Three Biopolymers and Origin of Life Scenarios
Abstract
1. Introduction
2. Results
2.1. How Far Can the “Chemical Era” Go?
2.2. Prebiotic “Worlds”
2.2.1. RNA-Only
Unit of Evolution Content
Autonomous Advent
Evolutionary Prospect
Extant Perspective
Critical View
- Nonenzymatic replication—A paper from the Szostak group [53] maintains: “it is not yet possible to copy, in an effective and prebiotically plausible manner, RNA templates long enough to encode ribozymes that might enable RNA-catalyzed self-replication processes”. Moreover, the present nonenzymatic mechanisms seem to contain fundamental pitfalls; to achieve template generality in replication, a specific primer should randomly exist in the pool for each replicated oligonucleotide, a requirement whose probability of realization is infinitesimal. Additionally, the current rate obtained for monomers addition is limited, while in the case of one pot template-directed replication via ligation, the expected competition at each elongation step [57], between the 64 possible combinatorial triplets or the 264 quartets, is bound to jam the process.
- The replicase problem—The most efficient RNA replicases generated in vitro so far had sequences of about 200 mer long. Their complexity is likely to represent an intrinsic difficulty, that is, a limited efficiency of the polymerase ribozyme caused by its weak affinity for its substrates [32,58]. Even if one assumes a much simpler ribozyme of merely 40 mer, as calculated by Joyce [59] “to represent all of these sequence combinations… would require 27 kg of RNA, which seems highly implausible”. The probability of spontaneous emergence of a complex RNA entity of about 200 mer is thus infinitesimal, even under significant relaxation of the requirement for sequence conservation.
- Cross-replication—Assuming that, despite the improbability, a single RNA replicase would have randomly emerged, the likelihood that a second ribozyme could appear at the same venue and point in time, to template-copy the first replicase, is close to none.
- Folded vs. unfolded replicases—This conundrum emanates from the fact that folded RNA ribozymes contain double-stranded regions that need to be melted in order to be copied. The requirement, in the context of the prebiotic RNA replicase, was therefore ambiguous—it had to retain its structure and function under the same environmental conditions in which the copied replicase was required to melt. To reconcile this difficulty, it was suggested that RNA replicases must have existed in a delicate balance between the folded state necessary for catalysis, and the unfolded state necessary for template activity [60]. Equilibrium, however, requires a large number of replicases, which is improbable considering the complexity of this ribozyme and more so, if it corresponds to the initial unit of evolution that should have been as simple as possible.
- Continuity problem—The transition from an RNA-only to an RNA–protein world, which must have predated LUCA, is a step function in the mathematical sense. It does not conform to the “continuity principle” that goes back to Darwin [1]. This transition was hypothesized to occur by proteins taking over catalysis [56], but such a scenario is not without difficulty, as referred to by Lanier and Williams [61]: “There is no evidence to our knowledge that Darwinian processes can revise the Molecular Toolbox or radically alter the Universal Gene Set. Available evidence suggests takeovers are unlikely by Darwinian processes”.
2.2.2. RNA–DNA
Unit of Evolution Content
Autonomous Advent
Extant Perspective
Evolutionary Prospect
Critical View
- Same as items 1–6 for the RNA-only unit of evolution.
- Thermodynamics—The scenario suggested for the direct formation of an RNA–DNA unit of evolution relies on a spontaneous heterogeneity-to-homogeneity process [65], which would have led from disorder to order, i.e., lowered the entropy of the system. Such a process could have taken place only at a specific temperature range that, dependent on the involved enthalpies, would have reduced the free energy of the system. The relevant temperature range was not determined, leaving the question of the feasibility of such a process unanswered.
2.2.3. RNA–Protein
Unit of Evolution Content
Autonomous Advent
Advent of the Proto-Ribosome
- Autonomous materialization of a simple noncoded proto-ribosome that could catalyze peptide bond formation between two random amino acids, i.e., proto-LSU.
- Autonomous advent of a minimal coded proto-ribosome that was capable of processively translating a code written in an RNA string into a code-directed polypeptide.
Advent of the Proto-tRNA
Aminoacylation of Proto-tRNAs and the Origin of the Genetic Code
Advent of the Proto-Polymerase
Evolutionary Prospect
Extant Perspective
Critical View
- Complexity—Although each of the RNA and protein components composing the RNA–protein unit of evolution seems to have an acceptable probability to materialize spontaneously, the whole process is extremely complex compared with the simple emergence of an RNA world, and the likelihood of its occurrence is significantly lower.
- Experimental verification—The capability of a noncoded dimeric proto-ribosome to assemble spontaneously and catalyze the synthesis of peptides was already shown experimentally [82,83]. The self-assembly of L-shaped proto-tRNAs from 12 mer RNA strands, as well as their self-aminoacylation, are experimentally testable. However, the possibility of lending experimental support to the self-assembly of the minimal coded proto-ribosome model [85], through the merger of the DPR with the L-shaped proto-SSU and the bridging element (Figure 1e), seems questionable. The proto-SSU sequence is less conserved than the other components of the model, while the bridging element contains a single-stranded segment of 20 nucleotides (LSU nucleotides 1925–1944), which may form different globular structures under varying conditions. The probability of obtaining each of these two moieties in the laboratory in their ancestral form is, therefore, minute, and an attempt to combine them with the DPR to yield a peptide-forming molecular machine seems extremely challenging.
- Stereochemical affinity?—According to the present scenario, the emergence of the genetic code and of the proto-tRNA self-aminoacylation are conditional on the verity of Woese’s stereochemical theory [100]. Up till now, this hypothesis has been tested mainly on aptamers [114] and within the ribosome [115], giving inconclusive results [99]. Initial docking simulations seem to indicate an enhanced preference of certain early-appearing amino acids towards their cognate coding triplet located in the tRNA acceptor stem (unpublished results), but credibility requires experimental support.
2.2.4. DNA–Protein
Unit of Evolution Content
Autonomous Advent
Extant Perspective
3. Discussion
- An initial RNA–protein system could have continuously evolved into the current RNA–DNA–protein world, without the need to go through a discontinuous step of transferring most of the catalysis from RNA to proteins, as the RNA world hypothesis entails.
- The chemical versatility and efficiency of enzymes would have been beneficial in promoting the emergence of life.
- The self-aminoacylation mechanism [98] discussed here requires only a “soft“ version of Woese’s stereochemical affinity. It suffices that a certain amino acid would have higher affinity towards its cognate coding triplets, compared to that exhibited by the limited number of the other contemporaneous amino acids, to allow the formation of conceivable percentage of correctly aminoacylated tRNAs; thus, of correctly translated proteins.
- The occurrence of the statistically-challenging step in the RNA–protein scenario, i.e., the advent of a proto-polymerase via the prebiotic translation of a random RNA chain that accidentally encoded it, seems to be inevitable in any scenario. Being the only pathway whereby the proto-polymerase could have established a constant presence in a primordial unit of evolution prior to LUCA implies that this step would have occurred whether the route to LUCA went through an RNA world or directly into an RNA–protein world.
- The scenario presented here for the autonomous advent of the protein-RNA unit of evolution is, in principle, experimentally verifiable. The capability of a noncoded dimeric proto-ribosome to assemble spontaneously and catalyze the synthesis of peptides was already demonstrated experimentally [82,83], while the self-assembly of L-shaped proto-tRNAs from 12 mer RNA strands, as well as their self-aminoacylation, are testable [98]. Obtaining a coded proto-ribosome and an enzyme possessing some replicative abilities should require wide-scale in vitro directed evolution experiments and a great deal of luck, but the attempt is doable. In contrast, the means for verifying “proteins taking over catalysis by RNA” [56], as implied by the RNA world hypothesis, are not present [61].
- The present scenario for the advent of an RNA–protein unit of evolution bypasses the “chicken and egg” conundrum posed by an enzymatic prebiotic aminoacylation, which requires that proto-tRNAs would be specifically aminoacylated by synthetases that needed specifically aminoacylated tRNAs for their formation. Autonomously formed and self-aminoacylated proto-tRNAs, being part of the RNA–protein unit of evolution, could have participated in the synthesis of the initial synthetase by translating a random RNA string encoding it.
- A situation where the two central types of polymerases in the living cell, i.e., the ribosome that polymerizes amino acids and the polymerase that polymerizes nucleotides, are formed from different polymers with distinct environmental sensitivities is advantageous. Under stress conditions that specifically affect proteins, such as a denaturating agent that melts proteins but not RNA, a ribosome could have recovered the content of the unit of evolution by producing additional proteins, while a ribosome-analog enzyme would be disabled. Symmetrically, under RNA stress conditions, the enzyme polymerase may still function to regenerate the corrupted RNA components [29]. This notion seems to provide a proper answer to a long-standing question, first posed by Crick [21], concerned with the preference of nature to adhere to a ribosome which is a ribozyme, rather than transferring the translation process to a more efficient protein.
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wolf, Y.I.; Koonin, E.V. On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization. Biol. Direct. 2007, 2, 14. [Google Scholar] [CrossRef]
- Fox, G.E. Origin and evolution of the ribosome. Cold Spring Harb. Perspect. Biol. 2010, 2, a003483. [Google Scholar] [CrossRef]
- Bowman, J.C.; Hud, N.V.; Williams, L.D. The ribosome challenge to the RNA world. J. Mol. Evol. 2015, 80, 143–161. [Google Scholar] [CrossRef]
- Koonin, E.V. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 2003, 1, 127–136. [Google Scholar] [CrossRef]
- Harris, J.K.; Kelley, S.T.; Spiegelman, G.B.; Pace, N.R. The genetic core of the universal ancestor. Genome Res. 2003, 13, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Charlebois, R.L.; Doolittle, W.F. Computing prokaryotic gene ubiquity: Rescuing the core from extinction. Genome Res. 2004, 14, 2469–2477. [Google Scholar] [CrossRef]
- Glass, J.I.; Merryman, C.; Wise, K.S.; Hutchison, C.A., 3rd; Smith, H.O. Minimal Cells-Real and Imagined. Cold Spring Harb. Perspect. Biol. 2017, 9, a023861. [Google Scholar] [CrossRef]
- Smith, J.M. The Problems of Biology; Oxford University Press: Oxford, UK, 1986. [Google Scholar]
- Koonin, E.V.; Martin, W. On the origin of genomes and cells within inorganic compartments. Trends Genet. 2005, 21, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.L.; Urey, H.C. Organic compound synthesis on the primitive Earth: Several questions about the origin of life have been answered, but much remains to be studied. Science 1959, 130, 245. [Google Scholar] [CrossRef]
- Chang, S. Prebiotic synthesis in planetary environments. In The Chemistry of Life’s Origin; Greenberg, J.M., Mendoza-Gómez, C.X., Pirronello, V., Eds.; Springer: Dordrecht, The Netherlands, 1993; Volume 416, pp. 259–299. [Google Scholar]
- Ménez, B.; Pisapia, C.; Andreani, M.; Jamme, F.; Vanbellingen, Q.P.; Brunelle, A.; Richard, L.; Dumas, P.; Réfrégiers, M. Abiotic synthesis of amino acids in the recesses of the oceanic lithosphere. Nature 2018, 564, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Botta, O.; Bada, J.L. Extraterrestrial Organic Compounds in meteorites. Surv. Geophys. 2002, 23, 411–467. [Google Scholar] [CrossRef]
- Ferris, J.P.; Hill, A.R., Jr.; Liu, R.; Orgel, L.E. Synthesis of long prebiotic oligomers on mineral surfaces. Nature 1996, 381, 59–61. [Google Scholar] [CrossRef] [PubMed]
- Powner, M.W.; Gerland, B.; Sutherland, J.D. Synthesis of Activated Pyrimidine Ribonucleotides in Prebiotically Plausible Conditions. Nature 2009, 459, 239–242. [Google Scholar] [CrossRef]
- Powner, M.W.; Sutherland, J.D.; Szostak, J.W. Chemoselective Multicomponent One-Pot Assembly of Purine Precursors in Water. J. Am. Chem. Soc. 2010, 132, 16677–16688. [Google Scholar] [CrossRef]
- Mast, C.B.; Schink, S.; Gerland, U.; Braun, D. Escalation of polymerization in a thermal gradient. Proc. Natl. Acad. Sci. USA 2013, 110, 8030–8035. [Google Scholar] [CrossRef] [PubMed]
- Jerome, C.A.; Kim, H.J.; Mojzsis, S.J.; Benner, S.A.; Biondi, E. Catalytic Synthesis of Polyribonucleic Acid on Prebiotic Rock Glasses. Astrobiology 2022, 22, 629–636. [Google Scholar] [CrossRef]
- Rich, A. On the problems of evolution and biochemical information transfer. In Horizons in Biochemistry; Kasha, M., Pullman, B., Eds.; Academic Press: New York, NY, USA, 1962; pp. 103–126. [Google Scholar]
- Woese, C. The Genetic Code; Harper & Row: New York, NY, USA, 1967. [Google Scholar]
- Crick, F.H.C. The origin of the genetic code. J. Mol. Biol. 1968, 38, 367–379. [Google Scholar] [CrossRef]
- Orgel, L.E. Evolution of the genetic apparatus. J. Mol. Biol. 1968, 38, 381–393. [Google Scholar] [CrossRef]
- Gilbert, W. The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Bernhardt, H.S. The RNA world hypothesis: The worst theory of the early evolution of life (except for all the others). Biol. Direct 2012, 7, 23. [Google Scholar] [CrossRef]
- Robertson, M.P.; Joyce, G.F. The origins of the RNA world. Cold Spring Harb. Perspect. Biol. 2012, 4, a003608. [Google Scholar] [CrossRef]
- Eigen, M. Self-organization of matter and the evolution of biological macromolecules. Naturwissenschaften 1971, 58, 465–523. [Google Scholar] [CrossRef] [PubMed]
- Kunin, V. A system of two polymerases—A model for the origin of life. Orig. Life Evol. Biosph. 2000, 30, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. The cosmological model of eternal inflation and the transition from chance to biological evolution in the history of life. Biol. Direct 2007, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Agmon, I. The dimeric proto-ribosome: Structural details and implications on the origin of life. Int. J. Mol. Sci. 2009, 10, 2921–2934. [Google Scholar] [CrossRef]
- Rouch, D.A. Evolution of the first genetic cells and the universal genetic code: A hypothesis based on macromolecular coevolution of RNA and proteins. J. Theor. Biol. 2014, 357, 220–244. [Google Scholar] [CrossRef] [PubMed]
- Agmon, I.; Mor, T. A Model for the Emergence of Coded Life. In Theory and Practice of Natural Computing; TPNC 2015. Lecture Notes in Computer Science; Dediu, A.H., Magdalena, L., Martín-Vide, C., Eds.; Springer: Cham, Switzerland, 2015; Volume 9477. [Google Scholar]
- Tagami, S.; Li, P. The origin of life: RNA and protein co-evolution on the ancient earth. Dev. Growth Differ. 2023, 65, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Szathmáry, E. The origin of the genetic code: Amino acids as cofactors in an RNA world. Trends Genet. 1999, 15, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, A.; Nakashima, T.; Tokuriki, N.; Hosokawa, M.; Nogami, H.; Arioka, S.; Urabe, I.; Yomo, T. Evolvability of random polypeptides through functional selection within a small library. Protein Eng. 2002, 15, 619–626. [Google Scholar] [CrossRef]
- LaBean, T.H.; Butt, T.R.; Kauffman, S.A.; Schultes, E.A. Protein Folding Absent Selection. Genes 2011, 2, 608–626. [Google Scholar] [CrossRef]
- James, K.D.; Ellington, A.D. Catalysis in the RNA world. In The Molecular Origins of Life: Assembling Pieces of the Puzzle; Cambridge University Press: Cambridge, UK, 1998; Volume 28, pp. 269–294. [Google Scholar]
- Mavelli, F. Stochastic simulations of minimal cells: The Ribocell model. BMC Bioinform. 2012, 13, S10. [Google Scholar] [CrossRef] [PubMed]
- Briones, C.; Stich, M.; Manrubia, S.C. The dawn of the RNA World: Toward functional complexity through ligation of random RNA oligomers. RNA 2009, 15, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Strunk, G.; Ederhof, T. Machines for automated evolution experiments in vitro based on the serial-transfer concept. Biophys. Chem. 1997, 66, 193–202. [Google Scholar] [CrossRef]
- Joyce, G.F. Forty years of in vitro evolution. Angew. Chem. Int. Ed. Engl. 2007, 46, 6420–6436. [Google Scholar] [CrossRef] [PubMed]
- AbouHaidar, M.G.; Ivanov, I.G. Non-enzymatic RNA hydrolysis promoted by the combined catalytic activity of buffers and magnesium ions. Z. Naturforsch. 1999, 54, 542–548. [Google Scholar] [CrossRef]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef]
- Lindahl, T. Instability and Decay of the Primary Structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef]
- Bashkin, J.K. DNA Enzymes: New-Found Chemical Reactivity. Curr. Biol. 1997, 7, 286–288. [Google Scholar] [CrossRef]
- Leu, K.; Obermayer, B.; Rajamani, S.; Gerland, U.; Chen, I.A. The Prebiotic Evolutionary Advantage of Transferring Genetic Information from RNA to DNA. Nucleic Acids Res. 2011, 39, 8135–8147. [Google Scholar] [CrossRef]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Radzicka, R.; Wolfenden, A. proficient enzyme. Science 1995, 267, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Szostak, J.W. RNA-catalysed synthesis of complementary-strand RNA. Nature 1989, 339, 519–522. [Google Scholar] [CrossRef] [PubMed]
- Unrau, P.J.; Bartel, D.P. RNA-Catalyzed Nucleotide Synthesis. Nature 1998, 395, 260–263. [Google Scholar] [CrossRef] [PubMed]
- Joyce, G.F.; Orgel, L.E. Progress toward understanding the origin of the RNA world. In The RNA World; Will, C.L., Lührmann, R., Gesteland, R.F., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2006; pp. 23–57. [Google Scholar]
- Attwater, J.; Wochner, A.; Holliger, P. In-Ice Evolution of RNA Polymerase Ribozyme Activity. Nat. Chem. 2013, 5, 1011–1018. [Google Scholar] [CrossRef]
- Horning, D.P.; Joyce, G.F. Amplification of RNA by an RNA polymerase ribozyme. Proc. Natl. Acad. Sci. USA 2016, 113, 9786–9791. [Google Scholar] [CrossRef]
- Zhou, L.; O’Flaherty, D.K.; Szostak, J.W. Template-directed copying of RNA by non-enzymatic ligation. Angew. Chem. Int. Ed. Engl. 2020, 59, 15682–15687. [Google Scholar] [CrossRef]
- Salditt, A.; Keil, L.M.R.; Horning, D.P.; Mast, C.B.; Joyce, G.F.; Braun, D. Thermal Habitat for RNA Amplification and Accumulation. Phys. Rev. Lett. 2020, 125, 048104. [Google Scholar] [CrossRef]
- He, C.; Gállego, I.; Laughlin, B.; Grover, M.A.; Hud, N.V. A viscous solvent enables information transfer from gene-length nucleic acids in a model prebiotic replication cycle. Nat. Chem. 2017, 9, 318–324. [Google Scholar] [CrossRef]
- Cairns-Smith, A.G. Takeover mechanisms and early biochemical evolution. Biosystems 1977, 9, 105–109. [Google Scholar] [CrossRef]
- Prywes, N.; Blain, J.C.; Del Frate, F.; Szostak, J.W. Nonenzymatic copying of RNA templates containing all four letters is catalyzed by activated oligonucleotides. eLife 2016, 5, e17756. [Google Scholar] [CrossRef]
- Martin, L.L.; Unrau, P.J.; Müller, U.F. RNA synthesis by in vitro selected ribozymes for recreating an RNA world. Life 2015, 5, 247–268. [Google Scholar] [CrossRef]
- Joyce, G.F. Bit by bit: The Darwinian basis of life. PLoS Biol. 2012, 10, e1001323. [Google Scholar] [CrossRef] [PubMed]
- Green, R.; Szostak, J.W. Selection of a ribozyme that functions as a superior template in a self-copying reaction. Science 1992, 258, 1910–1915. [Google Scholar] [CrossRef] [PubMed]
- Lanier, K.A.; Williams, L.D. The Origin of Life: Models and Data. J. Mol. Evol. 2017, 84, 85–92. [Google Scholar] [CrossRef]
- Dworkin, J.P.; Lazcano, A.; Miller, S.L. The roads to and from the RNA world. J. Theor. Biol. 2003, 222, 127–134. [Google Scholar] [CrossRef]
- Xu, J.; Chmela, V.; Green, N.; Russell, D.A.; Janick, M.; Góra, R.W.; Szabla, R.; Bond, A.D.; Sutherland, J.D. Selective prebiotic formation of RNA pyrimidine and DNA purine nucleosides. Nature 2020, 582, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Trevino, S.G.; Zhang, N.; Elenko, M.P.; Luptak, A.; Szostak, J.W. Evolution of Functional Nucleic Acids in the Presence of Nonheritable Backbone Heterogeneity. Proc. Natl. Acad. Sci. USA 2011, 108, 13492–13497. [Google Scholar] [CrossRef]
- Gavette, J.V.; Stoop, M.; Hud, N.V.; Krishnamurthy, R. RNA-DNA Chimeras in the Context of an RNA World Transition to an RNA/DNA World. Angew. Chem. 2016, 55, 13204–13209. [Google Scholar] [CrossRef]
- Xu, L.; Wang, W.; Zhang, L.; Chong, J.; Huang, X.; Wang, D. Impact of template backbone heterogeneity on RNA polymerase II transcription. Nucleic Acids Res. 2015, 43, 2232–2241. [Google Scholar] [CrossRef][Green Version]
- Wuyts, J.; Van de Peer, Y.; De Wachter, R. Distribution of substitution rates and location of insertion sites in the tertiary structure of ribosomal RNA. Nucleic Acids Res. 2001, 29, 5017–5028. [Google Scholar] [CrossRef]
- Agmon, I.; Bashan, A.; Zarivach, R.; Yonath, A. Symmetry at the active site of the ribosome: Structural and functional implications. Biol. Chem. 2005, 386, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Fox, G.E.; Naik, A.K. The Evolutionary history of the ribosome. In The Genetic Code and the Origin of Life; de Pouplana, L.R., Ed.; Landes Bioscience: Georgetown, TX, USA, 2004; Chapter 6; pp. 92–105. [Google Scholar]
- Hury, J.; Nagaswamy, U.; Larios-Sanz, M.; Fox, G.E. Ribosome origins: The relative age of 23S rRNA domains. Orig. Life Evol. Biosph. 2006, 36, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Agmon, I.; Bashan, A.; Yonath, A. On ribosome conservation and evolution. Isr. J. Ecol. Evol. 2006, 52, 359–374. [Google Scholar] [CrossRef]
- Bokov, K.; Steinberg, S.V. A hierarchical model for evolution of 23S ribosomal RNA. Nature 2009, 457, 977–980. [Google Scholar] [CrossRef]
- Agmon, I.; Davidovich, C.; Bashan, A.; Yonath, A. Identification of the Prebiotic Translation Apparatus within the Contemporary Ribosome. 2009. Available online: https://precedings.nature.com/documents/-2921/version/1 (accessed on 18 March 2009).
- Hsiao, C.; Mohan, S.; Kalahar, B.K.; Williams, L.D. Peeling the onion: Ribosomes are ancient molecular fossils. Mol. Biol. Evol. 2009, 26, 2415–2425. [Google Scholar] [CrossRef]
- Petrov, A.S.; Gulen, B.; Norris, A.M.; Kovacs, N.A.; Bernier, C.R.; Lanier, K.A.; Fox, G.E.; Harvey, S.C.; Wartell, R.M.; Hud, N.V.; et al. History of the Ribosome and the Origin of Translation. Proc. Natl. Acad. Sci. USA 2015, 112, 15396–15401. [Google Scholar] [CrossRef]
- Nissen, P.; Hansen, J.; Ban, N.; Moore, P.B.; Steitz, T.A. The structural basis of ribosome activity in peptide bond synthesis. Science 2000, 289, 920–930. [Google Scholar] [CrossRef]
- Cannone, J.J.; Subramanian, S.; Schnare, M.N.; Collett, J.R.; D’Souza, L.M.; Du, Y.; Feng, B.; Lin, N.; Madabusi, L.V.; Müller, K.M.; et al. The comparative RNAWeb CRWSite: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinform. 2002, 3, 2. [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Davis, J.H.; Tonelli, M.; Scott, L.G.; Jaeger, L.; Williamson, J.R.; Butcher, S.E. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J. Mol. Biol. 2005, 351, 371–382. [Google Scholar] [CrossRef]
- Agmon, I.C. Could a proto-ribosome emerge spontaneously in the prebiotic world? Molecules 2016, 21, E1701. [Google Scholar] [CrossRef] [PubMed]
- Agmon, I. Sequence complementarity at the ribosomal Peptidyl Transferase Centre implies self-replicating origin. FEBS Lett. 2017, 591, 3252–3258. [Google Scholar] [CrossRef] [PubMed]
- Bose, T.; Fridkin, G.; Davidovich, C.; Krupkin, M.; Dinger, N.; Falkovich, A.H.; Peleg, Y.; Agmon, I.; Bashan, A.; Yonath, A. Origin of life: Protoribosome forms peptide bonds and links RNA and protein dominated worlds. Nucleic Acids Res. 2022, 50, 1815–1828. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Wang, Y. Protein-free ribosomal RNA scaffolds can assemble poly-lysine oligos from charged tRNA fragments. Biochem. Biophys. Res. Commun. 2021, 544, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Zarivach, R.; Bashan, A.; Berisio, R.; Harms, J.; Auerbach, T.; Schluenzen, F.; Bartels, H.; Baram, D.; Pyetan, E.; Sittner, A.; et al. Functional aspects of ribosomal architecture: Symmetry, chirality and regulation. J. Phys. Org. Chem. 2004, 17, 901–912. [Google Scholar] [CrossRef]
- Agmon, I. Hypothesis: Spontaneous Advent of the Prebiotic Translation System via the Accumulation of L-Shaped RNA Elements. Int. J. Mol. Sci. 2018, 19, 4021. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, H.; Waser, J. Evolution of early mechanisms of translation of genetic information into polypeptides. Nature 1982, 298, 585–586. [Google Scholar] [CrossRef] [PubMed]
- Schimmel, P.; Henderson, B. Possible role of aminoacyl-RNA complexes in noncoded peptide synthesis and origin of coded synthesis. Proc. Natl. Acad. Sci. USA 1994, 91, 11283–11286. [Google Scholar] [CrossRef]
- Di Giulio, M. The origin of the tRNA molecule: Implications for the origin of protein synthesis. J. Theor. Biol. 2004, 226, 89–93. [Google Scholar] [CrossRef]
- Jenner, L.B.; Demeshkina, N.; Yusupova, G.; Yusupov, M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 2010, 17, 555–560. [Google Scholar] [CrossRef]
- Jühling, F.; Mörl, M.; Hartmann, R.K.; Sprinzl, M.; Stadler, P.F.; Pütz, J. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009, 37, D159–D162. [Google Scholar] [CrossRef]
- Schimmel, P.; Giegé, R.; Moras, D.; Yokoyama, S. An operational RNA code for amino acids and possible relationship to genetic code. Proc. Natl. Acad. Sci. USA 1993, 90, 8763–8768. [Google Scholar] [CrossRef]
- Maizels, N.; Weiner, A.M. Phylogeny from function: Evidence from the molecular fossil record that tRNA originated in replication, not translation. Proc. Natl. Acad. Sci. USA 1994, 91, 6729–6734. [Google Scholar] [CrossRef]
- Eigen, M.; Winkler-Oswatitsch, R. Transfer-RNA, an early gene? Naturwissenschaften 1981, 68, 282–292. [Google Scholar] [CrossRef]
- Di Giulio, M. On the origin of the transfer RNA molecule. J. Theor. Biol. 1992, 159, 199–214. [Google Scholar] [CrossRef]
- Dick, T.P.; Schamel, W.A. Molecular evolution of transfer RNA from two precursor hairpins: Implications for the origin of protein synthesis. J. Mol. Evol. 1995, 1, 1–9. [Google Scholar]
- Nagaswamy, U.; Fox, G.E. RNA ligation and the origin of tRNA. Orig. Life Evol. Biosph. 2003, 33, 199–209. [Google Scholar] [CrossRef]
- Widmann, J.; Di Giulio, M.; Yarus, M.; Knight, R. tRNA creation by hairpin duplication. J. Mol. Evol. 2005, 61, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Agmon, I. Prebiotic Assembly of Cloverleaf tRNA, Its Aminoacylation and the Origin of Coding, Inferred from Acceptor Stem Coding-Triplets. Int. J. Mol. Sci. 2022, 23, 15756. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S.; Wolf, Y.I. Evolutionary Genomics of Defense Systems in Archaea and Bacteria. Annu. Rev. Microbiol. 2017, 71, 233–261. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R. On the evolution of the genetic code. Proc. Natl. Acad. Sci. USA 1965, 54, 1546–1552. [Google Scholar] [CrossRef]
- Lehmann, J.; Reichel, A.; Buguin, A.; Libchaber, A. Efficiency of a self-aminoacylating ribozyme: Effect of the length and base-composition of its 3′ extension. RNA 2007, 13, 1191–1197. [Google Scholar] [CrossRef][Green Version]
- Krzyzaniak, A.; Barciszewski, J.; Sałanski, P.; Jurczak, J. The non-enzymatic specific amino-acylation of transfer RNA at high pressure. Int. J. Biol. Macromol. 1994, 16, 153–158. [Google Scholar] [CrossRef]
- Corliss, J.B.; Baross, J.A.; Hoffman, S.E. An hypothesis concerning the relationships between submarine hot springs and the origin of life on earth. Oceanol. Acta 1981, 4, 59–69. [Google Scholar]
- Agmon, I.; Fayerverker, I.; Mor, T. Coding triplets in the tRNA acceptor-TΨC arm and their role in present and past tRNA recognition. FEBS Lett. 2021, 595, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Rodin, S.N.; Ohno, S. Four primordial modes of tRNA-synthetase recognition, determined by the (G, C) operational code. Proc. Natl. Acad. Sci. USA 1997, 94, 5183–5188. [Google Scholar] [CrossRef] [PubMed]
- Gavrilova, L.P.; Koteliansky, V.E.; Spirin, A.S. Ribosomal protein S12 and ‘non-enzymatic’ translocation. FEBS Lett. 1974, 45, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Koonin, E.V.; Aravind, L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct. Biol. 2003, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Hordijk, W.; Hein, J.; Steel, M. Autocatalytic sets and the origin of life. Entropy 2010, 12, 1733–1742. [Google Scholar] [CrossRef]
- Härtlein, M.; Cusack, S. Structure, function and evolution of seryl-tRNA synthetases: Implications for the evolution of aminoacyl-tRNA synthetases and the genetic code. J. Mol. Evol. 1995, 40, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Hartman, H. Speculations on the evolution of the genetic code IV. The evolution of the aminoacyl-tRNA synthetases. Orig. Life Evol. Biosph. 1995, 25, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Giegé, R.; Sissler, M.; Florentz, C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998, 26, 5017–5035. [Google Scholar] [CrossRef] [PubMed]
- Orgel, L.E. β-Turns and the evolution of protein synthesis. In The Organization and Expression of the Eukaryotic Genome, Proceedings of the International Symposium, Teheran, Iran, 3–6 May 1976; Bradbury, E.M., Javaherian, K., Eds.; Academic Press: London, UK, 1997; pp. 499–504. [Google Scholar]
- Kovacs, N.A.; Petrov, A.S.; Lanier, K.A.; Williams, L.D. Frozen in Time: The History of Proteins. Mol. Biol. Evol. 2017, 34, 1252–1260. [Google Scholar] [CrossRef] [PubMed]
- Yarus, M.; Caporaso, J.G.; Knight, R. Origins of the Genetic Code: The Escaped Triplet Theory. Annu. Rev. Biochem. 2005, 74, 179–198. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Wang, L. Imprints of the genetic code in the ribosome. Proc. Natl. Acad. Sci. USA 2010, 107, 8298–8303. [Google Scholar] [CrossRef]
- Powner, M.W.; Sutherland, J.D.; Szostak, J.W. The Origin of nucleotides. Synlett 2011, 14, 1956–1964. [Google Scholar] [CrossRef]
- Lathe, R. Fast tidal cycling and the origin of life. Icarus 2004, 168, 18–22. [Google Scholar] [CrossRef]
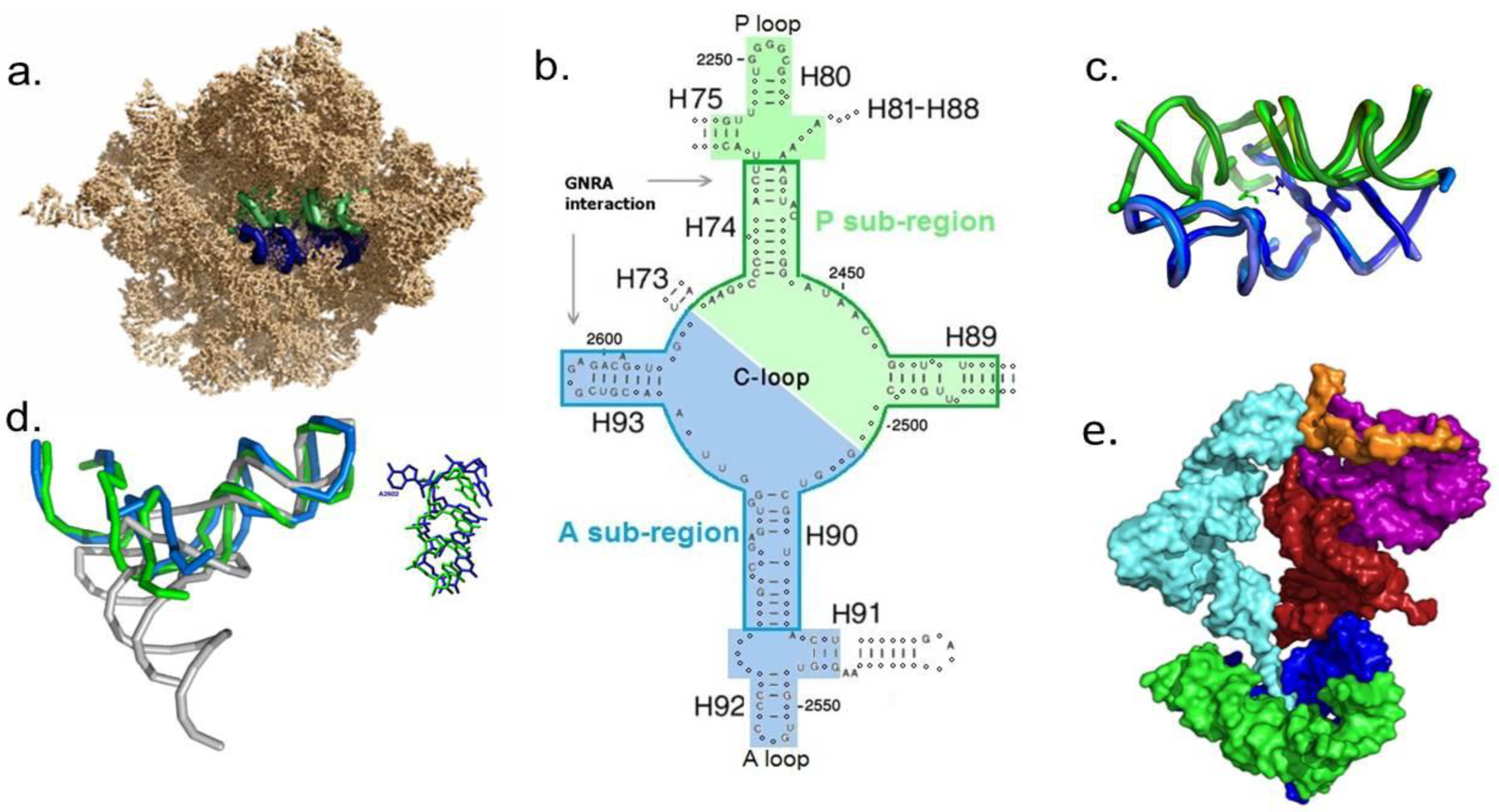
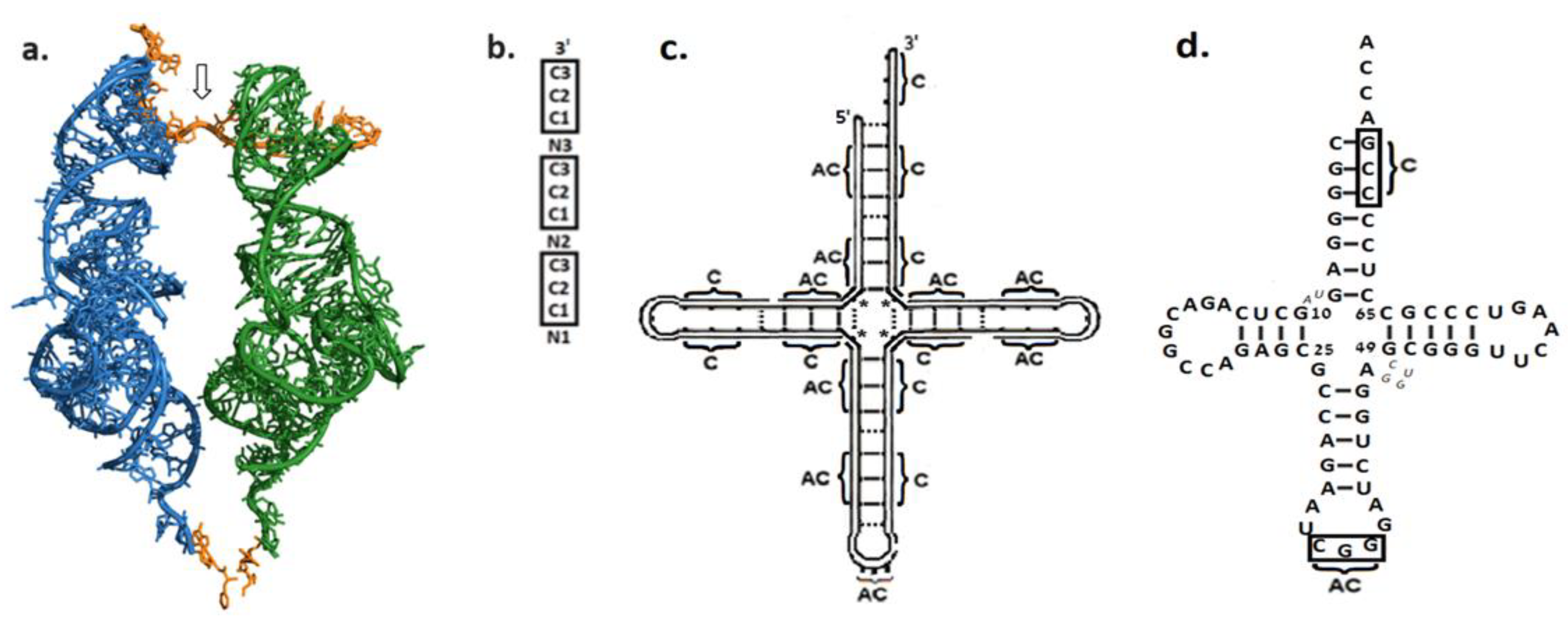
| Hypothetical Set | Polymers Content of the “Unit of Evolution” | Score as Genetic Material | Score as Catalyst | Rank = Genetic × Catalytic Scores |
|---|---|---|---|---|
| 1 | Protein | 0 * (Protein) | 2 * (Protein) | 0 * |
| 2 | DNA | 2 * (DNA) | 0 * (DNA) | 0 * |
| 3 | RNA | 1 * (RNA) | 1 * (RNA) | 1 * |
| 4 | RNA–DNA | 2 * (DNA) | 1 * (RNA) | 2 * |
| 5 | RNA–protein | 1 * (RNA) | 2 * (Protein) + 1 * (RNA) = 3 * | 3 * |
| 6 | DNA–protein | 2 * (DNA) | 2 * (Protein) | 4 * |
| 7 | RNA–DNA–protein | 2 * (DNA) | 2 * (Protein) + 1 * (RNA) = 3 * | 6 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agmon, I. Three Biopolymers and Origin of Life Scenarios. Life 2024, 14, 277. https://doi.org/10.3390/life14020277
Agmon I. Three Biopolymers and Origin of Life Scenarios. Life. 2024; 14(2):277. https://doi.org/10.3390/life14020277
Chicago/Turabian StyleAgmon, Ilana. 2024. "Three Biopolymers and Origin of Life Scenarios" Life 14, no. 2: 277. https://doi.org/10.3390/life14020277
APA StyleAgmon, I. (2024). Three Biopolymers and Origin of Life Scenarios. Life, 14(2), 277. https://doi.org/10.3390/life14020277







