Genomic Characterization of Probiotic Purple Nonsulfur Bacteria Cereibacter sphaeroides Strains S3W10 and SS15: Implications for Enhanced Shrimp Aquaculture
Abstract
1. Introduction
2. Material and Methods
2.1. Bacterial Strains
2.2. Whole-Genome Sequencing Analysis and Assembly
2.3. Genome Annotation and Visualization
2.4. Identification of Carbohydrate-Active enZyme (CAZyme)
2.5. Detection of Predicted Secondary Metabolite Biosynthetic Gene Clusters (BGCs)
2.6. Pan-Genome Analysis
2.7. The Average Nucleotide Identity (ANI) and Phylogenetic Analyses
3. Results and Discussion
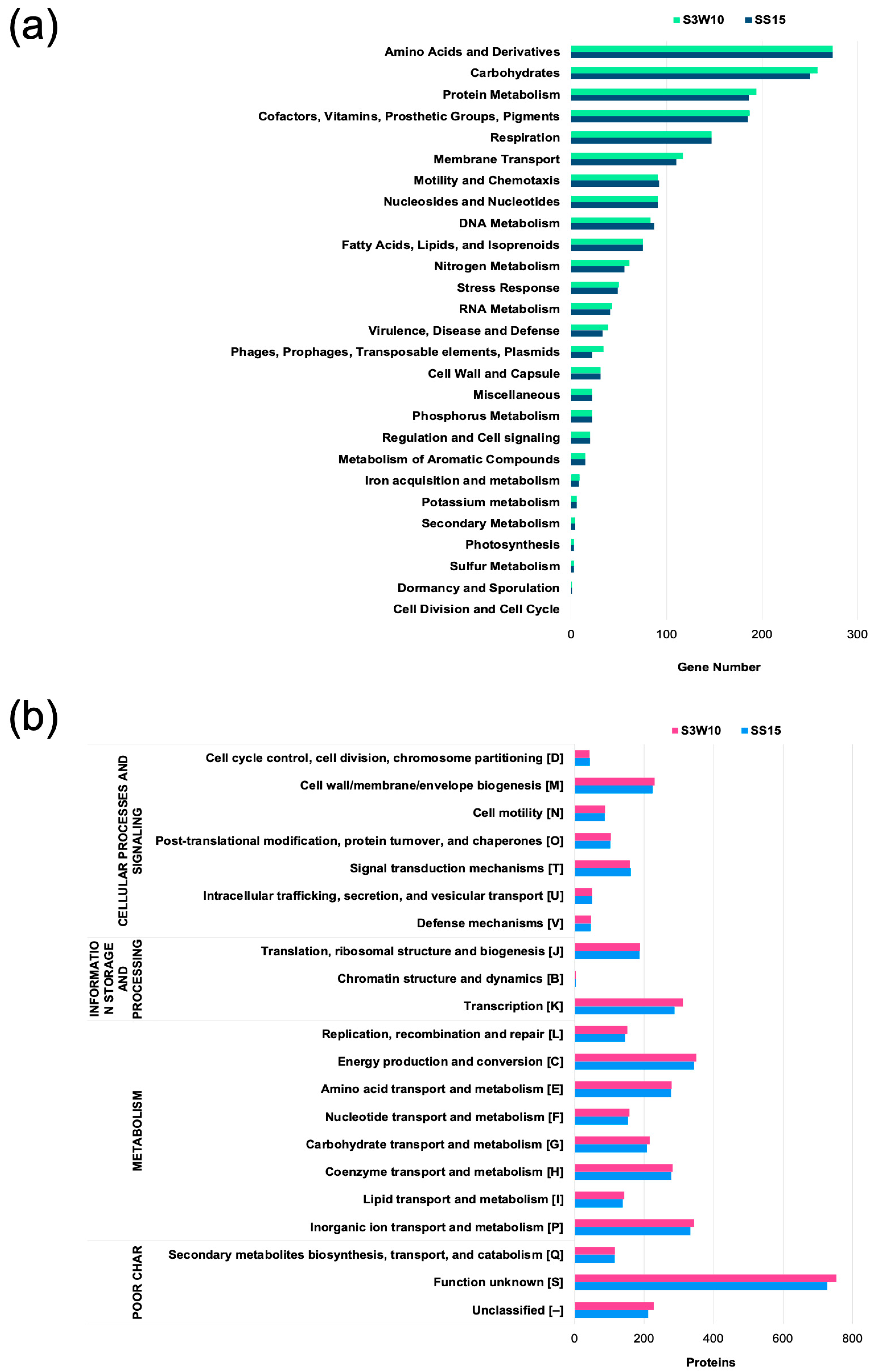
3.1. Genome Characteristics and Annotation of C. sphaeroides S3W10 and SS15 Strains
3.2. Mobile Genetic Element Analysis
3.3. Identification of Virulence Factors, Antibiotic Resistance Genes and Plasmids
3.4. Probiotic Properties of C. sphaeroides Strains
3.5. Carbohydrate-Active enZyme (CAZyme)
3.6. Secondary Metabolites Analysis
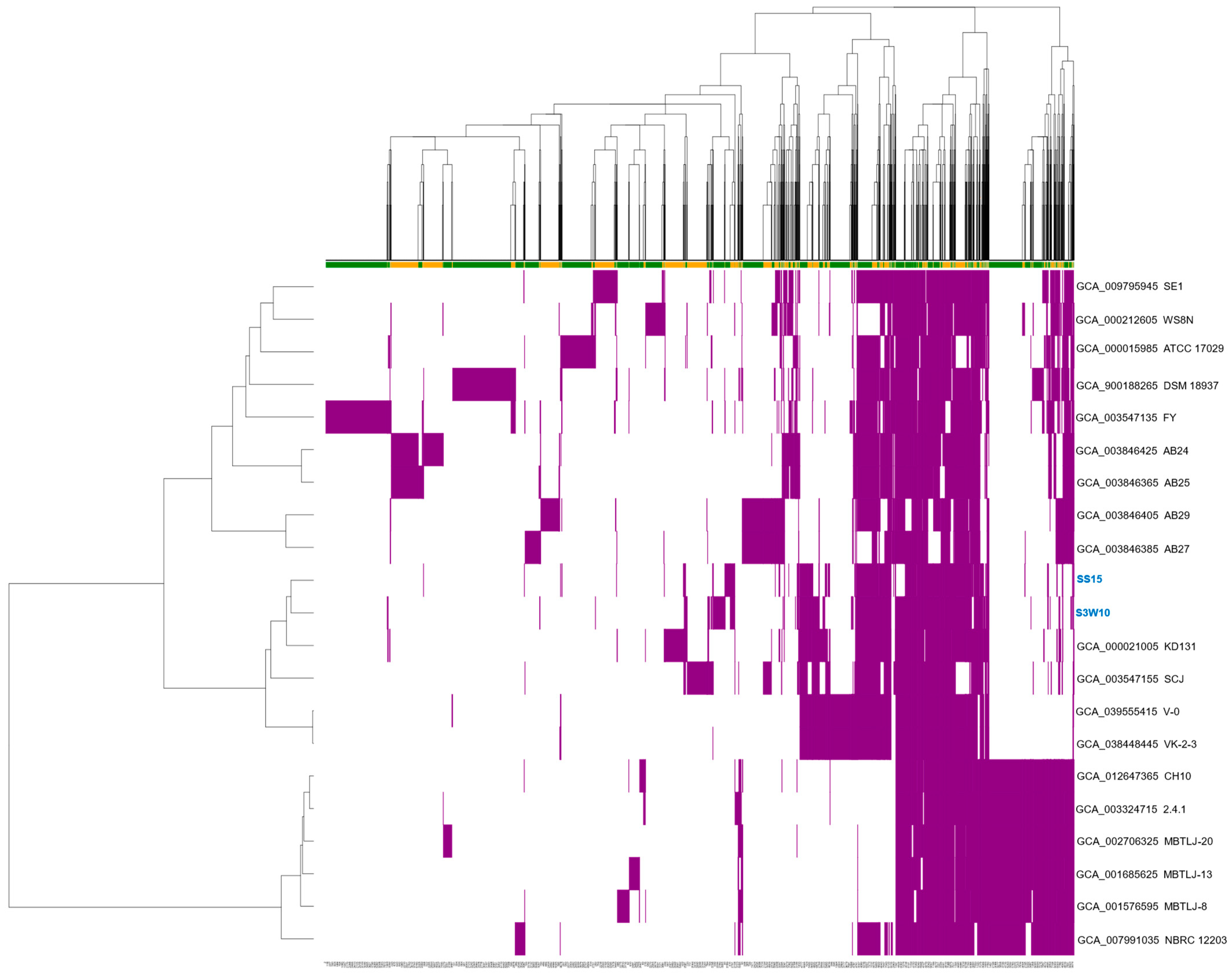
3.7. Pan-Genome of C. sphaeroides
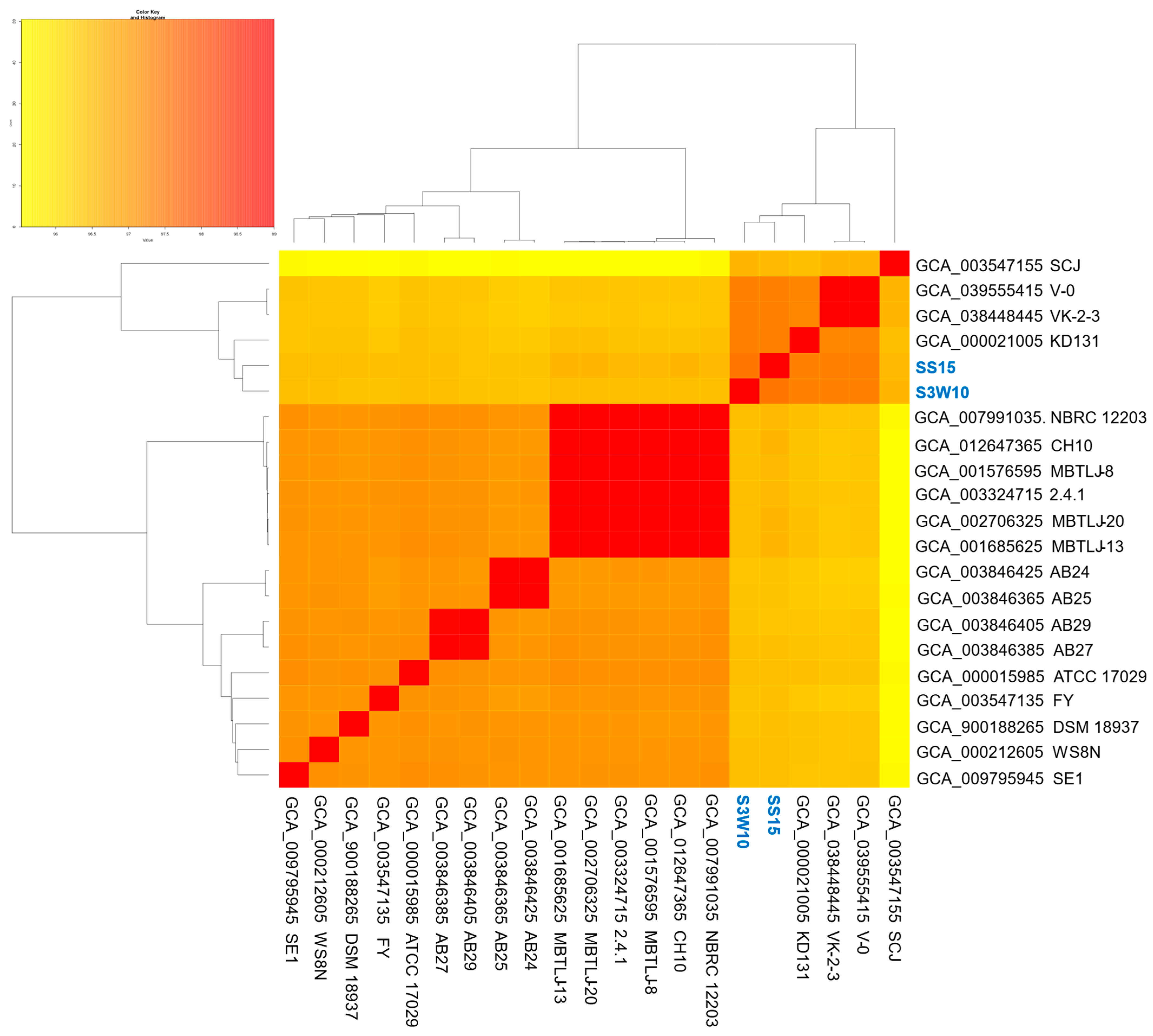
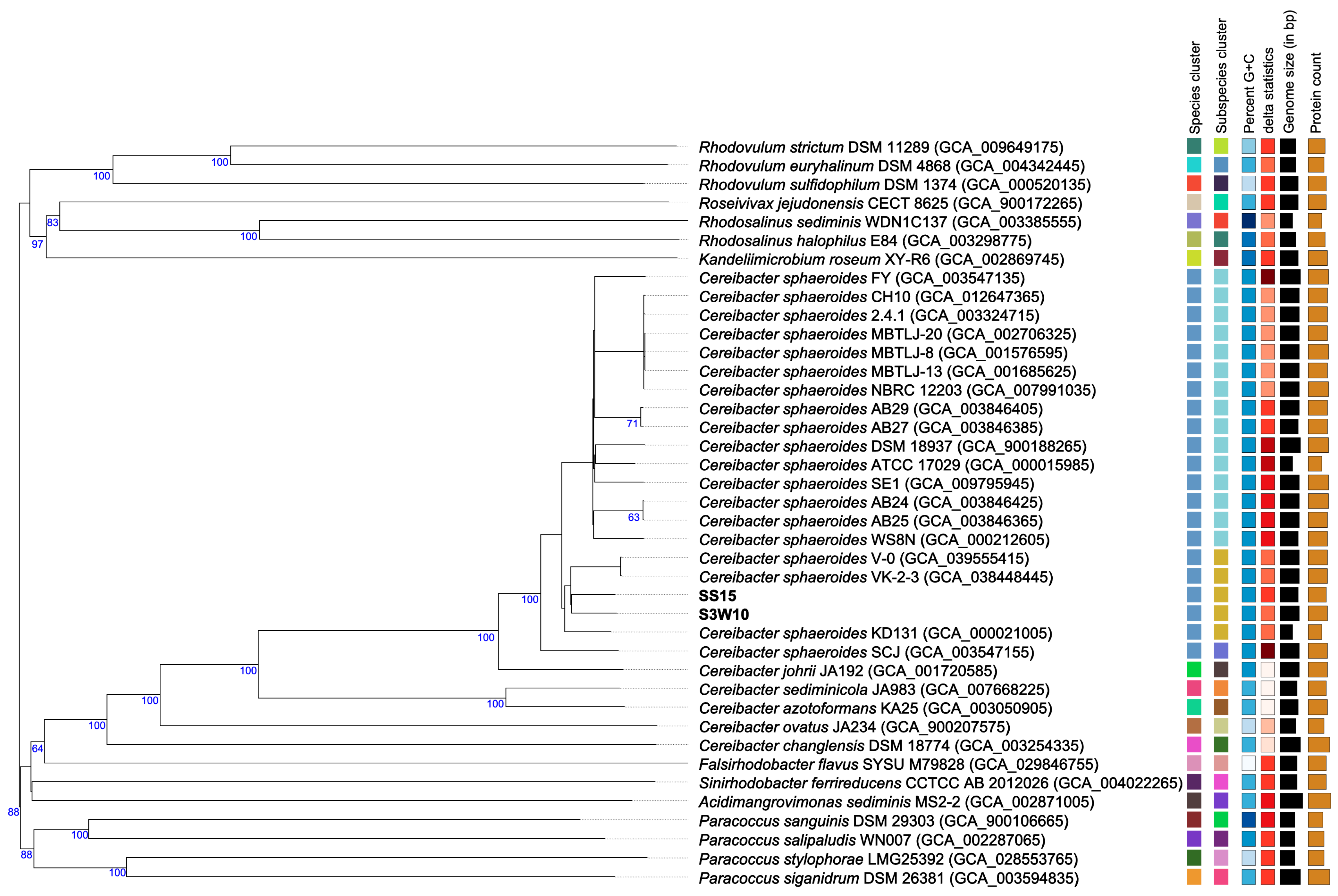
3.8. Phylogenomic of C. sphaeroides Species
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dhar, K.; Venkateswarlu, K.; Megharaj, M. Anoxygenic phototrophic purple non-sulfur bacteria: Tool for bioremediation of hazardous environmental pollutants. World J. Microbiol. Biotechnol. 2023, 39, 283. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sha, X.; Li, R.; Li, Y.; Khaleque, H.N.; Zhang, Y.; Bohu, T.; Bai, Z.; Zhuang, X. Comparative genome analysis provides molecular evidence for reclassification of the photosynthetic bacterium Rhodobacter sphaeroides EBL0706 as a strain of Luteovulum azotoformans. Microorganisms 2021, 9, 1754. [Google Scholar] [CrossRef] [PubMed]
- Chumpol, S.; Kantachote, D.; Rattanachuay, P.; Vuddhakul, V.; Nitoda, T.; Kanzaki, H. In vitro and in vivo selection of probiotic purple nonsulphur bacteria with an ability to inhibit shrimp pathogens: Acute hepatopancreatic necrosis disease-causing Vibrio parahaemolyticus and other vibrios. Aquac. Res. 2016, 48, 3182–3197. [Google Scholar] [CrossRef]
- Alloul, A.; Wille, M.; Lucenti, P.; Bossier, P.; Stappen, G.V.; Vlaeminck, S.E. Purple bacteria as added-value protein ingredient in shrimp feed: Penaeus vannamei growth performance, and tolerance against Vibrio and ammonia stress. Aquaculture 2021, 530, 735788. [Google Scholar] [CrossRef]
- Miyasaka, H.; Koga, A.; Tani, Y.; Maki, T.; Hayashi, S.; Yamamoto, S.; Desk, S. The effects of a marine photosynthetic bacteria Rhodovulum sulfidophilum on the growth and survival rate of Marsupenaeus japonicus (Kuruma shrimp). SDRP-JAFFS 2021, 3, 245–249. [Google Scholar] [CrossRef]
- Bhattacharya, P.; Ninan, K.N. Social cost-benefit analysis of intensive versus traditional shrimp farming: A case study from India. Nat. Resour. Forum. 2011, 35, 321–333. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, S.-A.; Jo, N.G.; Bae, J.-H.; Nguyen, M.T.; Jo, Y.M.; Han, N.S. Phenotypic and genomic analyses of bacteriocin-producing probiotic Enterococcus faecium EFEL8600 isolated from Korean soy-Meju. Front. Microbiol. 2023, 14, 1237442. [Google Scholar] [CrossRef]
- Ghattargi, V.C.; Gaikwad, M.A.; Meti, B.S.; Nimonkar, Y.; Dixit, K.; Prakash, O.; Shouche, Y.S.; Pawar, S.; Dhotre, D. Comparative genome analysis reveals key genetic factors associated with probiotic property in Enterococcus faecium strains. BMC Genom. 2018, 19, 652. [Google Scholar] [CrossRef]
- Akbari, V.; Shahali, A.; Soltani, R. Probiotic Lactobacillus and the potential risk of spreading antibiotic resistance: A systematic review. Res. Pharm. Sci. 2023, 18, 468–477. [Google Scholar]
- Su, H.; Hu, X.; Xu, W.; Xu, Y.; Wen, G.; Cao, Y. Diversity, abundances and distribution of antibiotic resistance genes and virulence factors in the South China Sea revealed by metagenomic sequencing. Sci. Total Environ. 2022, 814, 152803. [Google Scholar] [CrossRef]
- Chukamnerd, A.; Jeenkeawpiam, K.; Chusri, S.; Pomwised, R.; Singkhamanan, K.; Surachat, K. BacSeq: A user-friendly automated pipeline for whole-genome sequence analysis of bacterial genomes. Microorganisms 2023, 11, 1769. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Brown, C.L.; Mullet, J.; Hindi, F.; Stoll, J.E.; Gupta, S.; Choi, M.; Keenum, I.; Vikesland, P.; Pruden, A.; Zhang, L. mobileOG-db: A manually curated database of protein families mediating the life cycle of bacterial mobile genetic elements. Appl. Environ. Microbiol. 2022, 88, e00991-22. [Google Scholar] [CrossRef]
- Starikova, E.V.; Tikhonova, P.O.; Prianichnikov, N.A.; Rands, C.M.; Zdobnov, E.M.; Ilina, E.N.; Govorun, V.M. Phigaro: High-throughput prophage sequence annotation. Bioinformatics 2020, 36, 3882–3884. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Group, S.F.U.R.C.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S. Islandviewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef] [PubMed]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef]
- Russel, J.; Pinilla-Redondo, R.; Mayo-Muñoz, D.; Shah, S.A.; Sørensen, S.J. CRISPRCasTyper: Automated identification, annotation, and classification of CRISPR-Cas loci. CRISPR J. 2020, 3, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- Chivian, D.; Jungbluth, S.P.; Dehal, P.S.; Wood-Charlson, E.M.; Canon, R.S.; Allen, B.H.; Clark, M.M.; Gu, T.; Land, M.L.; Price, G.A. Metagenome-assembled genome extraction and analysis from microbiomes using KBase. Nat. Protoc. 2023, 18, 208–238. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; Van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Summo, M.; Meyer, D.F. PanExplorer: A web-based tool for exploratory analysis and visualization of bacterial pan-genomes. Bioinformatics 2022, 38, 4412–4414. [Google Scholar] [CrossRef]
- Perrin, A.; Rocha, E.P. PanACoTA: A modular tool for massive microbial comparative genomics. NAR Genom. Bioinform. 2021, 3, lqaa106. [Google Scholar]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.d.S.; Calaça, P.R.d.A.; Porto, A.L.F.; Souza, P.R.E.d.; Freitas, N.S.A.d.; Soares, M.T.C.V. What differentiates probiotic from pathogenic bacteria? The genetic mobility of Enterococcus faecium offers new molecular insights. OMICS J. Integr. Biol. 2020, 24, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Hyde, J.R.; Armond, T.; Herring, J.A.; Hope, S.; Grose, J.H.; Breakwell, D.P.; Pickett, B.E. Diversity and conservation of the genome architecture of phages infecting the Alphaproteobacteria. Microbiol. Spectr. 2024, 12, e02827-23. [Google Scholar] [CrossRef] [PubMed]
- Surekhamol, I.S.; Deepa, G.; Pai, S.S.; Sreelakshmi, B.; Varghese, S.M.; Singh, I.S.B. Isolation and characterization of broad spectrum bacteriophages lytic to Vibrio harveyi from shrimp farms of Kerala, India. Lett. Appl. Microbiol. 2014, 58, 197–204. [Google Scholar] [CrossRef]
- Tan, C.W.; Rukayadi, Y.; Hasan, H.; Abdul-Mutalib, N.-A.; Jambari, N.N.; Hara, H.; Thung, T.Y.; Lee, E.; Radu, S. Isolation and characterization of six Vibrio parahaemolyticus lytic bacteriophages from seafood samples. Front. Microbiol. 2021, 12, 616548. [Google Scholar] [CrossRef]
- Humphrey, S.; Marouli, A.; Thümmler, K.; Mullin, M.; Pritchard, L.; Wall, D.M. Genomic characterization of prophage elements in Clostridium clostridioforme: An understudied component of the intestinal microbiome. Microbiology 2024, 170, 001486. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S. Evolutionary plasticity and functional versatility of CRISPR systems. PLoS Biol. 2022, 20, e3001481. [Google Scholar] [CrossRef] [PubMed]
- Teklemariam, A.D.; Al-Hindi, R.R.; Qadri, I.; Alharbi, M.G.; Ramadan, W.S.; Ayubu, J.; Al-Hejin, A.M.; Hakim, R.F.; Hakim, F.F.; Hakim, R.F. The battle between bacteria and bacteriophages: A conundrum to their immune system. Antibiotics 2023, 12, 381. [Google Scholar] [CrossRef] [PubMed]
- Chumpol, S.; Kantachote, D.; Nitoda, T.; Kanzaki, H. Administration of purple nonsulfur bacteria as single cell protein by mixing with shrimp feed to enhance growth, immune response and survival in white shrimp (Litopenaeus vannamei) cultivation. Aquaculture 2018, 489, 85–95. [Google Scholar] [CrossRef]
- Chumpol, S.; Kantachote, D.; Nitoda, T.; Kanzaki, H. The roles of probiotic purple nonsulfur bacteria to control water quality and prevent acute hepatopancreatic necrosis disease (AHPND) for enhancement growth with higher survival in white shrimp (Litopenaeus vannamei) during cultivation. Aquaculture 2017, 473, 327–336. [Google Scholar] [CrossRef]
- Mendes, V.; Maranha, A.; Lamosa, P.; da Costa, M.S.; Empadinhas, N. Biochemical characterization of the maltokinase from Mycobacterium bovis BCG. BMC Biochem. 2010, 11, 21. [Google Scholar] [CrossRef]
- Kovačić, F.; Granzin, J.; Wilhelm, S.; Kojić-Prodić, B.; Batra-Safferling, R.; Jaeger, K.E. Structural and functional characterisation of TesA—A novel lysophospholipase a from Pseudomonas aeruginosa. PLoS ONE 2013, 8, e69125. [Google Scholar] [CrossRef] [PubMed]
- Freiding, S.; Gutsche, K.A.; Ehrmann, M.A.; Vogel, R.F. Genetic screening of Lactobacillus sakei and Lactobacillus curvatus strains for their peptidolytic system and amino acid metabolism, and comparison of their volatilomes in a model system. Syst. Appl. Microbiol. 2011, 34, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Soto, M.A.; Desai, D.; Bannon, C.; LaRoche, J.; Bertrand, E.M. Cobalamin producers and prokaryotic consumers in the Northwest Atlantic. Environ. Microbiol. 2023, 25, 1300–1313. [Google Scholar] [CrossRef] [PubMed]
- González-Pedrajo, B.; de la Mora, J.; Ballado, T.; Camarena, L.; Dreyfus, G. Characterization of the flgG operon of Rhodobacter sphaeroides WS8 and its role in flagellum biosynthesis. Biochim. Biophys. Acta-Gene Struct. Expr. 2002, 1579, 55–63. [Google Scholar] [CrossRef]
- Xie, G.; Zhu, Y.; Zhong, Z.; Du, Q.; Wu, Y.; Xing, K.; Zhang, M.; Shu, H. Functional genomic characterization unveils probiotic features of Bacillus cereus G1-11 isolated from the gut of the hybrid grouper (Epinephelus fuscoguttatus♀× E. lanceolatus♂). LWT 2023, 184, 115088. [Google Scholar] [CrossRef]
- Guan, N.; Liu, L. Microbial response to acid stress: Mechanisms and applications. Appl. Microbiol. Biotechnol. 2020, 104, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Roncarati, D.; Scarlato, V. Regulation of heat-shock genes in bacteria: From signal sensing to gene expression output. FEMS Microbiol. Rev. 2017, 41, 549–574. [Google Scholar] [CrossRef]
- Choquet, G.; Jehan, N.; Pissavin, C.; Blanco, C.; Jebbar, M. OusB, a broad-specificity ABC-type transporter from Erwinia chrysanthemi, mediates uptake of glycine betaine and choline with a high affinity. Appl. Environ. Microbiol. 2005, 71, 3389–3398. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Pericone, C.D.; Лысенкo, E.; Goldfine, H.; Weiser, J.N. Multiple mechanisms for choline transport and utilization in Haemophilus Influenzae. Mol. Microbiol. 2003, 50, 537–548. [Google Scholar] [CrossRef]
- Ballal, A.; Apte, S.K. Differential expression of the two kdp operons in the nitrogen-fixing cyanobacterium Anabaena sp. strain L-31. Appl. Environ. Microbiol. 2005, 71, 5297–5303. [Google Scholar] [CrossRef]
- Nanatani, K.; Shijuku, T.; Takano, Y.; Zulkifli, L.; Yamazaki, T.; Tominaga, A.; Souma, S.; Onai, K.; Morishita, M.; Ishiura, M. Comparative analysis of kdp and ktr mutants reveals distinct roles of the potassium transporters in the model cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 2015, 197, 676–687. [Google Scholar] [CrossRef] [PubMed]
- Zeller, T.; Klug, G. Detoxification of hydrogen peroxide and expression of catalase genes in Rhodobacter. Microbiology 2004, 150, 3451–3462. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Farmer, R.M.; Tabita, F.R. Phosphoribulokinase mediates nitrogenase-induced carbon dioxide fixation gene repression in Rhodobacter Sphaeroides. Microbiology 2015, 161, 2184–2191. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-H.; Zheng, D.; Feng, N.; Liu, T.; Liu, Y.; Gong, S.; Cui, H.; Xiang, H. The effects of gibberellins and mepiquat chloride on nitrogenase activity in Bradyrhizobium Japonicum. Acta Physiol. Plant. 2014, 37, 1723. [Google Scholar] [CrossRef]
- Martin, D.E.; Reinhold-Hurek, B. Distinct roles of P (II)--like signal transmitter proteins and amtB in regulation of nif gene expression, nitrogenase activity, and posttranslational modification of NifH in Azoarcus sp. strain BH72. J. Bacteriol. 2002, 184, 2251–2259. [Google Scholar] [CrossRef] [PubMed]
- Drepper, T.; Raabe, K.; Giaourakis, D.; Gendrullis, M.; Masepohl, B.; Klipp, W. The Hfq-like protein NrfA of the phototrophic purple bacterium Rhodobacter capsulatus controls nitrogen fixation via regulation of nifA and anfA expression. FEMS Microbiol. Lett. 2002, 215, 221–227. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dong, S.; Liu, Y.-J.; Zhou, H.; Xiao, Y.; Xu, J.; Cui, Q.; Wang, X.; Feng, Y. Structural insight into a GH1 β-glucosidase from the oleaginous microalga, Nannochloropsis oceanica. Int. J. Biol. Macromol. 2021, 170, 196–206. [Google Scholar] [CrossRef]
- Huang, Z.; Ni, G.; Zhao, X.; Wang, F.; Qu, M. Characterization of a GH8 Β-1,4-glucanase from Bacillus subtilis B111 and its saccharification potential for agricultural straws. J. Microbiol. Biotechnol. 2021, 31, 1446–1454. [Google Scholar] [CrossRef]
- Mousslim, M.; Pagano, A.; Andreotti, N.; Garrouste, F.; Thuault, S.; Peyrot, V.; Parat, F.; Luis, J.; Culcasi, M.; Thétiot-Laurent, S. Peptide screen identifies a new NADPH oxidase inhibitor: Impact on cell migration and invasion. Eur. J. Pharmacol. 2017, 794, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Janeček, Š.; Svensson, B.; MacGregor, E.A. α-Amylase: An enzyme specificity found in various families of glycoside hydrolases. Cell Mol. Life Sci. 2014, 71, 1149–1170. [Google Scholar] [CrossRef] [PubMed]
- Vera, C.; Guerrero, C.; Aburto, C.; Cordova, A.; Illanes, A. Conventional and non-conventional applications of β-galactosidases. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140271. [Google Scholar] [CrossRef]
- Oliveira, F.S.; da Silva Rodrigues, R.; De Carvalho, A.F.; Nero, L.A. Genomic analyses of Pediococcus pentosaceus ST65ACC, a bacteriocinogenic strain isolated from artisanal raw-milk cheese. Probiotics Antimicrob. Proteins 2023, 15, 630–645. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Giri, S.S.; Sukumaran, V.; Oviya, M. Potential probiotic Lactobacillus plantarum VSG3 improves the growth, immunity, and disease resistance of tropical freshwater fish, Labeo rohita. Fish Shellfish. Immunol. 2013, 34, 660–666. [Google Scholar] [CrossRef] [PubMed]
- Allameh, S.K.; Ringø, E.; Yusoff, F.; Daud, H.M.; Ideris, A. Dietary supplement of Enterococcus faecalis on digestive enzyme activities, short-chain fatty acid production, immune system response and disease resistance of Javanese carp (Puntius gonionotus, Bleeker 1850). Aquac. Nutr. 2017, 23, 331–338. [Google Scholar] [CrossRef]
- Cui, T.; Rao, Y.-Z.; Li, J.; Ren, C.; Tang, D.; Lin, T.S.; Ji, J.; Chen, R.; Yan, A. Two distinct C-type lysozymes in goldfish: Molecular characterization, antimicrobial potential, and transcriptional regulation in response to opposing effects of bacteria/lipopolysaccharide and dexamethasone/leptin. Int. J. Mol. Sci. 2020, 21, 501. [Google Scholar] [CrossRef]
- Nuntapong, N.; Phromkunthong, W.; Suanyuk, N.; Corlay, D. Natural pigment from Paracoccus carotinifaciens (Panaferd®-AX) enhanced colour and immune system of Pacific white shrimp (Litopenaeus vannamei). Aquac. Res. 2022, 53, 5925–5936. [Google Scholar] [CrossRef]
- Koga, A.; Goto, M.; Hayashi, S.; Yamamoto, S.; Miyasaka, H. Probiotic effects of a marine purple non-sulfur bacterium, Rhodovulum sulfidophilum KKMI01, on Kuruma shrimp (Marsupenaeus japonicus). Microorganisms 2022, 10, 244. [Google Scholar] [CrossRef] [PubMed]
- Laining, A.; Trismawanti, I.; Kamaruddin, K.; Makmur, M. Carotenoid-enriched diet for pre-maturation stage of pond-reared tiger shrimp, Penaeus monodon part I. the effects on growth, pigmentation and whole body nutrient content. Indones. Aquac. J. 2017, 12, 59–66. [Google Scholar] [CrossRef]
- Su, A.; Chi, S.; Liu, Y.; Tan, S.; Shan, Q.; Chen, Z.; Meng, Y. Metabolic redesign of Rhodobacter sphaeroides for lycopene production. J. Agric. Food Chem. 2018, 66, 5879–5885. [Google Scholar] [CrossRef] [PubMed]
- Schimming, O.; Fleischhacker, F.; Nollmann, F.I.; Bode, H.B. Yeast homologous recombination cloning leading to the novel peptides ambactin and xenolindicin. Chembiochem 2014, 15, 1290–1294. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wang, Z.; Chen, J.; Zhan, Y.; Wang, T.; Xia, L.; Wang, S.; Hua, Z.; Zhang, J. Supplementation of the diet with salecan attenuates the symptoms of colitis induced by dextran sulphate sodium in mice. Br. J. Nutr. 2014, 111, 1822–1829. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Cheng, R.; Li, J.; Wang, Y.; Zhu, B.; Ma, S.; Zhang, W.; Dong, W.; Wang, S.; Zhang, J. Identification of substituent groups and related genes involved in salecan biosynthesis in Agrobacterium sp. ZX09. Appl. Microbiol. Biotechnol. 2016, 101, 585–598. [Google Scholar] [CrossRef]
- Shen, K.; Bao, L.; Liu, M.; Li, W.; Zhou, Q.; Ding, J.; Peng, F.; Hu, B.; Wen, C.; Kumar, V.; et al. Dietary supplementation of Β-1, 3-glucan improves the intestinal health of white shrimp (Litopenaeus vannamei) by modulating intestinal microbiota and inhibiting inflammatory response. Front. Immunol. 2023, 14, 1119902. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Tettelin, H.; Riley, D.; Cattuto, C.; Medini, D. Comparative genomics: The bacterial pan-genome. Curr. Opin. Microbiol. 2008, 11, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Xu, X.; Huang, Z.; Xiao, Y.; Yu, K.; Jiang, M.; Yin, S.; Zheng, M.; Meng, H.; Han, Y.; et al. Genomic characteristics and pan-genome analysis of Rhodococcus equi. Front. Cell. Infect. Microbiol. 2022, 12, 807610. [Google Scholar] [CrossRef]


| Attribute | S3W10 | SS15 |
|---|---|---|
| Genome size (bp) | 4,597,894 | 4,413,552 |
| Contigs | 130 | 74 |
| DNA G + C content (%) | 69.2 | 69.2 |
| Protein coding sequences (CDSs) | 4524 | 4294 |
| ANIb (%) with SS15 | 98.29 | 100 |
| rRNA | 3 | 3 |
| tRNA | 52 | 51 |
| tmRNA | 1 | 1 |
| RAST subsystems | 338 | 336 |
| Gene | Product/Description | Gene Number | |
|---|---|---|---|
| S3W10 | SS15 | ||
| Adhesion | |||
| flgK | Flagellar hook-associated protein FlgK | 1 | 1 |
| flgL | Bacterial flagellin C-terminal helical region | 1 | 1 |
| fliC | Flagellin subunit protein | 2 | 2 |
| flgG | Flagellar basal-body rod protein FlgG | 3 | 3 |
| lspA | Lipoprotein signal peptidase | 1 | 1 |
| msrA | Peptide methionine sulfoxide reductase | 1 | 1 |
| Acid stress | |||
| atpA, atpD, atpG, atpH, atpC | FoF1-type ATP synthase (alpha, beta, gamma, delta, epsilon) subunit | 2, 2, 3, 1, 2 | 2, 2, 3, 1, 2 |
| atpE | FoF1-type ATP synthase subunit K | 2 | 2 |
| aspS | Aspartyl-tRNA synthetase | 1 | 1 |
| Heat stress | |||
| dnaK | DnaK chaperone protein | 3 | 2 |
| dnaJ | DnaJ chaperone protein | 2 | 2 |
| grpE | Chaperonin GroEL (HSP60 family) | 1 | 1 |
| groES | Co-chaperonin GroES (HSP10) | 1 | 1 |
| ibpB | Small heat shock protein (HSP20) family | 1 | 1 |
| hslV, hslU | ATP-dependent protease HslVU (ClpYQ) | 1, 1 | 1, 1 |
| Osmotic pressure stress | |||
| kdpA, kdpB, kdpC | K+-transporting ATPase, A, B, C chain | 1, 1, 1 | 1, 1, 1 |
| betA, betB | Involved in the biosynthesis of the osmoprotectant glycine betaine | 1, 2 | 1, 2 |
| otsA, otsB | Involved in the osmoprotection via the biosynthesis of trehalose | 1, 1 | 1, 1 |
| Oxidative stress | |||
| sodB, sodC | Destroys radicals that are normally produced within the cells and which are toxic to biological systems | 1, 1 | 1, 1 |
| katE | Catalase (COG0753) | 1 | 1 |
| tlpA | Alkyl hydroperoxide reductase | 1 | 1 |
| trxA | Thioredoxin reductase | 1 | 1 |
| msrA | Repair enzyme for proteins that have been inactivated by oxidation | 1 | 1 |
| Bile resistance | |||
| arsB | Bile acid sodium symporter | 1 | 1 |
| Photosynthesis | |||
| pufL | Photosynthetic reaction center L subunit | 1 | 1 |
| pufA, pufB | Antenna complexes are light-harvesting systems, which transfer the excitation energy to the reaction centers | 1, 1 | 1, 1 |
| rbcL | Ribulose 1,5-bisphosphate carboxylase large subunit | 1 | 1 |
| Lactate synthesis | |||
| ldhA | Lactate dehydrogenase or related 2-hydroxyacid dehydrogenase | 1 | 1 |
| Vitamin biosynthesis | |||
| cobS, cobN, cobU, cobT | cob/cbi operon for cobalamin (vitamin B12) biosynthesis | 1, 1, 1, 1 | 1, 1, 1, 1 |
| pdxA, pdxJ, pdxH, pdxK | pdx operon for vitamin B6 biosynthesis | 2, 1, 1, 1 | 2, 1, 1, 1 |
| Organic matter degradation | |||
| treS | Trehalose synthase/amylase TreS | 1 | 1 |
| tesA | GDSL-like Lipase/Acylhydrolase family | 1 | 1 |
| degP | Peptidase S1C family with trypsin-like peptidase domain (COG0265) | 3 | 3 |
| pepN | Aminopeptidase N | 1 | 1 |
| dcp | PFAM peptidase M3A and M3B, thimet oligopeptidase F | 1 | 1 |
| pepF | TIGRFAM oligoendopeptidase, pepF M3 family | 1 | 1 |
| Carotenoid biosynthesis | |||
| crtF | Demethylspheroidene O-methyltransferase | 1 | 1 |
| crtE | Encodes geranylgeranyl diphosphate (GGPP) synthase, which synthesizes GGPP, a precursor for carotenoid biosynthesis | 1 | 1 |
| crtC | Involved in the biosynthesis of carotenoids spheroidene and spirilloxanthin | 1 | 1 |
| crtB | Encodes phytoene synthase, which converts GGPP into phytoene | 1 | 1 |
| crtI | Converts phytoene into all-trans-neurosporene as the major product, through intermediates phytofluene and zeta-carotene | 1 | 2 |
| crtA | Spheroidene monooxygenase, involved in the hydroxylation of spheroidene, contributing to carotenoid diversity | 1 | 1 |
| tspO | Part of the TspO MBR family, involved in carotenoid biosynthesis | 1 | 1 |
| Exopolysaccharide (EPS) biosynthesis | |||
| wza | Polysaccharide biosynthesis/export protein | 1 | 1 |
| galU | UTP-glucose-1-phosphate uridylyltransferase | 1 | 1 |
| galE, manC | Involved in sugar precursor synthesis for EPS | 4, 1 | 4, 1 |
| Nitrogen removal and recycling | |||
| amtB | Ammonia channel protein AmtB | 1 | 1 |
| gdhA | Glu, Leu, Phe, Val dehydrogenases family | 1 | 1 |
| glnA | Glutamine synthetase | 1 | 1 |
| norB | Nitric oxide reductase large subunit | 1 | 1 |
| nirK | Nitrite reductase | 1 | 1 |
| cysI | Nitrite and sulfite reductase 4Fe-4S | 1 | 1 |
| ureA, ureB, ureC | Urease gamma, beta, alpha subunit | 1, 1, 1 | 1, 1, 1 |
| ureE, ureF | Urease accessory protein (UreE, UreF) | 1, 1 | 1, 1 |
| nifH | Nitrogenase subunit NifH | 2 | 1 |
| nifD | Nitrogenase molybdenum-iron protein, alpha chains | 2 | 2 |
| - | COG3381: Nitrate reductase delta subunit | 1 | 1 |
| Phosphorus removal and recycling | |||
| ppx | Inorganic pyrophosphatase/exopolyphosphatase | 1 | 1 |
| ppk2 | Polyphosphate kinase 2, PPK2 family | 1 | 1 |
| pstB | ABC-type phosphate transport system, ATPase component | 1 | 1 |
| pstA, pstC | ABC-type phosphate transport system, permease component | 1, 1 | 1, 1 |
| pstS | ABC-type phosphate transport system, periplasmic component | 1 | 1 |
| Hydrogen sulfide detoxification | |||
| cysK | Cysteine synthase | 1 | 1 |
| cysE | Serine acetyltransferase | 1 | 1 |
| cysI | Sulfite reductase, beta subunit (hemoprotein) | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klaysubun, C.; Chaichana, N.; Suwannasin, S.; Singkhamanan, K.; Yaikhan, T.; Kantachote, D.; Pomwised, R.; Wonglapsuwan, M.; Surachat, K. Genomic Characterization of Probiotic Purple Nonsulfur Bacteria Cereibacter sphaeroides Strains S3W10 and SS15: Implications for Enhanced Shrimp Aquaculture. Life 2024, 14, 1691. https://doi.org/10.3390/life14121691
Klaysubun C, Chaichana N, Suwannasin S, Singkhamanan K, Yaikhan T, Kantachote D, Pomwised R, Wonglapsuwan M, Surachat K. Genomic Characterization of Probiotic Purple Nonsulfur Bacteria Cereibacter sphaeroides Strains S3W10 and SS15: Implications for Enhanced Shrimp Aquaculture. Life. 2024; 14(12):1691. https://doi.org/10.3390/life14121691
Chicago/Turabian StyleKlaysubun, Chollachai, Nattarika Chaichana, Sirikan Suwannasin, Kamonnut Singkhamanan, Thunchanok Yaikhan, Duangporn Kantachote, Rattanaruji Pomwised, Monwadee Wonglapsuwan, and Komwit Surachat. 2024. "Genomic Characterization of Probiotic Purple Nonsulfur Bacteria Cereibacter sphaeroides Strains S3W10 and SS15: Implications for Enhanced Shrimp Aquaculture" Life 14, no. 12: 1691. https://doi.org/10.3390/life14121691
APA StyleKlaysubun, C., Chaichana, N., Suwannasin, S., Singkhamanan, K., Yaikhan, T., Kantachote, D., Pomwised, R., Wonglapsuwan, M., & Surachat, K. (2024). Genomic Characterization of Probiotic Purple Nonsulfur Bacteria Cereibacter sphaeroides Strains S3W10 and SS15: Implications for Enhanced Shrimp Aquaculture. Life, 14(12), 1691. https://doi.org/10.3390/life14121691






