Combining Genetic and Phenotypic Analyses for Detecting Bread Wheat Genotypes of Drought Tolerance through Multivariate Analysis Techniques
Abstract
1. Introduction
2. Materials and Methods
2.1. Genomic DNA Extraction and SSR Markers
2.2. Plant Materials and Experimental Design
2.3. Measurements of Traits
2.3.1. Agro-Physio-Biochemical Traits
- -
- The estimation of Chl in leaves was performed using a colorimetric method by measuring absorbances at 663 nm and 646 nm with 80% acetone as the solvent. Approximately 0.5 g of leaf tissue was crushed in liquid nitrogen, and about 100 mg of the crushed tissue was taken. Then, 2 mL of acetone was added to the sample, which was then left in a dark place in the refrigerator for 48 h. Afterward, the sample was centrifuged, and the extract obtained was used for spectrophotometer readings to estimate Chl. The calculations for estimating Chl were based on the equations provided by Lichtenthaler and Wellburn [44].
- -
- To measure the activity of antioxidant enzymes, including SOD, CAT, POD, and PPO, fresh leaf samples weighing 0.5 g were utilized. The extraction of these enzymes involved crushing the leaves in liquid nitrogen and suspending them in a buffer containing 50 mM potassium phosphate buffer (pH 7.8) and 1% (w/v) polyvinyl polypyrrolidone. Afterward, the samples were subjected to centrifugation at 14,000× g for 10 min at 4 °C. The resulting supernatant, as described in references [45,46,47], was used as an enzyme extract for the subsequent tests assessing the activity of CAT, POD, PPO, and SOD.
- -
- The determination of DPPH radical scavenging ability involved assessing the decrease in absorbance at 517 nm [48]. The analyses were conducted using a UV–vis spectrophotometer in 3 mL cuvettes. To facilitate the analysis, a freshly prepared stock solution of DPPH (3.94 mg/100 mL methanol) radicals in methanol was utilized. Subsequently, 3 mL of the DPPH working solution was mixed with 0.5 mL of the extract and left in darkness for 30 min. The presence of an antioxidant agent in the reaction medium led to the disappearance of the purple color associated with DPPH radicals. In parallel, a reference sample consisting of 0.5 mL of the solvent was prepared. The maximal absorption of the newly prepared DPPH radical solution was observed at 517 nm. All analyses were performed in 3 replicates, and the absorbance was recorded at 517 nm. The blank sample referred to the reaction mixture that lacked any test compounds [49,50]. DPPH scavenging effect (%) = [A0 − A1)/A0] × 100.
- -
- The quantification of TPC was conducted using the Folin–Ciocalteau method, as previously described by Sarker and Oba [51]. In this procedure, extracts (100 μL) or a series of standards (12.5, 25, 50, 100, 150, and 200 μg mL−1 gallic acid) were added. Following reagent mixing and the ensuing reaction, 300 μL of the solution was transferred to a 96-well plate, and the absorbance was measured at 740 nm. The obtained results were expressed as the equivalent amount of gallic acid standard (mg GAE/g FW).
- -
- To quantify the contents of GB, leaf samples were ground using liquid nitrogen to ensure proper homogenization, as described by Grieve and Grattan [52]. Subsequently, 1 mg of the sample was transferred to a glass tube, and 1.5 mL of 2 N H2SO4 was added to it. The mixture was then placed in a water bath at 60 °C for ten minutes to extract Glycine betaine. After centrifugation at 3500× rpm for 10 min, the supernatants were collected for further analysis. To analyze the GB concentration, 125 μL of the supernatant sample was combined with 50 μL of cold Potassium tri-iodide KI-I2, which was prepared by dissolving 15.7 g of iodine and 20 g of potassium iodide in 100 mL of distilled water. This mixture was left at 0–4 °C for 16 h and then centrifuged at 10,000× rpm for 15 min. The upper liquid was discarded, leaving behind small crystals in the chamber of the tube. These crystals were dissolved by adding 1.4 mL of 1,2-dichloroethane and incubating the solution for 2–2.5 h. The samples were then examined using a spectrophotometer at 365 nm (U-2000, Hitachi Instruments, Tokyo, Japan). To determine the GB concentration, a standard curve was prepared using stock solutions of betaine with concentrations of 1, 2, 4, 6, and 8 μL. These stock solutions were used to calculate the GB concentration in the samples.
- -
- For proline content, the estimation of proline was performed using the protocol described by Boctor [53] with certain modifications. Initially, the sample was ground using liquid nitrogen. Then, 100 mg of the sample was taken and mixed with 500 μL of 3% Sulpho salicylic acid. The mixture was vortexed and placed on ice for five minutes, followed by centrifugation at the highest speed for five minutes at room temperature. Next, 200 μL of the supernatant was combined with 200 μL of 3% Sulphosalicylic acid, 400 μL of glacial acetic acid, and 400 μL of ninhydrin acid. The reaction components were vortexed thoroughly and placed in a water bath at 100 °C for one hour. To halt the reaction, the tubes were then placed on ice. In the final step, 1 mL of toluene was added to the reaction mixture. The solution was vigorously shaken by hand and left undisturbed for five minutes to allow the components to separate into two layers. The top layer was extracted, and the absorbance was measured using a spectrometer at 520 nm, with toluene serving as the blank.
2.3.2. Quality Traits
2.4. Statistical Analysis
- -
- Genotyping Analysis: SSR bands were scored (present (1) or absent (0)) to create a binary matrix. The genetic dissimilarity (matrix of pairwise) between genotypes was calculated using the coefficient of Jaccard dissimilarity. Agglomerative HC analysis was implemented using the unweighted pair group average method (UPGAM).
- -
- Phenotypic analysis: ANOVA (split-plot design) and genetic parameters for 30 traits were implemented using SAS v9.2 software (SAS Institute, Inc., Cary, NC, USA). The variance (mean squares) of data for 30 traits was used to compute variance components that are used to compute genetic parameters (genetic variance (σ2G), residual variance (σ2e), phenotypic variance (σ2Ph), heritability (h2 %), genotypic coefficient of variability (G.C.V. %), phenotypic coefficient of variability (Ph.C.V. %), genetic advance (GA), and genetic gain (GG)), as described by Al-Ashkar et al. [14]. Principal component analysis (PCA) was carried out based on data provided by the correlation matrix to find out the variables contributing the most to the variance and the components loading the most on the variables. PCA is useful for trait reduction, dealing with the problem of multicollinearity, and identifying important traits that are located in the first two components, and its outcomes were used to detect the drought tolerance index, which was used in SMLR (Stepwise multiple linear regression), PC (path coefficient), HC (hierarchical cluster), and LD (liner discriminant) analyses. PCA eliminated five traits that exhibited high multicollinearity. Twenty-five out of thirty traits (index) were used in SMLR to determine the key traits that contribute to enhancing and developing the variable of interest (GY), after which PC analysis was used to divide variation into direct and indirect effects. The effective indices (nine out of twenty-five traits) were used in the HC analysis to evaluate the genetic dissimilarity matrix between thirteen genotypes, characterized into three tolerance groups using Euclidean distance and Ward’s method of agglomeration. LD Analysis was employed to validate the genotype tolerance categories (the nine indices used as quantitative variables) with the three categories (as qualitative variables). Statistical analyses (PCA, SMLR, PC, HC, and DFA) were implemented through XLSTAT statistical package software (vers. 2019.1, Excel Add-ins soft SARL, New York, NY, USA).
3. Results
3.1. Screening Genetic Diversity of Drought Tolerance Genotypes
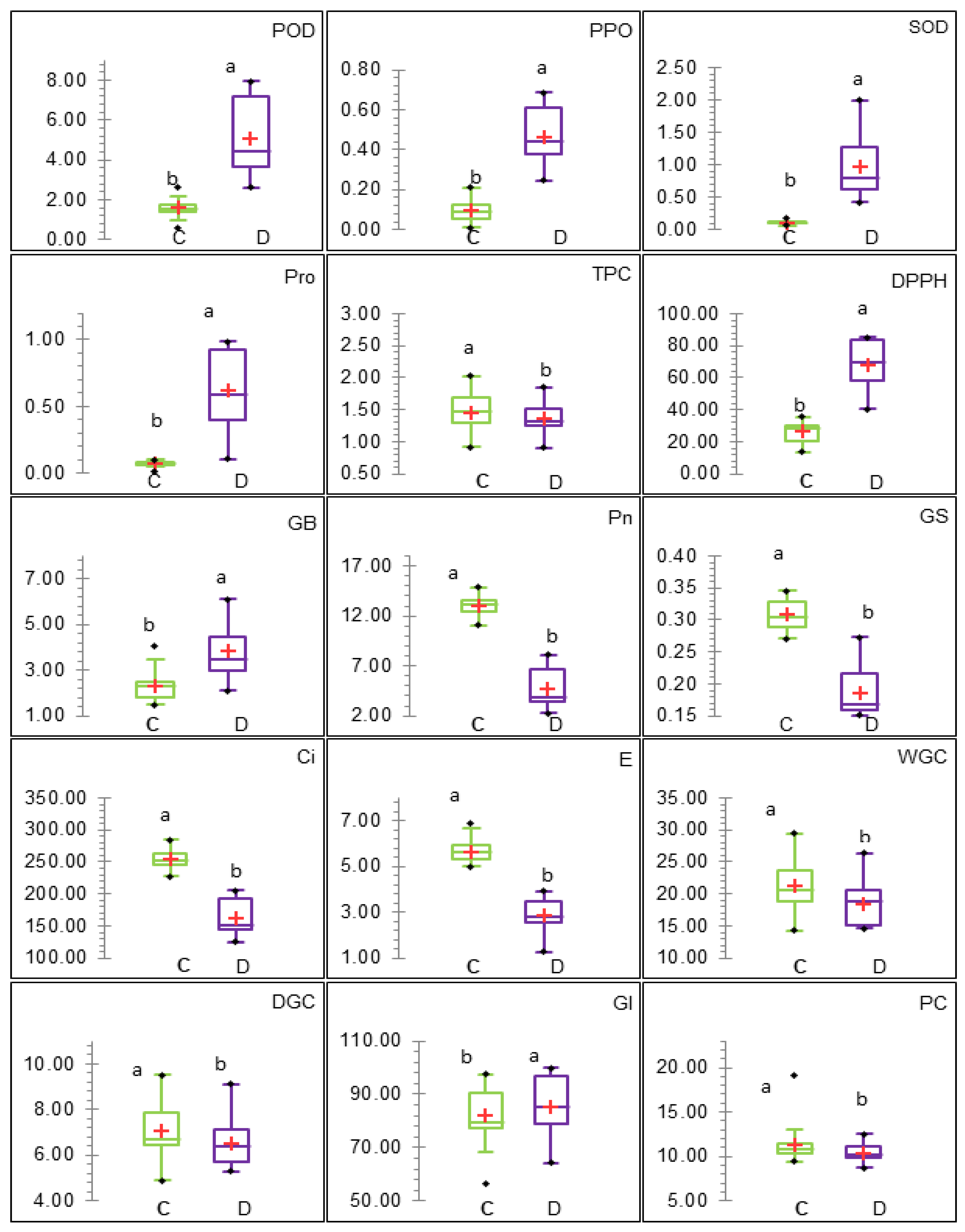
3.2. Phenotypic Analysis of Genotypes and Traits
3.2.1. ANOVA, Genetic Parameters, and Genotype Performance
3.2.2. Multidimensional Analyses in the Classification of Drought-Tolerant Genotypes
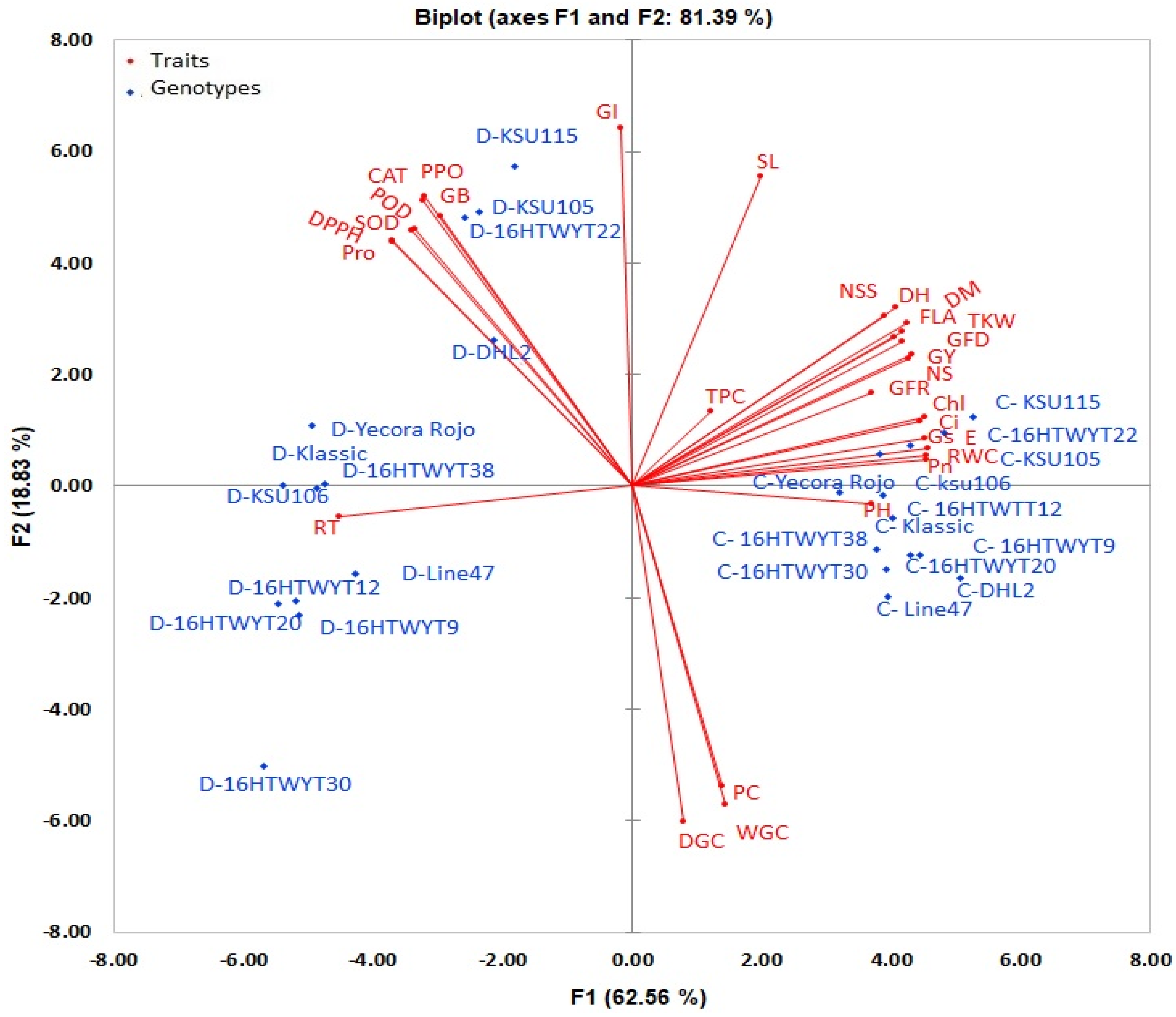
Principal Component Analysis (PCA)
SMLR and PC Analyses for the Performance of Yield Trait
Hierarchical Clustering and Linear Discriminant Analyses

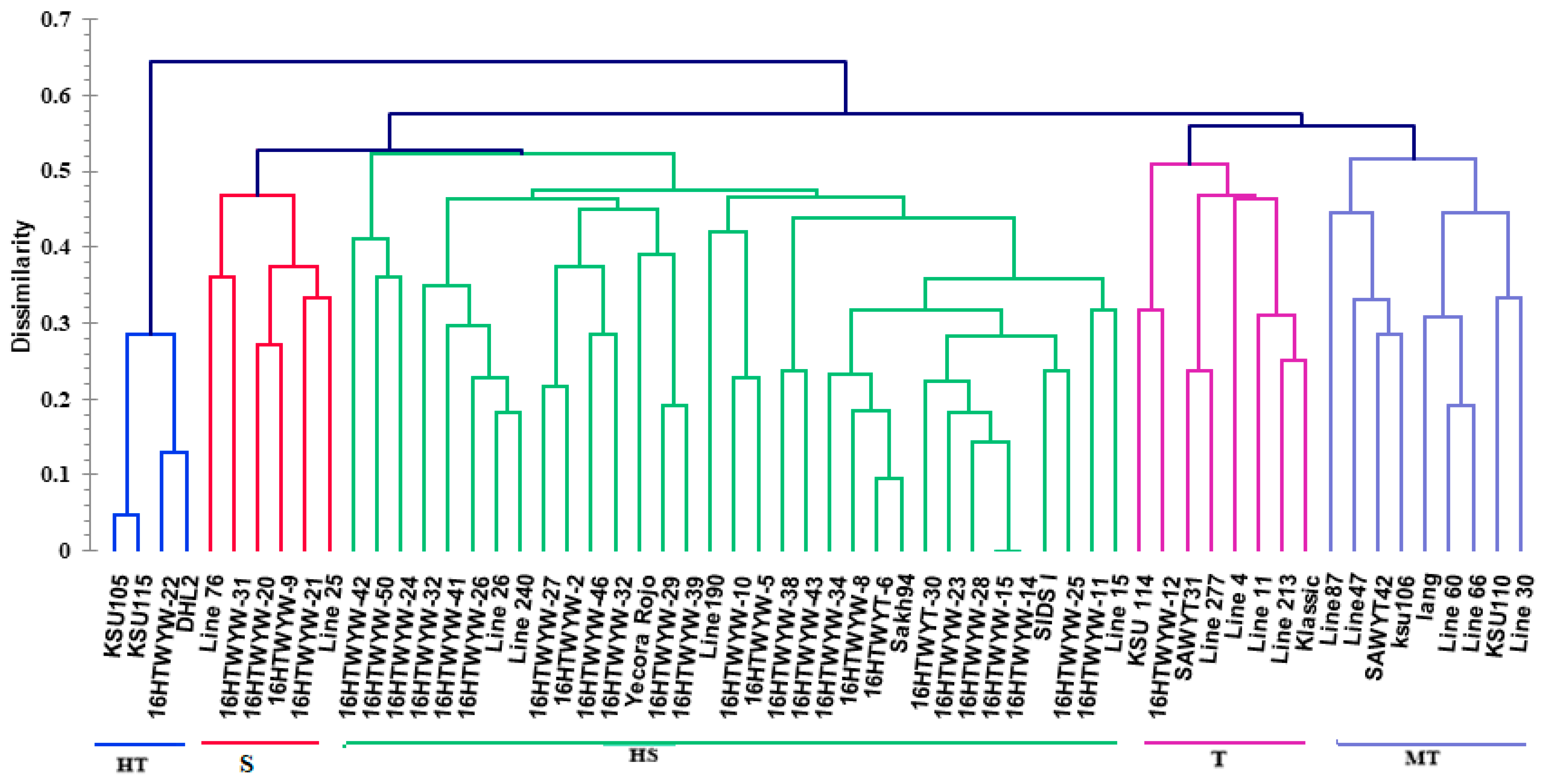
3.3. Genotypic Analysis Based on SSR Markers
3.3.1. Hierarchical Clustering of Genotypes Based on SSR Markers
3.3.2. Association of SSR Markers with Agro-Physio-Biochemical Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Al-Ashkar, I.; Alotaibi, M.; Refay, Y.; Ghazy, A.; Zakri, A.; Al-Doss, A. Selection criteria for high-yielding and early-flowering bread wheat hybrids under heat stress. PLoS ONE 2020, 15, e0236351. [Google Scholar] [CrossRef]
- Al-Ashkar, I.; Al-Suhaibani, N.; Abdella, K.; Sallam, M.; Alotaibi, M.; Seleiman, M.F. Combining Genetic and Multidimensional Analyses to Identify Interpretive Traits Related to Water Shortage Tolerance as an Indirect Selection Tool for Detecting Genotypes of Drought Tolerance in Wheat Breeding. Plants 2021, 10, 931. [Google Scholar] [CrossRef]
- Abdelsalam, N.R.; Kandil, E.E.; Al-Msari, M.A.F.; Al-Jaddadi, M.A.M.; Ali, H.M.; Salem, M.Z.M.; Elshikh, M.S. Effect of foliar application of NPK nanoparticle fertilization on yield and genotoxicity in wheat (Triticum aestivum L.). Sci. Total Environ. 2019, 653, 1128–1139. [Google Scholar] [CrossRef]
- Shewry, P.R.; Hey, S.J. The contribution of wheat to human diet and health. Food Energy Secur. 2015, 4, 178–202. [Google Scholar] [CrossRef]
- Barakat, M.N.; Al-Doss, A.A.; Moustafa, K.A.; Motawei, M.I.; Alamri, M.S.; Mergoum, M.; Sallam, M.S.; Al-Ashkar, I.M. Molecular detection of QTLs for flour quality traits in two doubled haploid populations in spring wheat under heat stress. Cereal Res. Commun. 2020, 48, 525–532. [Google Scholar] [CrossRef]
- Al-Ashkar, I.; Sallam, M.; Almutairi, K.F.; Shady, M.; Ibrahim, A.; Alghamdi, S.S. Detection of High-Performance Wheat Genotypes and Genetic Stability to Determine Complex Interplay between Genotypes and Environments. Agronomy 2023, 13, 585. [Google Scholar] [CrossRef]
- Mwadzingeni, L.; Shimelis, H.; Dube, E.; Laing, M.D.; Tsilo, T.J. Breeding wheat for drought tolerance: Progress and technologies. J. Integr. Agr. 2016, 15, 935–943. [Google Scholar] [CrossRef]
- Barakat, M.; Al-Doss, A.; El-Hendawy, S.; Al-Suhaibani, N.; Abdella, K.; Al-Ashkar, I. Deciphering novel QTL for spectral reflectance indices in spring wheat. Cereal Res. Commun. 2021, 49, 649–661. [Google Scholar] [CrossRef]
- Mondal, S.; Singh, R.P.; Crossa, J.; Huerta-Espino, J.; Sharma, I.; Chatrath, R.; Singh, G.P.; Sohu, V.S.; Mavi, G.S.; Sukuru, V.S.P.; et al. Earliness in wheat: A key to adaptation under terminal and continual high temperature stress in South Asia. Field Crop. Res. 2013, 151, 19–26. [Google Scholar] [CrossRef]
- Wasaya, A.; Manzoor, S.; Yasir, T.A.; Sarwar, N.; Mubeen, K.; Ismail, I.A.; Raza, A.; Rehman, A.; Hossain, A.; EL Sabagh, A. Evaluation of Fourteen Bread Wheat (Triticum aestivum L.) Genotypes by Observing Gas Exchange Parameters, Relative Water and Chlorophyll Content, and Yield Attributes under Drought Stress. Sustainability 2021, 13, 4799. [Google Scholar] [CrossRef]
- Raza, S.; Saleem, M.; Khan, I.; Jamil, M.; Ijaz, M.; Khan, M.A. Evaluating the drought stress tolerance efficiency of wheat (Triticum aestivum L.) cultivars. Russ. J. Agric. Socio-Econ. Sci. 2012, 12, 41–46. [Google Scholar]
- Hasanuzzaman, M.; Bhuyan, M.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Al-Ashkar, I.; Alderfasi, A.; El-Hendawy, S.; Al-Suhaibani, N.; El-Kafafi, S.; Seleiman, M.F. Detecting Salt Tolerance in Doubled Haploid Wheat Lines. Agronomy 2019, 9, 211. [Google Scholar] [CrossRef]
- Naseer, M.A.; Hussain, S.; Nengyan, Z.; Ejaz, I.; Ahmad, S.; Farooq, M.; Ren, X. Shading under drought stress during grain filling attenuates photosynthesis, grain yield and quality of winter wheat in the Loess Plateau of China. J. Agron. Crop Sci. 2022, 208, 255–263. [Google Scholar] [CrossRef]
- Li, M.; Yang, Y.; Raza, A.; Yin, S.; Wang, H.; Zhang, Y.; Dong, J.; Wang, G.; Zhong, C.; Zhang, H.; et al. Heterologous expression of Arabidopsis thaliana rty gene in strawberry (Fragaria × ananassa Duch.) improves drought tolerance. BMC Plant Biol. 2021, 21, 57. [Google Scholar] [CrossRef] [PubMed]
- Al-Ashkar, I.; Alderfasi, A.; Ben Romdhane, W.; Seleiman, M.F.; El-Said, R.A.; Al-Doss, A. Morphological and Genetic Diversity within Salt Tolerance Detection in Eighteen Wheat Genotypes. Plants 2020, 9, 287. [Google Scholar] [CrossRef] [PubMed]
- Gambetta, G.A.; Herrera, J.C.; Dayer, S.; Feng, Q.; Hochberg, U.; Castellarin, S.D. The physiology of drought stress in grapevine: Towards an integrative definition of drought tolerance. J. Exp. Bot. 2020, 71, 4658–4676. [Google Scholar] [CrossRef] [PubMed]
- Al-Ashkar, I.; Romdhane, W.B.; El-Said, R.A.; Ghazy, A.; Attia, K.; Al-Doss, A. Agro-Physiologic Responses and Stress-Related Gene Expression of Four Doubled Haploid Wheat Lines under Salinity Stress Conditions. Biology 2021, 10, 56. [Google Scholar] [CrossRef] [PubMed]
- Krystkowiak, K.; Adamski, T.; Surma, M.; Kaczmarek, Z. Relationship between phenotypic and genetic diversity of parental genotypes and the specific combining ability and heterosis effects in wheat. Euphytica 2009, 165, 419–434. [Google Scholar] [CrossRef]
- Hashem, M.; Sandhu, K.S.; Ismail, S.M.; Borner, A.; Sallam, A. Validation and marker-assisted selection of DArT-genomic regions associated with wheat yield-related traits under normal and drought conditions. Front. Genet. 2023, 14, 1195566. [Google Scholar] [CrossRef]
- Ainsworth, C.; Beynon, J.; Buchanan-Wollaston, V. Techniques in Plant Biology: The Practical Manual; University of London: London, UK, 1996. [Google Scholar]
- Al-Faifi, S.A.; Migdadi, H.M.; Algamdi, S.S.; Khan, M.A.; Ammar, M.H.; Al-Obeed, R.S.; Al-Thamra, M.I.; El-Harty, E.H.; Jakse, J. Development, characterization and use of genomic SSR markers for assessment of genetic diversity in some Saudi date palm cultivars. Electron. J. Biotechn. 2016, 21, 18–25. [Google Scholar] [CrossRef]
- Mahmood, U.; Hussain, S.; Hussain, S.; Ali, B.; Ashraf, U.; Zamir, S.; Al-Robai, S.A.; Alzahrani, F.O.; Hano, C.; El-Esawi, M.A. Morpho-Physio-Biochemical and Molecular Responses of Maize Hybrids to Salinity and Waterlogging during Stress and Recovery Phase. Plants 2021, 10, 1345. [Google Scholar] [CrossRef] [PubMed]
- Belete, Y.; Shimelis, H.; Laing, M.; Mathew, I. Genetic diversity and population structure of bread wheat genotypes determined via phenotypic and SSR marker analyses under drought-stress conditions. J. Crop Improv. 2021, 35, 303–325. [Google Scholar] [CrossRef]
- Eivazi, A.R.; Naghavi, M.R.; Hajheidari, M.; Pirseyedi, S.M.; Ghaffari, M.R.; Mohammadi, S.A.; Majidi, I.; Salekdeh, G.H.; Mardi, M. Assessing wheat (Triticum easativum L.) genetic diversity using quality traits, amplified fragment length polymorphisms, simple sequence repeats and proteome analysis. Ann. Appl. Biol. 2008, 152, 81–91. [Google Scholar] [CrossRef]
- Zhang, D.D.; Bai, G.H.; Zhu, C.S.; Yu, J.M.; Carver, B.F. Genetic Diversity, Population Structure, and Linkage Disequilibrium in US Elite Winter Wheat. Plant Genome 2010, 3, 117–127. [Google Scholar] [CrossRef]
- Mohi-Ud-Din, M.; Hossain, M.A.; Rohman, M.M.; Uddin, M.N.; Haque, M.S.; Dessoky, E.S.; Alqurashi, M.; Aloufi, S. Assessment of Genetic Diversity of Bread Wheat Genotypes for Drought Tolerance Using Canopy Reflectance-Based Phenotyping and SSR Marker-Based Genotyping. Sustainability 2022, 14, 9818. [Google Scholar] [CrossRef]
- El-Rawy, M.A.; Hassan, M.I. Assessment of Genetic Diversity in Durum and Bread Wheat Genotypes Based on Drought Tolerance and SSR Markers. Plant Breed. Biotechnol. 2021, 9, 89–103. [Google Scholar] [CrossRef]
- Ateş Sönmezoğlu, Ö.; Terzi, B. Characterization of some bread wheat genotypes using molecular markers for drought tolerance. Physiol. Mol. Biol. Plants 2018, 24, 159–166. [Google Scholar] [CrossRef]
- Semahegn, Y.; Shimelis, H.; Laing, M.; Mathew, I. Evaluation of bread wheat (Triticum aestivum L.) genotypes for yield and related traits under drought stress conditions. Acta Agric. Scand. Sect. B—Soil Plant Sci. 2020, 70, 474–484. [Google Scholar]
- Mir, R.R.; Zaman-Allah, M.; Sreenivasulu, N.; Trethowan, R.; Varshney, R.K. Integrated genomics, physiology and breeding approaches for improving drought tolerance in crops. Theor. Appl. Genet. 2012, 125, 625–645. [Google Scholar] [CrossRef]
- Oguz, M.C.; Aycan, M.; Oguz, E.; Poyraz, I.; Yildiz, M. Drought Stress Tolerance in Plants: Interplay of Molecular, Biochemical and Physiological Responses in Important Development Stages. Physiologia 2022, 2, 180–197. [Google Scholar] [CrossRef]
- Zeng, L.; Shannon, M.C.; Grieve, C.M. Evaluation of salt tolerance in rice genotypes by multiple agronomic parameters. Euphytica 2002, 127, 235–245. [Google Scholar] [CrossRef]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8018. [Google Scholar] [CrossRef]
- Morgan, J.M.; Tan, M.K. Chromosomal location of a wheat osmoregulation gene using RFLP analysis. Aust. J. Plant Physiol. 1996, 23, 803–806. [Google Scholar] [CrossRef]
- Quarrie, P. The scientific library of the Earls of Macclesfield. Notes Rec. Roy. Soc. 2006, 60, 5–24. [Google Scholar] [CrossRef][Green Version]
- Cattivelli, L.; Baldi, P.; Crosatti, C.; Di Fonzo, N.; Faccioli, P.; Grossi, M.; Mastrangelo, A.M.; Pecchioni, N.; Stanca, A.M. Chromosome regions and stress-related sequences involved in resistance to abiotic stress in Triticeae. Plant Mol. Biol. 2002, 48, 649–665. [Google Scholar] [CrossRef] [PubMed]
- Galiba, G. Mapping of genes regulating abiotic stress tolerance in cereals. Acta Agron. Hung. 2002, 50, 235–247. [Google Scholar] [CrossRef]
- Pestsova, E.; Ganal, M.W.; Roder, M.S. Isolation and mapping of microsatellite markers specific for the D genome of bread wheat. Genome 2000, 43, 689–697. [Google Scholar] [CrossRef]
- Röder, M.S.; Korzun, V.; Gill, B.S.; Ganal, M.W. The physical mapping of microsatellite markers in wheat. Genome 1998, 41, 278–283. [Google Scholar] [CrossRef]
- Röder, M.S.; Plaschke, J.; König, S.U.; Börner, A.; Sorrells, M.E.; Tanksley, S.D.; Ganal, M.W. Abundance, variability and chromosomal location of microsatellites in wheat. Molec. Gen. Genet. 1995, 246, 327–333. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning; CSH Press: Cold Spring, NY, USA, 2012. [Google Scholar]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef]
- Aebi, H. [13] Catalase in vitro. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1984; Volume 105, pp. 121–126. [Google Scholar]
- Chance, B.; Maehly, A.C. Preparation and assays of enzymes. Methods Enzymol. 1955, 2, 773–775. [Google Scholar]
- Duckworth, H.W.; Coleman, J.E. Physicochemical and kinetic properties of mushroom tyrosinase. J. Biol. Chem. 1970, 245, 1613–1625. [Google Scholar] [CrossRef] [PubMed]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a Free-Radical Method to Evaluate Antioxidant Activity. Food Sci. Technol.-Leb. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Gulcin, I. Antioxidants and antioxidant methods: An updated overview. Arch. Toxicol. 2020, 94, 651–715. [Google Scholar] [CrossRef] [PubMed]
- Özaslan, M.S.; Saglamtas, R.; Demir, Y.; Genç, Y.; Saraçoglu, I.; Gülçin, I. Isolation of Some Phenolic Compounds from Plantago subulata L. and Determination of Their Antidiabetic, Anticholinesterase, Antiepileptic and Antioxidant Activity. Chem. Biodivers. 2022, 19, e202200280. [Google Scholar] [CrossRef]
- Sarker, U.; Oba, S. Phenolic profiles and antioxidant activities in selected drought-tolerant leafy vegetable amaranth. Sci. Rep. 2020, 10, 18287. [Google Scholar] [CrossRef]
- Grieve, C.M.; Grattan, S.R. Rapid Assay for Determination of Water-Soluble Quaternary Ammonium-Compounds. Plant Soil 1983, 70, 303–307. [Google Scholar] [CrossRef]
- Boctor, F.N. An improved method for colorimetric determination of proline with isatin. Anal. Biochem. 1971, 43, 66–70. [Google Scholar] [CrossRef]
- American Association of Cereal Chemists; Approved Methods Committee. Approved Methods of the American Association of Cereal Chemists; AACC: St. Paul, MN, USA, 2000. [Google Scholar]
- Erdayani, E.; Nagarajan, R.; Grant, N.P.; Gill, K.S. Genome-wide analysis of the HSP101/CLPB gene family for heat tolerance in hexaploid wheat. Sci. Rep. 2020, 10, 3948. [Google Scholar] [CrossRef] [PubMed]
- Mourad, A.M.I.; Belamkar, V.; Baenziger, P.S. Molecular genetic analysis of spring wheat core collection using genetic diversity, population structure, and linkage disequilibrium. BMC Genom. 2020, 21, 434. [Google Scholar] [CrossRef] [PubMed]
- Amro, A.; Harb, S.; Farghaly, K.A.; Ali, M.M.; Mohammed, A.G.; Mourad, A.M.; Afifi, M.; Börner, A.; Sallam, A. Growth responses and genetic variation among highly ecologically diverse spring wheat genotypes grown under seawater stress. Front. Plant Sci. 2022, 13, 996538. [Google Scholar] [CrossRef]
- Tanin, M.J.; Saini, D.K.; Sandhu, K.S.; Pal, N.; Gudi, S.; Chaudhary, J.; Sharma, A. Consensus genomic regions associated with multiple abiotic stress tolerance in wheat and implications for wheat breeding. Sci. Rep. 2022, 12, 13680. [Google Scholar] [CrossRef]
- Mourad, A.M.; Alomari, D.Z.; Alqudah, A.M.; Sallam, A.; Salem, K.F. Recent advances in wheat (Triticum spp.) breeding. In Advances in Plant Breeding Strategies: Cereals; Springer: Cham, Switzerland, 2019; pp. 559–593. [Google Scholar]
- Mondal, S.; Sallam, A.; Sehgal, D.; Sukumaran, S.; Farhad, M.; Navaneetha Krishnan, J.; Kumar, U.; Biswal, A. Advances in breeding for abiotic stress tolerance in wheat. In Genomic Designing for Abiotic Stress Resistant Cereal Crops; Springer: Cham, Switzerland, 2021; pp. 71–103. [Google Scholar]
- Nepolean, T.; Kaul, J.; Mukri, G.; Mittal, S. Genomics-Enabled Next-Generation Breeding Approaches for Developing System-Specific Drought Tolerant Hybrids in Maize. Front. Plant Sci. 2018, 9, 361. [Google Scholar] [CrossRef]
- Al-Ashkar, I.; Al-Doss, A.; Ullah, N. Accelerating Crop Improvement through Speed Breeding. In Climate-Resilient Agriculture, Vol 1: Crop Responses and Agroecological Perspectives; Hasanuzzaman, M., Ed.; Springer International Publishing: Cham, Switzerland, 2023; pp. 821–847. [Google Scholar]
- Urrea-Gómez, R.; Ceballos, H.; Pandey, S.; Bahía Filho, A.F.; León, L.A. A greenhouse screening technique for acid soil tolerance in maize. Agron. J. 1996, 88, 806–812. [Google Scholar] [CrossRef]
- Boakye-Peprah, B.; Ofori, K.; Asante, I.; Parkes, E. Genetic variability of three cassava traits across three locations in Ghana. Afr. J. Plant Sci. 2013, 7, 265–267. [Google Scholar] [CrossRef]
- Falconer, D.S. Introduction to Quantitative Genetics; Pearson Education India: Delhi, India, 1996. [Google Scholar]
- Obala, J.; Saxena, R.K.; Singh, V.K.; Vechalapu, S.; Das, R.; Rathore, A.; Sameer-Kumar, C.V.; Saxena, K.; Tongoona, P.; Sibiya, J.; et al. Genetic variation and relationships of total seed protein content with some agronomic traits in pigeonpea (‘Cajanus cajan’(L.) Millsp.). Aust. J. Crop Sci. 2018, 12, 1859–1865. [Google Scholar] [CrossRef]
- Burton, G. Qualitative inheritance in grasses. Vol. 1. In Proceedings of the 6th International Grassland Congress, Pennsylvania State College, State College, PA, USA, 17–23 August 1952; pp. 17–23. [Google Scholar]
- Al-Ashkar, I.; Sallam, M.; Ghazy, A.; Ibrahim, A.; Alotaibi, M.; Ullah, N.; Al-Doss, A. Agro-Physiological Indices and Multidimensional Analyses for Detecting Heat Tolerance in Wheat Genotypes. Agronomy 2023, 13, 154. [Google Scholar] [CrossRef]
- Hosseini, S.; Sarvestani, Z.; Pirdashti, H.; Afkhami, A.; Hazrati, S. Estimation of heritability and genetic advance for screening some rice genotypes at salt stress conditions. Int. J. Agron. Plant Prod. 2012, 3, 475–482. [Google Scholar]
- Grzesiak, S.; Hordyńska, N.; Szczyrek, P.; Grzesiak, M.T.; Noga, A.; Szechyńska-Hebda, M. Variation among wheat (Triticum easativum L.) genotypes in response to the drought stress: I–selection approaches. J. Plant Interact. 2019, 14, 30–44. [Google Scholar] [CrossRef]
- Liu, C.Y.; Sukumaran, S.; Claverie, E.; Sansaloni, C.; Dreisigacker, S.; Reynolds, M. Genetic dissection of heat and drought stress QTLs in phenology-controlled synthetic-derived recombinant inbred lines in spring wheat. Mol. Breed. 2019, 39, 34. [Google Scholar] [CrossRef]
- Khan, A.S.; Ashfaq, M.; Asad, M.A. A correlation and path coefficient analysis for some yield components in bread wheat. Asian J. Plant Sci. 2003, 2, 582–584. [Google Scholar] [CrossRef]
- Bojarian, M.; Asadi-Gharneh, H.A.; Golabadi, M. Factor analysis, stepwise regression and path coefficient analyses of yield, yield-associated traits, and fruit quality in tomato. Int. J. Veg. Sci. 2019, 25, 542–553. [Google Scholar] [CrossRef]
- Yang, B.; Chen, N.; Dang, Y.; Wang, Y.; Wen, H.; Zheng, J.; Zheng, X.; Zhao, J.; Lu, J.; Qiao, L. Identification and validation of quantitative trait loci for chlorophyll content of flag leaf in wheat under different phosphorus treatments. Front. Plant Sci. 2022, 13, 1019012. [Google Scholar] [CrossRef] [PubMed]
- Tahmasebi, S.; Heidari, B.; Pakniyat, H.; McIntyre, C.L. Mapping QTLs associated with agronomic and physiological traits under terminal drought and heat stress conditions in wheat (Triticum aestivum L.). Genome 2016, 60, 26–45. [Google Scholar] [CrossRef] [PubMed]
- Annunziata, M.G.; Ciarmiello, L.F.; Woodrow, P.; Dell’Aversana, E.; Carillo, P. Spatial and Temporal Profile of Glycine Betaine Accumulation in Plants Under Abiotic Stresses. Front. Plant Sci. 2019, 10, 230. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.; Singh, N.; Haribhushan, A. Compatible solute engineering in plants for abiotic stress tolerance—Role of glycine betaine. Curr. Genom. 2013, 14, 157–165. [Google Scholar] [CrossRef]
- De Leon, T.B.; Linscombe, S.; Gregorio, G.; Subudhi, P.K. Genetic variation in Southern USA rice genotypes for seedling salinity tolerance. Front. Plant Sci. 2015, 6, 374. [Google Scholar] [CrossRef] [PubMed]
- Junaid, M.B.; El-Hendawy, S.; Al-Ashkar, I.; Al-Suhaibani, N.; Alotaibi, M. Integrating Agro-Morpho-Physiological Traits and SSR Markers for Detecting the Salt Tolerance of Advanced Spring Wheat Lines under Field Conditions. Agriculture 2023, 13, 2135. [Google Scholar] [CrossRef]
- El-Hendawy, S.; Elshafei, A.; Al-Suhaibani, N.; Alotabi, M.; Hassan, W.; Dewir, Y.H.; Abdella, K. Assessment of the salt tolerance of wheat genotypes during the germination stage based on germination ability parameters and associated SSR markers. J. Plant Interact. 2019, 14, 151–163. [Google Scholar] [CrossRef]
- ahmad Alkuddsi, Y.; Patil, S.; Manjula, S.; Nadaf, H.; Patil, B.C. Relationship between SSR-based molecular marker and cotton F 1 inter specific hybrids performance for seed cotton yield and fiber properties. Genom. Appl. Biol. 2013, 4, 22–34. [Google Scholar] [CrossRef][Green Version]
- Barakat, M.N.; Saleh, M.; Al-Doss, A.A.; Moustafa, K.A.; Elshafei, A.A.; Al-Qurainy, F.H. Identification of New SSR Markers Linked to Leaf Chlorophyll Content, Flag Leaf Senescence and Cell Membrane Stability Traits in Wheat under Water Stressed Condition. Acta Biol. Hung. 2015, 66, 93–102. [Google Scholar] [CrossRef]
- Khan, D.; Muhammad, I.; Shuaib, M.; Hussain, F.; Romman, M.; Azam, N.; Abidullah, S.; Zeb, A.; Rauf, A.; Bahadur, S. Investigation of grain yield and drought resistance in selected wheat lines based on molecular markers. Ukr. J. Ecol. 2021, 11, 44–50. [Google Scholar]
- Wang, S.; Jia, S.; Sun, D.; Wang, H.; Dong, F.; Ma, H.; Jing, R.; Ma, G. Genetic basis of traits related to stomatal conductance in wheat cultivars in response to drought stress. Photosynthetica 2015, 53, 299–305. [Google Scholar] [CrossRef]
- Kadam, S.; Singh, K.; Shukla, S.; Goel, S.; Vikram, P.; Pawar, V.; Gaikwad, K.; Khanna-Chopra, R.; Singh, N. Genomic associations for drought tolerance on the short arm of wheat chromosome 4B. Funct. Integr. Genom. 2012, 12, 447–464. [Google Scholar] [CrossRef] [PubMed]


| Source | DF | Pn | Gs | Ci | E | Chl | PC | GI | WGC | DGC | PH | DH | DM | SL | NS | NSS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rep | 2 | 0.365 | 0.0002 | 25.906 | 0.277 | 0.004 | 0.631 | 12.747 | 0.616 | 0.003 | 3.128 | 0.051 | 13.128 | 0.474 | 762.859 | 0.154 |
| I | 1 | 1365.716 ** | 0.292 ** | 166,828.1 ** | 149.66 ** | 30.688 * | 17.033 * | 211.207 ns | 139.361 ** | 5.600 ** | 4063.705 ** | 714.051 ** | 5008.013 ** | 11.538 ** | 213,938.782 ** | 149.53 ** |
| Error a | 2 | 0.382 | 0.00001 | 103.586 | 0.188 | 0.395 | 0.905 | 30.756 | 1.044 | 0.003 | 0.359 | 0.051 | 13.128 | 0.115 | 158.244 | 1.077 |
| G | 12 | 7.055 ** | 0.003 ** | 2131.897 ** | 0.96 ** | 0.964 ** | 6.609 ** | 555.003 ** | 43.507 ** | 4.933 ** | 167.513 ** | 32.218 ** | 150.427 ** | 14.421 ** | 3908.987 ** | 8.872 ** |
| I * G | 12 | 3.004 ** | 0.001 ** | 601.333 ** | 0.431 ** | 0.215 ** | 2.573 ** | 54.762 ** | 24.021 ** | 2.043 ** | 87.205 ** | 12.218 ** | 50.568 ** | 0.344 ** | 2395.226 ** | 2.65 ** |
| Errorb | 48 | 0.632 | 0.0003 | 74.685 | 0.138 | 0.055 | 0.721 | 12.296 | 1.077 | 0.021 | 10.244 | 0.051 | 0.517 | 0.517 | 425.44 | 1.004 |
| Genetic Parameters | ||||||||||||||||
| σ2G | 0.675 | 0.000 | 255.094 | 0.088 | 0.125 | 0.673 | 83.374 | 3.248 | 0.482 | 13.385 | 3.333 | 16.643 | 2.346 | 252.294 | 1.037 | |
| σ2e | 0.105 | 0.000 | 12.448 | 0.023 | 0.009 | 0.120 | 2.049 | 0.180 | 0.004 | 1.707 | 0.009 | 0.086 | 0.086 | 70.907 | 0.167 | |
| σ2Ph | 1.176 | 0.001 | 355.316 | 0.160 | 0.161 | 1.102 | 92.501 | 7.251 | 0.822 | 27.919 | 5.370 | 25.071 | 2.432 | 651.498 | 1.479 | |
| h2 % | 57.420 | 66.667 | 71.794 | 55.104 | 77.697 | 61.068 | 90.133 | 44.788 | 58.585 | 47.941 | 62.077 | 66.384 | 96.457 | 38.725 | 70.131 | |
| G.C.V. % | 9.191 | 7.347 | 7.662 | 6.948 | 13.036 | 7.579 | 10.916 | 9.083 | 10.185 | 4.249 | 2.593 | 3.698 | 15.601 | 3.269 | 6.507 | |
| Ph.C.V. % | 12.130 | 8.998 | 9.043 | 9.360 | 14.789 | 9.698 | 11.498 | 13.573 | 13.306 | 6.137 | 3.291 | 4.539 | 15.885 | 5.253 | 7.770 | |
| GA | 1.283 | 0.031 | 27.878 | 0.454 | 0.642 | 1.320 | 17.858 | 2.484 | 1.094 | 5.218 | 2.963 | 6.847 | 3.099 | 20.362 | 1.757 | |
| GG % | 14.348 | 12.358 | 13.374 | 10.625 | 23.671 | 12.200 | 21.348 | 12.523 | 16.059 | 6.060 | 4.209 | 6.206 | 31.563 | 4.190 | 11.226 | |
| Source | DF | GFD | GFR | TKW | GY | RT | FLA | CAT | POD | PPO | SOD | DPPH | TPC | Pro | RWC | GB |
| Rep | 2 | 0.051 | 0.304 | 3.962 | 0.012 | 0.279 | 2.665 | 0.005 | 0.94 | 0.007 | 0.034 | 0.096 | 0.004 | 0.006 | 0.279 | 0.079 |
| I | 1 | 1354.167 ** | 44.192 * | 1117.30 ** | 79.50 ** | 13,479.04 ** | 4185.28 ** | 216.430 ** | 230.043 ** | 2.619 ** | 14.878 ** | 34,491.283 ** | 0.180 ns | 5.865 ** | 13,479.05 ** | 46.066 * |
| Error a | 2 | 0.051 | 0.574 | 3.882 | 0.067 | 62.231 | 4.203 | 0.2 | 0.06 | 0.009 | 0.024 | 0.096 | 0.168 | 0.002 | 62.231 | 0.735 |
| G | 12 | 57.013 ** | 2.76 ** | 38.715 ** | 1.088 ** | 73.831 ** | 194.66 ** | 6.78 ** | 5.312 ** | 0.038 ** | 0.403 ** | 460.565 ** | 0.14 * | 0.163 ** | 73.831 ** | 3.371 ** |
| I * G | 12 | 25.333 ** | 1.045 ** | 13.241 ** | 0.416 ** | 34.313 ** | 54.192 ** | 3.208 ** | 4.641 ** | 0.02 ** | 0.222 ** | 190.791 ** | 0.08 ** | 0.062 ** | 34.313 ** | 1.688 ** |
| Error b | 48 | 0.051 | 0.531 | 1.645 | 0.078 | 3.248 | 14.302 | 0.067 | 0.139 | 0.001 | 0.006 | 8.877 | 0.045 | 0.009 | 3.248 | 0.185 |
| Genetic Parameters | ||||||||||||||||
| σ2G | 5.280 | 0.286 | 4.246 | 0.112 | 6.586 | 23.411 | 0.595 | 0.112 | 0.003 | 0.030 | 44.962 | 0.010 | 0.017 | 6.586 | 0.281 | |
| σ2e | 0.009 | 0.089 | 0.274 | 0.013 | 0.541 | 2.384 | 0.011 | 0.023 | 0.000 | 0.001 | 1.480 | 0.008 | 0.002 | 0.541 | 0.031 | |
| σ2Ph | 9.502 | 0.460 | 6.453 | 0.181 | 12.305 | 32.443 | 1.130 | 0.885 | 0.006 | 0.067 | 76.761 | 0.023 | 0.027 | 12.305 | 0.562 | |
| h2 % | 55.566 | 62.138 | 65.799 | 61.765 | 53.525 | 72.161 | 52.684 | 12.632 | 47.368 | 44.913 | 58.575 | 42.857 | 61.963 | 53.525 | 49.926 | |
| G.C.V. % | 5.638 | 3.259 | 4.890 | 4.983 | 14.227 | 12.019 | 35.450 | 10.005 | 19.542 | 32.210 | 14.267 | 7.109 | 37.975 | 3.131 | 17.304 | |
| Ph.C.V. % | 7.564 | 4.135 | 6.028 | 6.340 | 19.446 | 14.149 | 48.840 | 28.150 | 28.394 | 48.062 | 18.641 | 10.860 | 48.243 | 4.280 | 24.490 | |
| GA | 3.528 | 0.868 | 3.443 | 0.542 | 3.868 | 8.467 | 1.154 | 0.245 | 0.078 | 0.240 | 10.572 | 0.135 | 0.210 | 3.868 | 0.771 | |
| GG % | 8.658 | 5.293 | 8.171 | 8.067 | 21.441 | 21.033 | 53.006 | 7.325 | 27.707 | 44.467 | 22.493 | 9.588 | 61.580 | 4.719 | 25.188 | |
| PCA1 | PCA2 | PCA3 | PCA4 | |
|---|---|---|---|---|
| Eigenvalue | 18.769 | 5.648 | 1.504 | 1.016 |
| Variability (%) | 62.564 | 18.827 | 5.013 | 3.386 |
| Cumulative % | 62.564 | 81.391 | 86.404 | 89.790 |
| Eigenvectors: | ||||
| RWC | 0.974 | 0.004 | 0.005 | 0.0002 |
| RT | 0.974 | 0.004 | 0.0005 | 0.0002 |
| Chl | 0.931 | 0.020 | 0.003 | 0.0001 |
| CAT | 0.496 | 0.372 | 0.057 | 0.016 |
| POD | 0.549 | 0.299 | 0.069 | 0.000 |
| PPO | 0.485 | 0.383 | 0.085 | 0.001 |
| SOD | 0.647 | 0.275 | 0.034 | 0.005 |
| Pro | 0.534 | 0.301 | 0.013 | 0.0003 |
| TPC | 0.069 | 0.026 | 0.293 | 0.381 |
| DPPH | 0.646 | 0.273 | 0.036 | 0.008 |
| GB | 0.417 | 0.334 | 0.0001 | 0.002 |
| Pn | 0.97 | 0.003 | 0.003 | 0.001 |
| Gs | 0.959 | 0.010 | 0.012 | 0.003 |
| Ci | 0.958 | 0.021 | 0.002 | 0.000 |
| E | 0.978 | 0.007 | 0.000 | 0.001 |
| FAL | 0.776 | 0.100 | 0.0003 | 0.001 |
| DH | 0.776 | 0.146 | 0.008 | 0.005 |
| DM | 0.846 | 0.121 | 0.010 | 0.003 |
| GFD | 0.822 | 0.095 | 0.011 | 0.002 |
| GFR | 0.641 | 0.040 | 0.002 | 0.019 |
| PH | 0.645 | 0.001 | 0.029 | 0.033 |
| NS | 0.859 | 0.074 | 0.001 | 0.010 |
| SL | 0.187 | 0.440 | 0.058 | 0.108 |
| NSS | 0.717 | 0.133 | 0.040 | 0.014 |
| TWK | 0.817 | 0.108 | 0.003 | 0.006 |
| GY | 0.876 | 0.079 | 0.005 | 0.007 |
| WGC | 0.098 | 0.464 | 0.262 | 0.135 |
| DGC | 0.029 | 0.515 | 0.285 | 0.131 |
| GI | 0.001 | 0.589 | 0.001 | 0.109 |
| PC | 0.091 | 0.410 | 0.173 | 0.173 |
| Stepwise Regression | Path Coefficient | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Dependent Variable | Source | Partitioning the Correlation | R2 | ||||||
| Regression Coefficient | p-Value | R2 Par. | R2 Com. | Direct Effect | Indirect Effect | Correlation Value | Direct Effect | ||
| GY | Intercept | 18.876 | |||||||
| FLA | 14.247 | <0.0001 | 0.870 | 0.870 | 0.681 | 0.230 | 0.911 | 0.464 | |
| Gs | 0.790 | <0.0001 | 0.049 | 0.919 | 0.340 | −0.371 | −0.031 | 0.115 | |
| PH | 21.076 | 0.001 | 0.056 | 0.975 | 0.294 | −0.282 | 0.011 | 0.086 | |
| RT | −1.151 | 0.017 | 0.014 | 0.988 | −0.128 | 0.278 | 0.150 | 0.016 | |
| Total direct effect | 0.681 | ||||||||
| Total indirect effect | 0.307 | ||||||||
| Total R2 | 0.988 | 0.988 | |||||||
| Residual | 0.109 | 0.109 | |||||||
| FAL | Intercept | 0.309 | |||||||
| GB | −0.319 | <0.0001 | 0.827 | 0.827 | −0.661 | −0.240 | −0.901 | 0.437 | |
| PPO | −0.171 | 0.002 | 0.059 | 0.886 | −0.257 | 0.070 | −0.186 | 0.066 | |
| Chl | −0.380 | 0.005 | 0.048 | 0.935 | −0.214 | 0.114 | −0.099 | 0.046 | |
| GFD | −0.150 | 0.007 | 0.040 | 0.975 | −0.292 | 0.792 | 0.500 | 0.085 | |
| Total direct effect | 0.634 | ||||||||
| Total indirect effect | 0.341 | ||||||||
| Total R2 | 0.975 | 0.975 | |||||||
| Residual | 0.158 | 0.158 | |||||||
| Genotypes | Classification | Cross-Validation | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Prior | Posterior | Membership Probabilities | Posterior | Membership Probabilities | |||||
| Pr (MT) | Pr (S) | Pr (T) | MT | S | T | ||||
| 16HTWYT30 | S | S | 0.000 | 1.000 | 0.000 | MT | 1.000 | 0.000 | 0.000 |
| DHL2 | T | T | 0.000 | 0.000 | 1.000 | T | 0.000 | 0.000 | 1.000 |
| 16HTWYT20 | S | S | 0.000 | 1.000 | 0.000 | MT | 1.000 | 0.000 | 0.000 |
| 16HTWYT38 | MT | MT | 1.000 | 0.000 | 0.000 | MT | 1.000 | 0.000 | 0.000 |
| 16HTWYT9 | S | S | 0.000 | 1.000 | 0.000 | S | 0.000 | 1.000 | 0.000 |
| KSU105 | MT | MT | 1.000 | 0.000 | 0.000 | MT | 1.000 | 0.000 | 0.000 |
| 16HTWYT12 | S | S | 0.000 | 1.000 | 0.000 | MT | 1.000 | 0.000 | 0.000 |
| Yecora Rojo | MT | MT | 1.000 | 0.000 | 0.000 | MT | 1.000 | 0.000 | 0.000 |
| 16HTWYT22 | T | T | 0.000 | 0.000 | 1.000 | S | 0.000 | 1.000 | 0.000 |
| KSU115 | T | T | 0.000 | 0.000 | 1.000 | T | 0.000 | 0.000 | 1.000 |
| Klassic | MT | MT | 1.000 | 0.000 | 0.000 | MT | 1.000 | 0.000 | 0.000 |
| Line47 | S | S | 0.000 | 1.000 | 0.000 | S | 0.000 | 1.000 | 0.000 |
| ksu106 | MT | MT | 1.000 | 0.000 | 0.000 | MT | 1.000 | 0.000 | 0.000 |
| Traits | Treatments | Markres | R2 Par. | R2 Com. | p-Value * |
|---|---|---|---|---|---|
| GY | Control | Gwm337 | 0.336 | 0.336 | 0.038 |
| Drought | Wmc326 | 0.385 | 0.385 | 0.024 | |
| index | wmc326 | 0.425 | 0.425 | 0.016 | |
| FLA | Drought | Wmc326 | 0.460 | 0.460 | 0.11 |
| index | Wmc65 | 0.359 | 0.359 | 0.030 | |
| GB | Control | Wmc154 | 0.579 | 0.579 | 0.000 |
| Cfd1 | 0.204 | 0.783 | 0.012 | ||
| Drought | Wmc326 | 0.623 | 0.623 | 0.000 | |
| Wmc503 | 0.195 | 0.819 | 0.001 | ||
| Wmc65 | 0.114 | 0.933 | 0.001 | ||
| Cfd9 | 0.034 | 0.967 | 0.22 | ||
| index | Wmc326 | 0.456 | 0.456 | 0.000 | |
| Wmc65 | 0.219 | 0.675 | 0.000 | ||
| Wmc170 | 0.211 | 0.886 | 0.001 | ||
| Wmc249 | 0.049 | 0.935 | 0.012 | ||
| Wmc405 | 0.030 | 0.965 | 0.043 | ||
| Gs | Control | Wmc405 | 0.313 | 0.313 | 0.047 |
| PPO | Control | Cfd9 | 0.389 | 0.389 | 0.015 |
| Gwm369 | 0.222 | 0.611 | 0.038 | ||
| Drought | Wmc326 | 0.458 | 0.458 | 0.11 | |
| index | Cfd1 | 0.434 | 0.434 | 0.002 | |
| Gwm369 | 0.210 | 0.644 | 0.036 | ||
| Chl | Control | Wmc503 | 0.392 | 0.392 | 0.022 |
| Drought | Wmc326 | 0.418 | 0.418 | 0.017 | |
| index | Wmc326 | 0.401 | 0.401 | 0.005 | |
| Cfd9 | 0.228 | 0.629 | 0.033 | ||
| RT | Control | Gwm369 | 0.699 | 0.699 | 0.000 |
| Wmc326 | 0.159 | 0.857 | 0.006 | ||
| Wmc154 | 0.054 | 0.911 | 0.044 | ||
| Drought | Wmc326 | 0.321 | 0.321 | 0.000 | |
| Cfd18 | 0.306 | 0.628 | 0.000 | ||
| Cfd9 | 0.150 | 0.778 | 0.011 | ||
| Wmc18 | 0.108 | 0.886 | 0.008 | ||
| Wmc154 | 0.058 | 0.944 | 0.031 | ||
| index | Gwm369 | 0.546 | 0.546 | 0.017 | |
| Cfd1 | 0.256 | 0.801 | 0.000 | ||
| Wmc177 | 0.121 | 0.922 | 0.005 | ||
| GFD | Drought | Wmc326 | 0.356 | 0.356 | 0.031 |
| index | Wmc170 | 0.350 | 0.350 | 0.033 | |
| PH | Control | Wmc65 | 0.396 | 0.396 | 0.001 |
| Wmc154 | 0.334 | 0.730 | 0.000 | ||
| Wmc74 | 0.169 | 0.899 | 0.002 | ||
| Wmc18 | 0.048 | 0.920 | 0.028 | ||
| index | Wmc65 | 0.464 | 0.464 | 0.010 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sallam, M.; Ghazy, A.; Al-Doss, A.; Al-Ashkar, I. Combining Genetic and Phenotypic Analyses for Detecting Bread Wheat Genotypes of Drought Tolerance through Multivariate Analysis Techniques. Life 2024, 14, 183. https://doi.org/10.3390/life14020183
Sallam M, Ghazy A, Al-Doss A, Al-Ashkar I. Combining Genetic and Phenotypic Analyses for Detecting Bread Wheat Genotypes of Drought Tolerance through Multivariate Analysis Techniques. Life. 2024; 14(2):183. https://doi.org/10.3390/life14020183
Chicago/Turabian StyleSallam, Mohammed, Abdelhalim Ghazy, Abdullah Al-Doss, and Ibrahim Al-Ashkar. 2024. "Combining Genetic and Phenotypic Analyses for Detecting Bread Wheat Genotypes of Drought Tolerance through Multivariate Analysis Techniques" Life 14, no. 2: 183. https://doi.org/10.3390/life14020183
APA StyleSallam, M., Ghazy, A., Al-Doss, A., & Al-Ashkar, I. (2024). Combining Genetic and Phenotypic Analyses for Detecting Bread Wheat Genotypes of Drought Tolerance through Multivariate Analysis Techniques. Life, 14(2), 183. https://doi.org/10.3390/life14020183









