characterization of immortalized human breast cancer cell lines from breast cancer patient-derived xenografts (PDX). NPJ Breast Cancer 2021, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Richardson, R.M.; Nguyen, B.; Holt, S.E.; Broaddus, W.C.; Fillmore, H.L. Ectopic telomerase expression inhibits neuronal differentiation of NT2 neural progenitor cells. Neurosci. Lett. 2007, 421, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Yasuhara, T.; Maki, M.; Matsukawa, N.; Masuda, T.; Yu, S.J.; Ali, M.; Yu, G.; Xu, L.; Kim, S.U. Neural progenitor NT2N cell lines from teratocarcinoma for transplantation therapy in stroke. Prog. Neurobiol. 2008, 85, 318–334. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Sugimoto, N.; Eto, K. Ex vivo generation of platelet products from human iPS cells. Inflamm. Regen. 2020, 40, 30. [Google Scholar] [CrossRef] [PubMed]
- Niu, N.; Wang, L. In vitro human cell line models to predict clinical response to anticancer drugs. Pharmacogenomics 2015, 16, 273–285. [Google Scholar] [CrossRef]
- Soice, E.; Johnston, J. Immortalizing Cells for Human Consumption. Int. J. Mol. Sci. 2021, 22, 11660. [Google Scholar] [CrossRef]
- McClintock, B. The Behavior in Successive Nuclear Divisions of a Chromosome Broken at Meiosis. Proc. Natl. Acad. Sci. USA 1939, 25, 405–416. [Google Scholar] [CrossRef]
- Muller, H.J. The remaking of chromosomes. Collect. Net. 1938, 8, 198. [Google Scholar]
- Aubert, G.; Lansdorp, P.M. Telomeres and aging. Physiol. Rev. 2008, 88, 557–579. [Google Scholar] [CrossRef] [PubMed]
- Young, A.J. The role of telomeres in the mechanisms and evolution of life-history trade-offs and ageing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20160452. [Google Scholar] [CrossRef] [PubMed]
- Arnoult, N.; Karlseder, J. Complex interactions between the DNA-damage response and mammalian telomeres. Nat. Struct. Mol. Biol. 2015, 22, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Denchi, E.L.; de Lange, T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 2007, 448, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.-R. A highly conserved repetitive DNA sequence,(TTAGGG) n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626. [Google Scholar] [CrossRef] [PubMed]
- Baird, D.M.; Rowson, J.; Wynford-Thomas, D.; Kipling, D. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat. Genet. 2003, 33, 203–207. [Google Scholar] [CrossRef]
- Lansdorp, P.M.; Verwoerd, N.P.; Van De Rijke, F.M.; Dragowska, V.; Little, M.-T.; Dirks, R.W.; Raap, A.K.; Tanke, H.J. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 1996, 5, 685–691. [Google Scholar] [CrossRef]
- Zijlmans, J.M.J.; Martens, U.M.; Poon, S.S.; Raap, A.K.; Tanke, H.J.; Ward, R.K.; Lansdorp, P.M. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl. Acad. Sci. USA 1997, 94, 7423–7428. [Google Scholar] [CrossRef]
- Britt-Compton, B.; Rowson, J.; Locke, M.; Mackenzie, I.; Kipling, D.; Baird, D.M. Structural stability and chromosome-specific telomere length is governed by cis-acting determinants in humans. Hum. Mol. Genet. 2006, 15, 725–733. [Google Scholar] [CrossRef]
- Martens, U.M.; Zijlmans, J.M.J.; Poon, S.S.; Dragowska, W.; Yui, J.; Chavez, E.A.; Ward, R.K.; Lansdorp, P.M. Short telomeres on human chromosome 17p. Nat. Genet. 1998, 18, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Olovnikov, A.M. A theory of marginotomy: The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 1973, 41, 181–190. [Google Scholar] [CrossRef]
- Watson, J.D. Origin of concatemeric T7DNA. Nat. New Biol. 1972, 239, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Cooke, H.J.; Smith, B. Variability at the telomeres of the human X/Y pseudoautosomal region. In Cold Spring Harbor Symposia on Quantitative Biology; Cold Spring Harbor Laboratory Press: Long Island City, NY, USA, 2019; pp. 213–219. [Google Scholar]
- Greider, C.W.; Blackburn, E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985, 43, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Counter, C.M.; Avilion, A.A.; LeFeuvre, C.E.; Stewart, N.G.; Greider, C.W.; Harley, C.B.; Bacchetti, S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992, 11, 1921–1929. [Google Scholar] [CrossRef] [PubMed]
- Hemmatzadeh, F.; Keyvanfar, H.; Hasan, N.H.; Niap, F.; Bani Hassan, E.; Hematzade, A.; Ebrahimie, E.; McWhorter, A.; Ignjatovic, J. Interaction between Bovine leukemia virus (BLV) infection and age on telomerase misregulation. Vet. Res. Commun. 2015, 39, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Hastie, N.D.; Dempster, M.; Dunlop, M.G.; Thompson, A.M.; Green, D.K.; Allshire, R.C. Telomere reduction in human colorectal carcinoma and with ageing. Nature 1990, 346, 866–868. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, S.; Yan, Z.; Pei, M. A prospect of cell immortalization combined with matrix microenvironmental optimization strategy for tissue engineering and regeneration. Cell Biosci. 2019, 9, 7. [Google Scholar] [CrossRef]
- de Bardet, J.C.; Cardentey, C.R.; González, B.L.; Patrone, D.; Mulet, I.L.; Siniscalco, D.; Robinson-Agramonte, M.L.A. Cell Immortalization: In Vivo Molecular Bases and In Vitro Techniques for Obtention. BioTech 2023, 12, 14. [Google Scholar] [CrossRef]
- Choi, M.; Lee, C. Immortalization of primary keratinocytes and its application to skin research. Biomol. Ther. 2015, 23, 391. [Google Scholar] [CrossRef]
- Royds, J.a.; Iacopetta, B. p53 and disease: When the guardian angel fails. Cell Death Differ. 2006, 13, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Sanyal, S.; Bruzzone, R. Breaking Bad: How Viruses Subvert the Cell Cycle. Front. Cell Infect. Microbiol. 2018, 8, 396. [Google Scholar] [CrossRef] [PubMed]
- Batinac, T.; Gruber, F.; Lipozencić, J.; Zamolo-Koncar, G.; Stasić, A.; Brajac, I. Protein p53--structure, function, and possible therapeutic implications. Acta Dermatovenerol. Croat. 2003, 11, 225–230. [Google Scholar] [PubMed]
- Bryan, T.; Redder, R.R. SV40-lnduced Immortalization of Human Cells. Crit. Rev. Oncog. 1994, 5, 331–357. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, A.; McClellan, A.J.; Vartikar, J.; Marks, I.; Cantalupo, P.; Li, Y.; Whyte, P.; Rundell, K.; Brodsky, J.L.; Pipas, J.M. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol. Cell Biol. 1997, 17, 4761–4773. [Google Scholar] [CrossRef] [PubMed]
- Symonds, H.; Krall, L.; Remington, L.; Saenz-Robles, M.; Lowe, S.; Jacks, T.; Van Dyke, T. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell 1994, 78, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Orosz, D.E.; Woost, P.G.; Kolb, R.J.; Finesilver, M.B.; Jin, W.; Frisa, P.S.; Choo, C.-K.; Yau, C.-F.; Chan, K.-W.; Resnick, M.I. Growth, immortalization, and differentiation potential of normal adult human proximal tubule cells. Vitr. Cell. Dev. Biol. Anim. 2004, 40, 22–34. [Google Scholar] [CrossRef]
- Garcia-Mesa, Y.; Jay, T.R.; Checkley, M.A.; Luttge, B.; Dobrowolski, C.; Valadkhan, S.; Landreth, G.E.; Karn, J.; Alvarez-Carbonell, D. Immortalization of primary microglia: A new platform to study HIV regulation in the central nervous system. J. Neurovirol. 2017, 23, 47–66. [Google Scholar] [CrossRef]
- Darimont, C.; Macé, K. Immortalization of human preadipocytes. Biochimie 2003, 85, 1231–1233. [Google Scholar] [CrossRef]
- Longworth, M.S.; Laimins, L.A. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol. Mol. Biol. Rev. 2004, 68, 362–372. [Google Scholar] [CrossRef]
- Motoyama, S.; Ladines-Llave, C.A.; Luis Villanueva, S.; Maruo, T. The role of human papilloma virus in the molecular biology of cervical carcinogenesis. Kobe J. Med. Sci. 2004, 50, 9–19. [Google Scholar]
- Münger, K.; Phelps, W.C.; Bubb, V.; Howley, P.M.; Schlegel, R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 1989, 63, 4417–4421. [Google Scholar] [CrossRef] [PubMed]
- Scheffner, M.; Werness, B.A.; Huibregtse, J.M.; Levine, A.J.; Howley, P.M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 1990, 63, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Dyson, N.; Howley, P.M.; Münger, K.; Harlow, E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 1989, 243, 934–937. [Google Scholar] [CrossRef] [PubMed]
- Trakarnsanga, K.; Griffiths, R.E.; Wilson, M.C.; Blair, A.; Satchwell, T.J.; Meinders, M.; Cogan, N.; Kupzig, S.; Kurita, R.; Nakamura, Y. An immortalized adult human erythroid line facilitates sustainable and scalable generation of functional red cells. Nat. Commun. 2017, 8, 14750. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.-Y.; Yang, W.; Lee, E.-j.; Han, G.H.; Cho, H.; Chay, D.B.; Kim, J.-h. Establishment of five immortalized human ovarian surface epithelial cell lines via SV40 T antigen or HPV E6/E7 expression. PLoS ONE 2018, 13, e0205297. [Google Scholar] [CrossRef] [PubMed]
- Sieburg, M.; Tripp, A.; Ma, J.W.; Feuer, G. Human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2 tax oncoproteins modulate cell cycle progression and apoptosis. J. Virol. 2004, 78, 10399–10409. [Google Scholar] [CrossRef][Green Version]
- Imai, M.; Higuchi, M.; Kawamura, H.; Yoshita, M.; Takahashi, M.; Oie, M.; Matsuki, H.; Tanaka, Y.; Aoyagi, Y.; Fujii, M. Human T cell leukemia virus type 2 (HTLV-2) Tax2 has a dominant activity over HTLV-1 Tax1 to immortalize human CD4+ T cells. Virus Genes. 2013, 46, 39–46. [Google Scholar] [CrossRef]
- Usman, N.; Suarez, M. Adenoviruses. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2022. [Google Scholar]
- Quinlan, M.P.; Douglas, J.L. Immortalization of primary epithelial cells requires first- and second-exon functions of adenovirus type 5 12S. J. Virol. 1992, 66, 2020–2030. [Google Scholar] [CrossRef]
- Cone, R.D.; Grodzicker, T.; Jaramillo, M. A retrovirus expressing the 12S adenoviral E1A gene product can immortalize epithelial cells from a broad range of rat tissues. Mol. Cell Biol. 1988, 8, 1036–1044. [Google Scholar] [CrossRef]
- Tosato, G.; Cohen, J.I. Generation of Epstein-Barr Virus (EBV)-immortalized B cell lines. Curr. Protoc. Immunol. 2007, 7, 7.22.1–7.22.4. [Google Scholar] [CrossRef] [PubMed]
- Yap, H.Y.; Siow, T.S.; Chow, S.K.; Teow, S.Y. Epstein-Barr Virus- (EBV-) Immortalized Lymphoblastoid Cell Lines (LCLs) Express High Level of CD23 but Low CD27 to Support Their Growth. Adv. Virol. 2019, 2019, 6464521. [Google Scholar] [CrossRef] [PubMed]
- Moosmann, A.; Khan, N.; Cobbold, M.; Zentz, C.; Delecluse, H.J.; Hollweck, G.; Hislop, A.D.; Blake, N.W.; Croom-Carter, D.; Wollenberg, B.; et al. B cells immortalized by a mini-Epstein-Barr virus encoding a foreign antigen efficiently reactivate specific cytotoxic T cells. Blood 2002, 100, 1755–1764. [Google Scholar] [CrossRef] [PubMed]
- Middleton, T.; Gahn, T.A.; Martin, J.M.; Sugden, B. Immortalizing genes of Epstein-Barr virus. Adv. Virus Res. 1991, 40, 19–55. [Google Scholar] [CrossRef]
- Hawley, R.G.; Hawley, T.S.; Cantor, A.B. TLX1 (HOX11) immortalization of embryonic stem cell-derived and primary murine hematopoietic progenitors. Curr. Protoc. Stem Cell Biol. 2008, 7, 1F.7.1–1F.7.19. [Google Scholar] [CrossRef]
- Perkins, A.C.; Cory, S. Conditional immortalization of mouse myelomonocytic, megakaryocytic and mast cell progenitors by the Hox-2.4 homeobox gene. EMBO J. 1993, 12, 3835–3846. [Google Scholar] [CrossRef]
- Calvo, K.R.; Sykes, D.B.; Pasillas, M.; Kamps, M.P. Hoxa9 immortalizes a granulocyte-macrophage colony-stimulating factor-dependent promyelocyte capable of biphenotypic differentiation to neutrophils or macrophages, independent of enforced meis expression. Mol. Cell Biol. 2000, 20, 3274–3285. [Google Scholar] [CrossRef][Green Version]
- Pinto do Ó, P.; Richter, K.; Carlsson, L. Hematopoietic progenitor/stem cells immortalized by Lhx2 generate functional hematopoietic cells in vivo. Blood 2002, 99, 3939–3946. [Google Scholar] [CrossRef]
- Marinković, D.; Marinković, T. c-Myc misregulation triggers complex process of genomic instability. Genet. Belgrade 2018, 50, 731–745. [Google Scholar] [CrossRef]
- Vafa, O.; Wade, M.; Kern, S.; Beeche, M.; Pandita, T.K.; Hampton, G.M.; Wahl, G.M. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: A mechanism for oncogene-induced genetic instability. Mol. Cell 2002, 9, 1031–1044. [Google Scholar] [CrossRef]
- Klapproth, K.; Sander, S.; Marinkovic, D.; Baumann, B.; Wirth, T. The IKK2/NF-κB pathway suppresses MYC-induced lymphomagenesis. Blood J. Am. Soc. Hematol. 2009, 114, 2448–2458. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, L.; Ferrari, D.; Rota Nodari, L.; Amati, B.; Snyder, E.; Vescovi, A.L. Immortalization of human neural stem cells with the c-myc mutant T58A. PLoS ONE 2008, 3, e3310. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Oganesyan, D.; Mooney, R.; Rong, X.; Christensen, M.J.; Shahmanyan, D.; Perrigue, P.M.; Benetatos, J.; Tsaturyan, L.; Aramburo, S. L-MYC expression maintains self-renewal and prolongs multipotency of primary human neural stem cells. Stem Cell Rep. 2016, 7, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Gil, J.; Kerai, P.; Lleonart, M.; Bernard, D.; Cigudosa, J.C.; Peters, G.; Carnero, A.; Beach, D. Immortalization of primary human prostate epithelial cells by c-Myc. Cancer Res. 2005, 65, 2179–2185. [Google Scholar] [CrossRef]
- Ramirez, R.D.; Sheridan, S.; Girard, L.; Sato, M.; Kim, Y.; Pollack, J.; Peyton, M.; Zou, Y.; Kurie, J.M.; DiMaio, J.M. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004, 64, 9027–9034. [Google Scholar] [CrossRef]
- Shiomi, K.; Kiyono, T.; Okamura, K.; Uezumi, M.; Goto, Y.; Yasumoto, S.; Shimizu, S.; Hashimoto, N. CDK4 and cyclin D1 allow human myogenic cells to recapture growth property without compromising differentiation potential. Gene Ther. 2011, 18, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Meyerson, M.; Counter, C.M.; Eaton, E.N.; Ellisen, L.W.; Steiner, P.; Caddle, S.D.; Ziaugra, L.; Beijersbergen, R.L.; Davidoff, M.J.; Liu, Q. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 1997, 90, 785–795. [Google Scholar] [CrossRef]
- Nakayama, J.-i.; Tahara, H.; Tahara, E.; Saito, M.; Ito, K.; Nakamura, H.; Nakanishi, T.; Nakanishi, E.; Ide, T.; Ishikawa, F. Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nat. Genet. 1998, 18, 65–68. [Google Scholar] [CrossRef]
- Cong, Y.-S.; Wen, J.; Bacchetti, S. The human telomerase catalytic subunit hTERT: Organization of the gene and characterization of the promoter. Hum. Mol. Genet. 1999, 8, 137–142. [Google Scholar] [CrossRef]
- Zvereva, M.; Shcherbakova, D.; Dontsova, O. Telomerase: Structure, functions, and activity regulation. Biochemistry 2010, 75, 1563–1583. [Google Scholar] [CrossRef]
- Harrington, L.; Zhou, W.; McPhail, T.; Oulton, R.; Yeung, D.S.; Mar, V.; Bass, M.B.; Robinson, M.O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997, 11, 3109–3115. [Google Scholar] [CrossRef] [PubMed]
- MacNeil, D.E.; Bensoussan, H.J.; Autexier, C. Telomerase regulation from beginning to the end. Genes 2016, 7, 64. [Google Scholar] [CrossRef] [PubMed]
- Beattie, T.L.; Zhou, W.; Robinson, M.O.; Harrington, L. Reconstitution of human telomerase activity in vitro. Curr. Biol. 1998, 8, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Weinrich, S.L.; Pruzan, R.; Ma, L.; Ouellette, M.; Tesmer, V.M.; Holt, S.E.; Bodnar, A.G.; Lichtsteiner, S.; Kim, N.W.; Trager, J.B.; et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 1997, 17, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, F. Regulation mechanisms of mammalian telomerase. A review. Biochemistry 1997, 62, 1332–1337. [Google Scholar] [PubMed]
- Ly, H. Telomere dynamics in induced pluripotent stem cells: Potentials for human disease modeling. World J. Stem Cells 2011, 3, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, C.; Teixeira, M.T.; Förstemann, K.; Lingner, J. Telomerase: Biochemical considerations for enzyme and substrate. Trends Biochem. Sci. 2002, 27, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes-Rojas, C.; Shippen, D.E. Telomerase regulation. Mutat. Res. 2012, 730, 20–27. [Google Scholar] [CrossRef]
- Depcrynski, A.N.; Sachs, P.C.; Elmore, L.W.; Holt, S.E. Regulation of Telomerase Through Transcriptional and Posttranslational Mechanisms. In Telomeres and Telomerase in Cancer; Humana Press: Totowa, NJ, USA, 2009; pp. 47–85. [Google Scholar]
- Dwyer, J.; Li, H.; Xu, D.; Liu, J.P. Transcriptional regulation of telomerase activity: Roles of the the Ets transcription factor family. Ann. N. Y. Acad. Sci. 2007, 1114, 36–47. [Google Scholar] [CrossRef]
- Counter, C.M.; Gupta, J.; Harley, C.B.; Leber, B.; Bacchetti, S. Telomerase activity in normal leukocytes and in hematologic malignancies. Blood 1995, 85, 2315–2320. [Google Scholar] [CrossRef]
- Broccoli, D.; Young, J.W.; de Lange, T. Telomerase activity in normal and malignant hematopoietic cells. Proc. Natl. Acad. Sci. USA 1995, 92, 9082–9086. [Google Scholar] [CrossRef]
- Trybek, T.; Kowalik, A.; Gozdz, S.; Kowalska, A. Telomeres and telomerase in oncogenesis. Oncol. Lett. 2020, 20, 1015–1027. [Google Scholar] [CrossRef] [PubMed]
- Palanca-Wessels, M.C.; Barrett, M.T.; Galipeau, P.C.; Rohrer, K.L.; Reid, B.J.; Rabinovitch, P.S. Genetic analysis of long-term Barrett’s esophagus epithelial cultures exhibiting cytogenetic and ploidy abnormalities. Gastroenterology 1998, 114, 295–304. [Google Scholar] [CrossRef]
- Rambhatla, L.; Chiu, C.-P.; Glickman, R.D.; Rowe-Rendleman, C. In vitro differentiation capacity of telomerase immortalized human RPE cells. Investig. Ophthalmol. Vis. Sci. 2002, 43, 1622–1630. [Google Scholar]
- Arbiser, J.L.; Yeung, R.; Weiss, S.W.; Arbiser, Z.K.; Amin, M.B.; Cohen, C.; Frank, D.; Mahajan, S.; Herron, G.S.; Yang, J. The generation and characterization of a cell line derived from a sporadic renal angiomyolipoma: Use of telomerase to obtain stable populations of cells from benign neoplasms. Am. J. Pathol. 2001, 159, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Harada, H.; Nakagawa, H.; Oyama, K.; Takaoka, M.; Andl, C.D.; Jacobmeier, B.; von Werder, A.; Enders, G.H.; Opitz, O.G.; Rustgi, A.K. Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation. Mol. Cancer Res. 2003, 1, 729–738. [Google Scholar]
- Thi, M.M.; Urban-Maldonado, M.; Spray, D.C.; Suadicani, S.O. Characterization of hTERT-immortalized osteoblast cell lines generated from wild-type and connexin43-null mouse calvaria. Am. J. Physiol. Cell Physiol. 2010, 299, C994–C1006. [Google Scholar] [CrossRef]
- Lin, H.; Mensch, J.; Haschke, M.; Jäger, K.; Köttgen, B.; Dernedde, J.; Orsó, E.; Walter, M. Establishment and Characterization of hTERT Immortalized Hutchinson–Gilford Progeria Fibroblast Cell Lines. Cells 2022, 11, 2784. [Google Scholar] [CrossRef]
- Yang, J.; Chang, E.; Cherry, A.M.; Bangs, C.D.; Oei, Y.; Bodnar, A.; Bronstein, A.; Chiu, C.-P.; Herron, G.S. Human endothelial cell life extension by telomerase expression. J. Biol. Chem. 1999, 274, 26141–26148. [Google Scholar] [CrossRef]
- Lee, K.M.; Choi, K.H.; Ouellette, M.M. Use of exogenous hTERT to immortalize primary human cells. Cytotechnology 2004, 45, 33–38. [Google Scholar] [CrossRef]
- Toouli, C.D.; Huschtscha, L.I.; Neumann, A.A.; Noble, J.R.; Colgin, L.M.; Hukku, B.; Reddel, R.R. Comparison of human mammary epithelial cells immortalized by simian virus 40 T-Antigen or by the telomerase catalytic subunit. Oncogene 2002, 21, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Dalerba, P.; Guiducci, C.; Poliani, P.L.; Cifola, I.; Parenza, M.; Frattini, M.; Gallino, G.; Carnevali, I.; Di Giulio, I.; Andreola, S. Reconstitution of human telomerase reverse transcriptase expression rescues colorectal carcinoma cells from in vitro senescence: Evidence against immortality as a constitutive trait of tumor cells. Cancer Res. 2005, 65, 2321–2329. [Google Scholar] [CrossRef]
- Jaiswal, K.; Morales, C.; Feagins, L.; Gandia, K.; Zhang, X.; Zhang, H.-Y.; Hormi-Carver, K.; Shen, Y.; Elder, F.; Ramirez, R. Characterization of telomerase-immortalized, non-neoplastic, human Barrett’s cell line (BAR-T). Dis. Esophagus 2007, 20, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Harley, C.B. Telomerase is not an oncogene. Oncogene 2002, 21, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Earle, W.R. Production of malignancy in vitro. J. Natl. Cancer Inst. 1943, 4, 165–212. [Google Scholar]
- Gey, G. Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Res. 1952, 12, 264–265. [Google Scholar]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef] [PubMed]
- La Porta, C.A.M.; Zapperi, S. Explaining the dynamics of tumor aggressiveness: At the crossroads between biology, artificial intelligence and complex systems. Semin. Cancer Biol. 2018, 53, 42–47. [Google Scholar] [CrossRef]
- Wang, L.; Hou, Y.; Sun, Y.; Zhao, L.; Tang, X.; Hu, P.; Yang, J.; Zeng, Z.; Yang, G.; Cui, X.; et al. c-Ski activates cancer-associated fibroblasts to regulate breast cancer cell invasion. Mol. Oncol. 2013, 7, 1116–1128. [Google Scholar] [CrossRef]
- Karakasheva, T.A.; Lin, E.W.; Tang, Q.; Qiao, E.; Waldron, T.J.; Soni, M.; Klein-Szanto, A.J.; Sahu, V.; Basu, D.; Ohashi, S. IL-6 Mediates Cross-Talk between Tumor Cells and Activated Fibroblasts in the Tumor MicroenvironmentTargeting IL6R Signaling in Upper-GI Cancers. Cancer Res. 2018, 78, 4957–4970. [Google Scholar] [CrossRef]
- Tang, X.; Tu, G.; Yang, G.; Wang, X.; Kang, L.; Yang, L.; Zeng, H.; Wan, X.; Qiao, Y.; Cui, X. Autocrine TGF-β1/miR-200s/miR-221/DNMT3B regulatory loop maintains CAF status to fuel breast cancer cell proliferation. Cancer Lett. 2019, 452, 79–89. [Google Scholar] [CrossRef]
- Yang, G.; Rosen, D.G.; Mercado-Uribe, I.; Colacino, J.A.; Mills, G.B.; Bast, R.C., Jr.; Zhou, C.; Liu, J. Knockdown of p53 combined with expression of the catalytic subunit of telomerase is sufficient to immortalize primary human ovarian surface epithelial cells. Carcinogenesis 2007, 28, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.M.; Ng, W.M.; Tan, H.S.; Vinitha, D.; Yang, Z.; Fan, J.B.; Zou, Y.; Hui, J.H.; Lee, E.H.; Lim, B. Molecular basis of immortalization of human mesenchymal stem cells by combination of p53 knockdown and human telomerase reverse transcriptase overexpression. Stem Cells Dev. 2013, 22, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Casillas, M.A.; Brotherton, S.L.; Andrews, L.G.; Ruppert, J.M.; Tollefsbol, T.O. Induction of endogenous telomerase (hTERT) by c-Myc in WI-38 fibroblasts transformed with specific genetic elements. Gene 2003, 316, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Song, X.; Wang, Y.; Liu, F.; Wei, J. Combining oncolytic viruses with cancer immunotherapy: Establishing a new generation of cancer treatment. Front. Immunol. 2020, 11, 683. [Google Scholar] [CrossRef] [PubMed]
- Carnero, A.; Blanco-Aparicio, C.; Kondoh, H.; Lleonart, M.E.; Martinez-Leal, J.F.; Mondello, C.; Scovassi, A.I.; Bisson, W.H.; Amedei, A.; Roy, R.; et al. Disruptive chemicals, senescence and immortality. Carcinogenesis 2015, 36 (Suppl. S1), S19–S37. [Google Scholar] [CrossRef]
- Semmrich, M.; Marchand, J.-B.; Fend, L.; Rehn, M.; Silvestre, N.; Mårtensson, L.; Foloppe, J.; Teige, I.; Quéméneur, E.; Frendeus, B. 594 BT-001, an oncolytic vaccinia virus armed with a Treg-depleting human recombinant anti-CTLA4 antibody and GM-CSF to target the tumor microenvironment. J. Immunother. Cancer 2020, 8, A356. [Google Scholar] [CrossRef]
- Khoshdel-Rad, N.; Zahmatkesh, E.; Moeinvaziri, F.; Haghparast, N.; Baharvand, H.; Aghdami, N.; Moghadasali, R. Promoting maturation of human pluripotent stem cell-derived renal microtissue by incorporation of endothelial and mesenchymal cells. Stem Cells Dev. 2021, 30, 428–440. [Google Scholar] [CrossRef]
- Newbold, R.F.; Warren, W.; Medcalf, A.S.; Amos, J. Mutagenicity of carcinogenic methylating agents is associated with a specific DNA modification. Nature 1980, 283, 596–599. [Google Scholar] [CrossRef]
- Yasaei, H.; Gilham, E.; Pickles, J.C.; Roberts, T.P.; O’Donovan, M.; Newbold, R.F. Carcinogen-specific mutational and epigenetic alterations in INK4A, INK4B and p53 tumour-suppressor genes drive induced senescence bypass in normal diploid mammalian cells. Oncogene 2013, 32, 171–179. [Google Scholar] [CrossRef]
- Dimri, G.; Band, H.; Band, V. Mammary epithelial cell transformation: Insights from cell culture and mouse models. Breast Cancer Res. 2005, 7, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Bader, A.; Petters, O.; Keller, M.; Pavlica, S. Paracetamol treatment increases telomerase activity in rat embryonic liver cells. Pharmacol. Rep. 2011, 63, 1435–1441. [Google Scholar] [CrossRef] [PubMed]
- Tsuruga, Y.; Kiyono, T.; Matsushita, M.; Takahashi, T.; Kasai, H.; Matsumoto, S.; Todo, S. Establishment of immortalized human hepatocytes by introduction of HPV16 E6/E7 and hTERT as cell sources for liver cell-based therapy. Cell Transpl. 2008, 17, 1083–1094. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.; Mai, G.; Villiger, P.; Oberholzer, J.; Salmon, P.; Morel, P.; Bühler, L.; Trono, D. Treatment of acetaminophen-induced acute liver failure in the mouse with conditionally immortalized human hepatocytes. J. Hepatol. 2005, 43, 1031–1037. [Google Scholar] [CrossRef] [PubMed]
- Bode-Böger, S.M.; Martens-Lobenhoffer, J.; Täger, M.; Schröder, H.; Scalera, F. Aspirin reduces endothelial cell senescence. Biochem. Biophys. Res. Commun. 2005, 334, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, N.; Afshinpour, M.; Fakhr, S.S.; Kalkhoran, P.G.; Shadman-Manesh, V.; Adelian, S.; Beiranvand, S.; Rezaei-Tazangi, F.; Khorram, R.; Hamblin, M.R.; et al. Cancer stem cells in colorectal cancer: Signaling pathways involved in stemness and therapy resistance. Crit. Rev. Oncol. Hematol. 2023, 182, 103920. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Esvelt, K.M.; Church, G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods 2013, 10, 957–963. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Sánchez-Rivera, F.J.; Jacks, T. Applications of the CRISPR-Cas9 system in cancer biology. Nat. Rev. Cancer 2015, 15, 387–395. [Google Scholar] [CrossRef]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef]
- Zhang, H.; Yan, Z.; Li, M.; Peabody, M.; He, T.C. CRISPR clear? Dimeric Cas9-Fok1 nucleases improve genome-editing specificity. Genes. Dis. 2014, 1, 6–7. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, L.; Yu, X.; Zhang, R.; Yan, S.; Zeng, Z.; Shu, Y.; Zhao, C.; Wu, X.; Lei, J.; et al. CRISPR/Cas9-mediated reversibly immortalized mouse bone marrow stromal stem cells (BMSCs) retain multipotent features of mesenchymal stem cells (MSCs). Oncotarget 2017, 8, 111847–111865. [Google Scholar] [CrossRef] [PubMed]
- De Masi, C.; Spitalieri, P.; Murdocca, M.; Novelli, G.; Sangiuolo, F. Application of CRISPR/Cas9 to human-induced pluripotent stem cells: From gene editing to drug discovery. Hum. Genom. 2020, 14, 25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, L.; Zhang, J.; Wang, Y. Drug Inducible CRISPR/Cas Systems. Comput. Struct. Biotechnol. J. 2019, 17, 1171–1177. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.J.; Cho, J.; Kwak, J.; Sung, Y.H.; Kang, B.C. Immortalization of primary marmoset skin fibroblasts by CRISPR-Cas9-mediated gene targeting. Anim. Cells Syst. 2022, 26, 266–274. [Google Scholar] [CrossRef]
- Fleckenstein, E.; Uphoff, C.C.; Drexler, H.G. Effective treatment of mycoplasma contamination in cell lines with enrofloxacin (Baytril). Leukemia 1994, 8, 1424–1434. [Google Scholar] [PubMed]
- Hay, R.J.; Macy, M.L.; Chen, T.R. Mycoplasma infection of cultured cells. Nature 1989, 339, 487–488. [Google Scholar] [CrossRef]
- Zhao, M.; Sano, D.; Pickering, C.R.; Jasser, S.A.; Henderson, Y.C.; Clayman, G.L.; Sturgis, E.M.; Ow, T.J.; Lotan, R.; Carey, T.E.; et al. Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clin. Cancer Res. 2011, 17, 7248–7264. [Google Scholar] [CrossRef]
- MacLeod, R.A.; Drexler, H.G. Public repositories: Users reluctant to give materials. Nature 2006, 439, 912. [Google Scholar] [CrossRef]
- Lorsch, J.R.; Collins, F.S.; Lippincott-Schwartz, J. Cell Biology. Fixing problems with cell lines. Science 2014, 346, 1452–1453. [Google Scholar] [CrossRef]
- Nelson-Rees, W.A.; Daniels, D.W.; Flandermeyer, R.R. Cross-contamination of cells in culture. Science 1981, 212, 446–452. [Google Scholar] [CrossRef]
- Guo, L.; Wang, Z.; Li, J.; Li, J.; Cui, L.; Dong, J.; Meng, X.; Qian, C.; Wang, H. Immortalization effect of SV40T lentiviral vectors on canine corneal epithelial cells. BMC Vet. Res. 2022, 18, 181. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hu, R.; Wang, Z.; Guo, Y.; Wang, S.; Zou, H.; Peng, Q.; Jiang, Y. Establishment of Immortalized Yak Ruminal Epithelial Cell Lines by Lentivirus-Mediated SV40T and hTERT Gene Transduction. Oxid. Med. Cell Longev. 2022, 2022, 8128028. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Sang, Y.; Fang, C.; Shao, B.; Yang, L.; Yao, K.; Zhao, X.; Gou, J.; Wei, Y.; Yi, T. Immunotherapy of tumors with human telomerase reverse transcriptase immortalized human umbilical vein endothelial cells. Int. J. Oncol. 2015, 47, 1901–1911. [Google Scholar] [CrossRef]
- Abdallah, B.M.; Haack-Sørensen, M.; Burns, J.S.; Elsnab, B.; Jakob, F.; Hokland, P.; Kassem, M. Maintenance of differentiation potential of human bone marrow mesenchymal stem cells immortalized by human telomerase reverse transcriptase gene despite of extensive proliferation. Biochem. Biophys. Res. Commun. 2005, 326, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Giri, S.; Bader, A. Immortalization of Human Fetal Hepatocyte by Ectopic Expression of Human Telomerase Reverse Transcriptase, Human Papilloma Virus (E7) and Simian Virus 40 Large T (SV40 T) Antigen Towards Bioartificial Liver Support. J. Clin. Exp. Hepatol. 2014, 4, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Tanzil, G.; Arnold, P.; Gögele, C.; Hahn, J.; Breier, A.; Meyer, M.; Kohl, B.; Schröpfer, M.; Schwarz, S. SV40 Transfected Human Anterior Cruciate Ligament Derived Ligamentocytes-Suitable as a Human in Vitro Model for Ligament Reconstruction? Int. J. Mol. Sci. 2020, 21, 593. [Google Scholar] [CrossRef]
- Kaur, G.; Dufour, J.M. Cell lines: Valuable tools or useless artifacts. Spermatogenesis 2012, 2, 1–5. [Google Scholar] [CrossRef]



| Immortal Cell Type | The Method of Immortality | Strategy | The Effectiveness of the Method | Gene Transfer Method | Immortal Cell Characteristics | Immortality Result | Ref. | |
|---|---|---|---|---|---|---|---|---|
| 1 | Human vocal fold epithelial cells | Increased hTERT gene expression | Expression of the catalytic subunit of telomerase | Most effective and safe in most cell types | Retrovirus | They maintained their phenotypes with almost identical genotypes in cellular pathways and functioned properly in relation to ion and protein transport and cell signaling. They also maintained the ability of stable reproduction for more than 8 months. |
| [30] |
| 2 | Primary canine corneal epithelial cells | SV40 T antigen | Induction of viral oncogenes that inactivate cell cycle proteins | Usually effective but not safe (viral gene) | Lentivirus | They maintained their biological characteristics and had a stronger proliferation capacity than normal cells. They also maintained their diploid karyotype and serum-dependent ability. |
| [166] |
| 3 | Yak rumen epithelial cells | Increased hTERT gene expression + SV40 T antigen | Expression of the catalytic subunit of telomerase + Induction of viral oncogenes that inactivate cell cycle proteins | A combination of methods is usually effective, but there is caution in using viral genes | Lentivirus | They maintained the morphological and functional characteristics of the primary cells and also had functions related to the normal transport and absorption of short-chain fatty acids. Cell proliferation and karyotype were normal. |
| [167] |
| 4 | Human retinal pigment epithelial RPE-1 cells | Increased hTERT gene expression | Expression of the catalytic subunit of telomerase | Most effective and safe in most cell types | ERT2-Cre-ERT2AAVS1 integration plasmid | Life expectancy increased. It does not have transformed phenotypes and has a stable and normal karyotype. |
| [31] |
| 5 | HUVECs | Increased hTERT gene expression | Expression of the catalytic subunit of telomerase | Most effective and safe in most cell types | Lentivirus | Cells showed longer lifespan and maintained endothelial characteristics. They expressed the factors CD31, VEGFR-II, and alpha5 integrin. Antitumor immunity was also confirmed. |
| [168] |
| 6 | Human bone marrow mesenchymal stem cells | Increased hTERT gene expression | Expression of the catalytic subunit of telomerase | Most effective and safe in most cell types | Retrovirus | The restoration of telomerase activity, the increase in the life span of the cells, the characteristics of the stem cells of self-renewal, and the ability to differentiate into the mesoderm-type cell lineage were preserved. |
| [169] |
| 7 | Human fetal hepatocytes | Increased hTERT gene expression + SV40 T antigen + E7 | Expression of the catalytic subunit of telomerase + overexpression of certain genes for immortalization | A combination of methods is usually effective, but there is caution in using viral genes | Vector + E. coli plasmid | A stable cell line was obtained from human fetal liver cells that was able to secrete albumin–urea and consume glucose. |
| [170] |
| 8 | Human neural stem cells | Expression of the v-myc gene | Retrovirus | It produces stem cells with increased proliferation capacity and they do not change shape in laboratory conditions and are not tumorigenic in the body. |
| [95] | ||
| 9 | Ligamentocytes derived from human anterior cruciate ligament | SV40 T antigen | Induction of viral oncogenes that inactivate cell cycle proteins | Usually effective but not safe (viral gene) | Vector transfection | Transfected ligamentocytes maintain the phenotype and vital properties of the cell. |
| [171] |
| 10 | Bone marrow mesenchymal stromal cells | Increased hTERT gene expression | Expression of the catalytic subunit of telomerase | Most effective and safe in most cell types | Lentivirus | Proliferated longer than wild-type BMSCs. Phenotype and karyotype were not significantly different from non-transfected cells. The cells also maintained the normal morphology and neural differentiation characteristics of stem cells when cultured in induction media. |
| [156] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chalak, M.; Hesaraki, M.; Mirbahari, S.N.; Yeganeh, M.; Abdi, S.; Rajabi, S.; Hemmatzadeh, F. Cell Immortality: In Vitro Effective Techniques to Achieve and Investigate Its Applications and Challenges. Life 2024, 14, 417. https://doi.org/10.3390/life14030417
Chalak M, Hesaraki M, Mirbahari SN, Yeganeh M, Abdi S, Rajabi S, Hemmatzadeh F. Cell Immortality: In Vitro Effective Techniques to Achieve and Investigate Its Applications and Challenges. Life. 2024; 14(3):417. https://doi.org/10.3390/life14030417
Chicago/Turabian StyleChalak, Mahla, Mahdi Hesaraki, Seyedeh Nasim Mirbahari, Meghdad Yeganeh, Shaghayegh Abdi, Sarah Rajabi, and Farhid Hemmatzadeh. 2024. "Cell Immortality: In Vitro Effective Techniques to Achieve and Investigate Its Applications and Challenges" Life 14, no. 3: 417. https://doi.org/10.3390/life14030417
APA StyleChalak, M., Hesaraki, M., Mirbahari, S. N., Yeganeh, M., Abdi, S., Rajabi, S., & Hemmatzadeh, F. (2024). Cell Immortality: In Vitro Effective Techniques to Achieve and Investigate Its Applications and Challenges. Life, 14(3), 417. https://doi.org/10.3390/life14030417






