Effect of Divalent Metal Ions on the Ribonuclease Activity of the Toxin Molecule HP0894 from Helicobacter pylori
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Protein Samples
2.2. The Ribonuclease Activity (RNase) Activity of HP0894 and Interaction of Divalent Metal Ions
2.3. Circular Dichroism Spectroscopy (CD)
2.4. Isothermal Titration Calorimetry (ITC) Assay
2.5. Nuclear Magnetic Resonance (NMR) Spectroscopy
3. Results
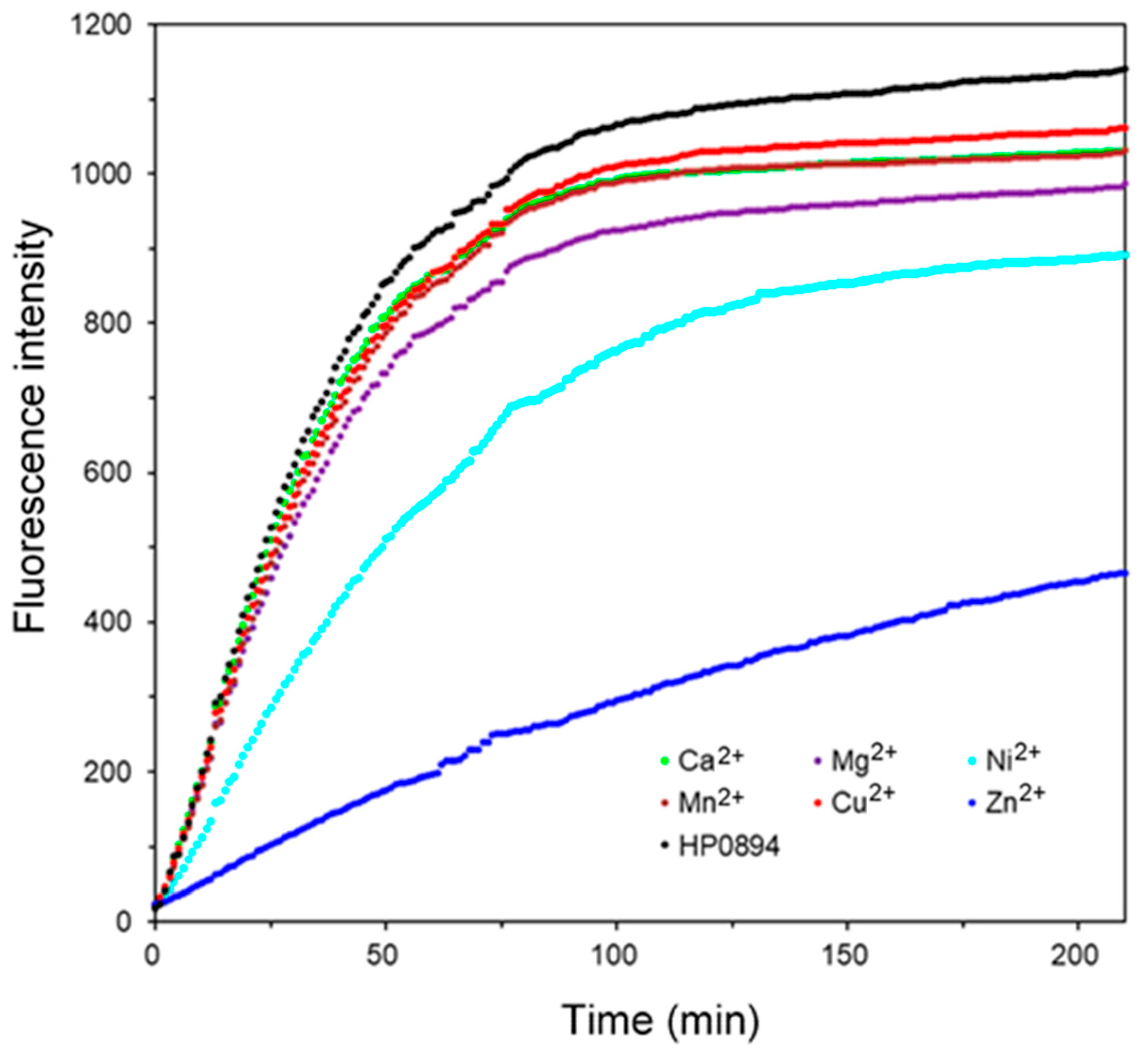
3.1. HP0894 Ribonuclease (RNase) Activity of HP0894 and Interference of Divalent Metal Ions
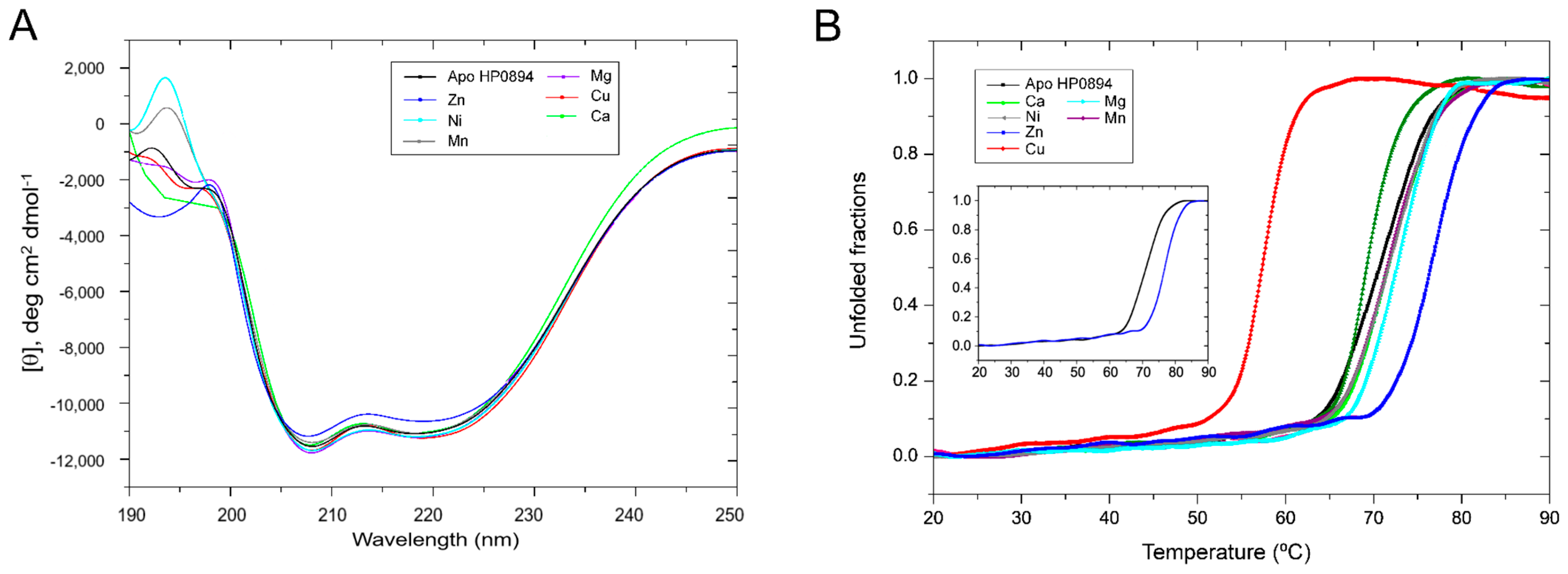
3.2. Effect of Metal Ions on the Secondary Structure of HP0894
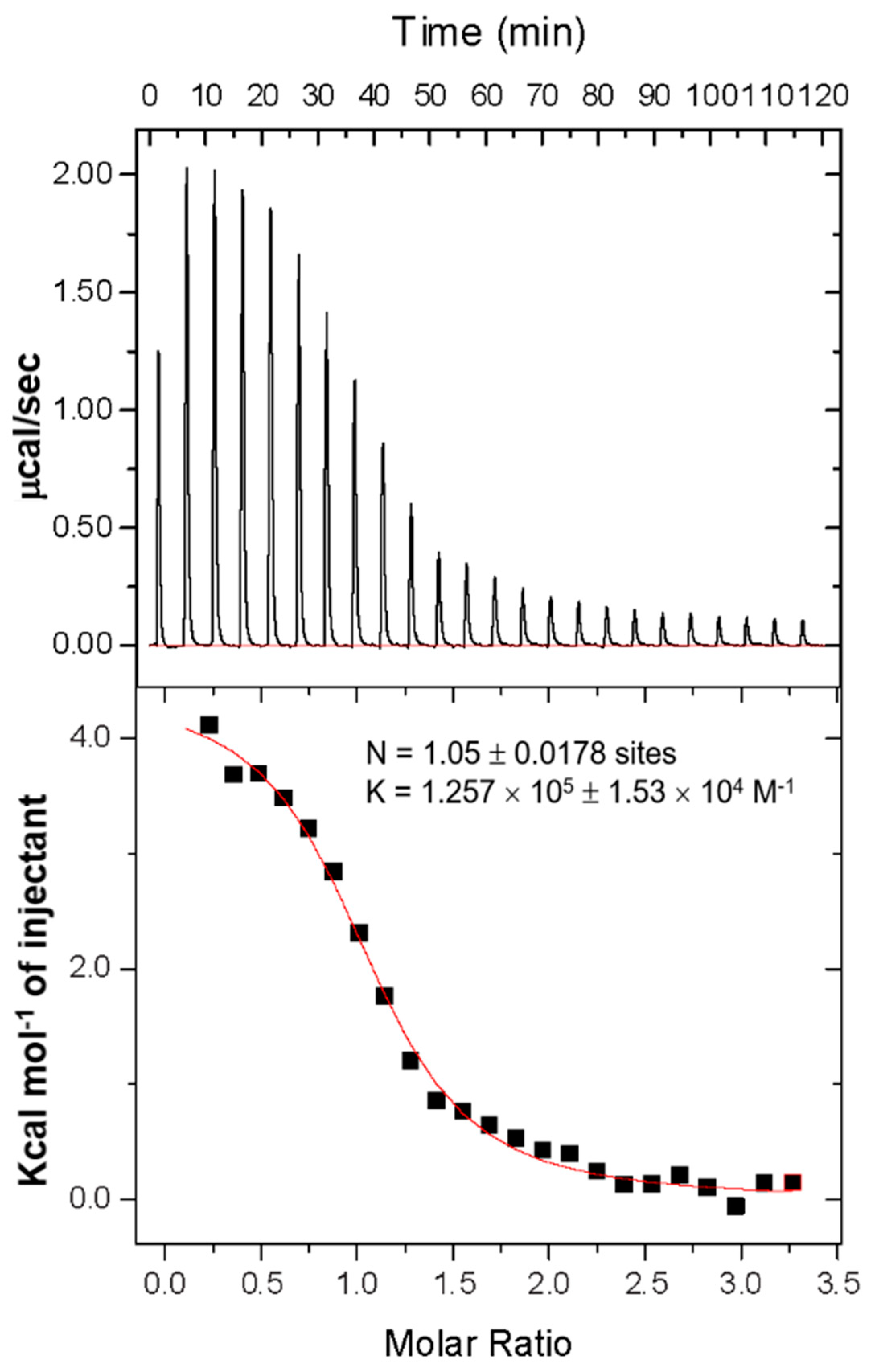
3.3. Thermal Analysis and Binding Stoichiometry Using Isothermal Titration Calorimetry
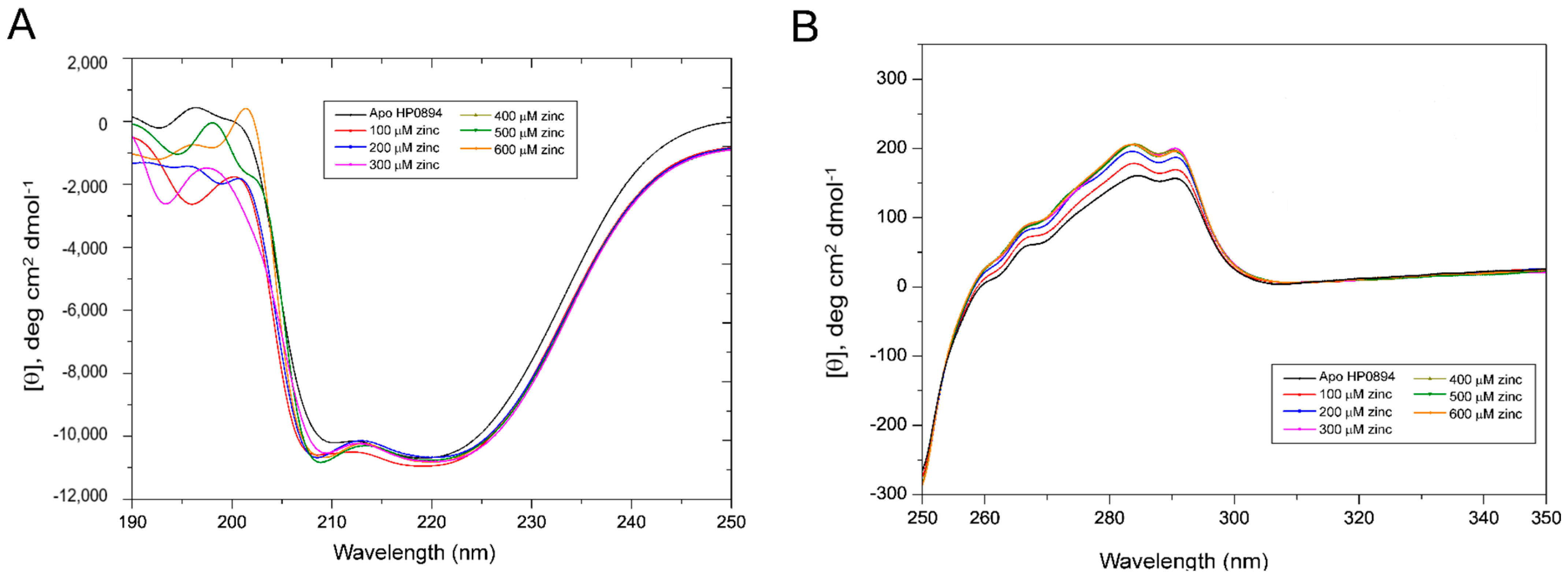
3.4. Zinc Titration and NMR Perturbation Experiments
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kamada, K.; Hanaoka, F. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol. Cell 2005, 19, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Neubauer, C.; Gao, Y.G.; Andersen, K.R.; Dunham, C.M.; Kelley, A.C.; Hentschel, J.; Gerdes, K.; Ramakrishnan, V.; Brodersen, D.E. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 2009, 139, 1084–1095. [Google Scholar] [CrossRef] [PubMed]
- Miallau, L.; Faller, M.; Chiang, J.; Arbing, M.; Guo, F.; Cascio, D.; Eisenberg, D. Structure and proposed activity of a member of the VapBC family of toxin-antitoxin systems. VapBC-5 from Mycobacterium tuberculosis. J. Biol. Chem. 2009, 284, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Buts, L.; Lah, J.; Dao-Thi, M.H.; Wyns, L.; Loris, R. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 2005, 30, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Condon, C. Shutdown decay of mRNA. Mol. Microbiol. 2006, 61, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 2000, 182, 561–572. [Google Scholar] [CrossRef]
- Maggi, S.; Yabre, K.; Ferrari, A.; Lazzi, C.; Kawano, M.; Rivetti, C.; Folli, C. Functional characterization of the type I toxin Lpt from Lactobacillus rhamnosus by fluorescence and atomic force microscopy. Sci. Rep. 2019, 9, 15208. [Google Scholar] [CrossRef]
- Thompson, M.K.; Nocedal, I.; Culviner, P.H.; Zhang, T.; Gozzi, K.R.; Laub, M.T. Escherichia coli SymE is a DNA-binding protein that can condense the nucleoid. Mol. Microbiol. 2022, 117, 851–870. [Google Scholar] [CrossRef]
- Jurėnas, D.; Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. Biology and evolution of bacterial toxin-antitoxin systems. Nat. Rev. Microbiol. 2022, 20, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Bukowski, M.; Rojowska, A.; Wladyka, B. Prokaryotic toxin-antitoxin systems-the role in bacterial physiology and application in molecular biology. Acta Biochim. Pol. 2011, 58, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Wagner, E.G. RNA antitoxins. Curr. Opin. Microbiol. 2007, 10, 117–124. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Inouye, M. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 2011, 9, 779–790. [Google Scholar] [CrossRef]
- Leplae, R.; Geeraerts, D.; Hallez, R.; Guglielmini, J.; Dreze, P.; Van Melderen, L. Diversity of bacterial type II toxin-antitoxin systems: A comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011, 39, 5513–5525. [Google Scholar] [CrossRef] [PubMed]
- Morales-Espinosa, R.; Delgado, G.; Serrano, L.R.; Castillo, E.; Santiago, C.A.; Hernández-Castro, R.; Gonzalez-Pedraza, A.; Mendez, J.L.; Mundo-Gallardo, L.F.; Manzo-Merino, J.; et al. High expression of Helicobacter pylori VapD in both the intracellular environment and biopsies from gastric patients with severity. PLoS ONE 2020, 15, e0230220. [Google Scholar] [CrossRef] [PubMed]
- Han, K.D.; Park, S.J.; Jang, S.B.; Lee, B.J. Solution structure of conserved hypothetical protein HP0892 from Helicobacter pylori. Proteins Struct. Funct. Bioinform. 2008, 70, 599–602. [Google Scholar] [CrossRef] [PubMed]
- Han, K.D.; Park, S.J.; Jang, S.B.; Son, W.S.; Lee, B.J. Solution structure of conserved hypothetical protein HP0894 from Helicobacter pylori. Proteins 2005, 61, 1114–1116. [Google Scholar] [CrossRef]
- Cárdenas-Mondragón, M.G.; Ares, M.A.; Panunzi, L.G.; Pacheco, S.; Camorlinga-Ponce, M.; Girón, J.A.; Torres, J.; De la Cruz, M.A. Transcriptional Profiling of Type II Toxin-Antitoxin Genes of Helicobacter pylori under Different Environmental Conditions: Identification of HP0967-HP0968 System. Front. Microbiol. 2016, 7, 1872. [Google Scholar] [CrossRef]
- Fineran, P.C.; Blower, T.R.; Foulds, I.J.; Humphreys, D.P.; Lilley, K.S.; Salmond, G.P. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. USA 2009, 106, 894–899. [Google Scholar] [CrossRef]
- Alonso, J.C. Toxin-Antitoxin Systems in Pathogenic Bacteria. Toxins 2021, 13, 74. [Google Scholar] [CrossRef]
- Pathak, C.; Im, H.; Yang, Y.-J.; Yoon, H.-J.; Kim, H.-M.; Kwon, A.-R.; Lee, B.-J. Crystal structure of apo and copper bound HP0894 toxin from Helicobacter pylori 26695 and insight into mRNase activity. Biochim. Biophys. Acta BBA-Proteins Proteom. 2013, 1834, 2579–2590. [Google Scholar] [CrossRef]
- Gerdes, K.; Christensen, S.K.; Løbner-Olesen, A. Prokaryotic toxin–antitoxin stress response loci. Nat. Rev. Microbiol. 2005, 3, 371–382. [Google Scholar] [CrossRef]
- Mutschler, H.; Gebhardt, M.; Shoeman, R.L.; Meinhart, A. A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol. 2011, 9, e1001033. [Google Scholar] [CrossRef]
- Dupont, C.L.; Yang, S.; Palenik, B.; Bourne, P.E. Modern proteomes contain putative imprints of ancient shifts in trace metal geochemistry. Proc. Natl. Acad. Sci. USA 2006, 103, 17822–17827. [Google Scholar] [CrossRef]
- Waldron, K.J.; Robinson, N.J. How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 2009, 7, 25–35. [Google Scholar] [CrossRef]
- Pang, J.; Guo, Q.; Lu, Z. The catalytic mechanism, metal dependence, substrate specificity, and biodiversity of ribonuclease H. Front. Microbiol. 2022, 13, 1034811. [Google Scholar] [CrossRef]
- Nicholson, A. Escherichia coli Ribonucleases: Paradigms for Understanding Cellular RNA Metabolism and Regulation, Ribonucleases: Structures and Functions; Academic, Press Inc.: New York, NY, USA, 1997; pp. 38–49. [Google Scholar]
- Katayanagi, K.; Miyagawa, M.; Matsushima, M.; Ishikawa, M.; Kanaya, S.; Ikehara, M.; Matsuzaki, T.; Morikawa, K. Three-dimensional structure of ribonuclease H from E. coli. Nature 1990, 347, 306–309. [Google Scholar] [CrossRef]
- Nowotny, M.; Yang, W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006, 25, 1924–1933. [Google Scholar] [CrossRef]
- Niemann, M.; Brecht, M.; Schlüter, E.; Weitzel, K.; Zacharias, M.; Göringer, H.U. TbMP42 is a structure-sensitive ribonuclease that likely follows a metal ion catalysis mechanism. Nucleic Acids Res. 2008, 36, 4465–4473. [Google Scholar] [CrossRef] [PubMed]
- Deutscher, M.; Marlor, C.W. Purification and characterization of Escherichia coli RNase T. J. Biol. Chem. 1985, 260, 7067–7071. [Google Scholar] [CrossRef] [PubMed]
- Boampong, K.; Smith, S.L.; Delahay, R.M. Rapid growth inhibitory activity of a YafQ-family endonuclease toxin of the Helicobacter pylori tfs4 integrative and conjugative element. Sci. Rep. 2020, 10, 18171. [Google Scholar] [CrossRef] [PubMed]
- Subedi, G.; Park, S.J. Mass Production and Characterization of Recombinant Human Grim-19 in Escherichia coli. Drug Targets Ther. 2023, 2, 71–79. [Google Scholar] [CrossRef]
- Han, K.-D.; Matsuura, A.; Ahn, H.-C.; Kwon, A.-R.; Min, Y.-H.; Park, H.-J.; Won, H.-S.; Park, S.J.; Kim, D.-Y.; Lee, B.-J. Functional identification of toxin-antitoxin molecules from Helicobacter pylori 26695 and structural elucidation of the molecular interactions. J. Biol. Chem. 2011, 286, 4842–4853. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef]
- Donghi, D.; Sigel, R.K. Metal ion-RNA interactions studied via multinuclear NMR. Methods Mol. Biol. 2012, 848, 253–273. [Google Scholar] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Johnsona, B.A.; Blevinsb, R.A. NMRView: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 1994, 4, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Schumann, F.H.; Riepl, H.; Maurer, T.; Gronwald, W.; Neidig, K.P.; Kalbitzer, H.R. Combined chemical shift changes and amino acid specific chemical shift mapping of protein-protein interactions. J. Biomol. NMR 2007, 39, 275–289. [Google Scholar] [CrossRef]
- Im, H.; Jang, S.B.; Pathak, C.; Yang, Y.J.; Yoon, H.J.; Yu, T.K.; Suh, J.Y.; Lee, B.J. Crystal structure of toxin HP0892 from Helicobacter pylori with two Zn(II) at 1.8 Å resolution. Protein Sci. 2014, 23, 819–832. [Google Scholar] [CrossRef]
- Capdevila, D.A.; Wang, J.; Giedroc, D.P. Bacterial Strategies to Maintain Zinc Metallostasis at the Host-Pathogen Interface. J. Biol. Chem. 2016, 291, 20858–20868. [Google Scholar] [CrossRef]
- Hogstrand, C.; Kille, P.; Nicholson, R.; Taylor, K. Zinc transporters and cancer: A potential role for ZIP7 as a hub for tyrosine kinase activation. Trends Mol. Med. 2009, 15, 101–111. [Google Scholar] [CrossRef]
- Maret, W. Inhibitory zinc sites in enzymes. BioMetals 2013, 26, 197–204. [Google Scholar] [CrossRef]
- Wei, Y.; Fu, D. Binding and transport of metal ions at the dimer interface of the Escherichia coli metal transporter YiiP. J. Biol. Chem. 2006, 281, 23492–23502. [Google Scholar] [CrossRef]
- Maret, W.; Li, Y. Coordination dynamics of zinc in proteins. Chem. Rev. 2009, 109, 4682–4707. [Google Scholar] [CrossRef]
- Maret, W. Zinc and the Zinc Proteome in Metallomics and the Cell; Springer: Berlin/Heidelberg, Germany, 2013; pp. 479–501. [Google Scholar]
- Debela, M.; Goettig, P.; Magdolen, V.; Huber, R.; Schechter, N.M.; Bode, W. Structural basis of the zinc inhibition of human tissue kallikrein 5. J. Mol. Biol. 2007, 373, 1017–1031. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hyun, J.-S.; Pun, R.; Park, S.J.; Lee, B.-J. Effect of Divalent Metal Ions on the Ribonuclease Activity of the Toxin Molecule HP0894 from Helicobacter pylori. Life 2024, 14, 225. https://doi.org/10.3390/life14020225
Hyun J-S, Pun R, Park SJ, Lee B-J. Effect of Divalent Metal Ions on the Ribonuclease Activity of the Toxin Molecule HP0894 from Helicobacter pylori. Life. 2024; 14(2):225. https://doi.org/10.3390/life14020225
Chicago/Turabian StyleHyun, Ja-Shil, Rabin Pun, Sung Jean Park, and Bong-Jin Lee. 2024. "Effect of Divalent Metal Ions on the Ribonuclease Activity of the Toxin Molecule HP0894 from Helicobacter pylori" Life 14, no. 2: 225. https://doi.org/10.3390/life14020225
APA StyleHyun, J.-S., Pun, R., Park, S. J., & Lee, B.-J. (2024). Effect of Divalent Metal Ions on the Ribonuclease Activity of the Toxin Molecule HP0894 from Helicobacter pylori. Life, 14(2), 225. https://doi.org/10.3390/life14020225






