Uncovering the Fungal Diversity and Biodeterioration Phenomenon on Archaeological Carvings of the Badami Cave Temples: A Microcosm Study
Abstract
:1. Introduction
2. Material and Methods
2.1. Description of the Studied Stone Carvings
2.2. Mycological Analyses
2.2.1. Fungal Isolation
2.2.2. Identification of Culturable Fungi
Morphological Analysis
Molecular Analysis
2.3. Biodeterioration Assays
2.3.1. Determination of Carbonate Solubilization
2.3.2. pH Changes of Substrate
2.3.3. Organic Acid Analysis
3. Results
3.1. Morphology and Molecular Diversity of Culturable Fungi
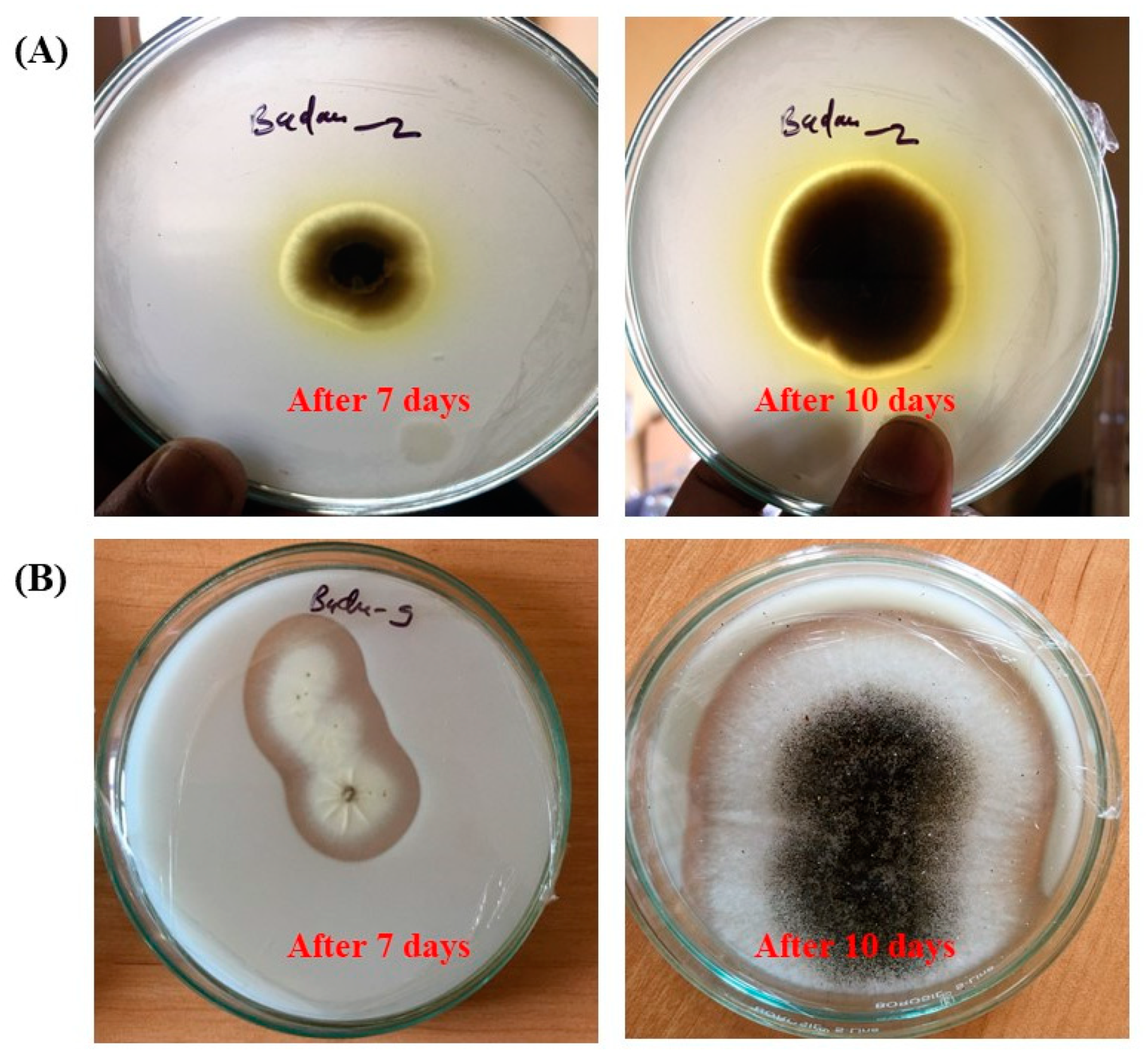
3.2. Biodeterioration Potential of Fungal Isolates
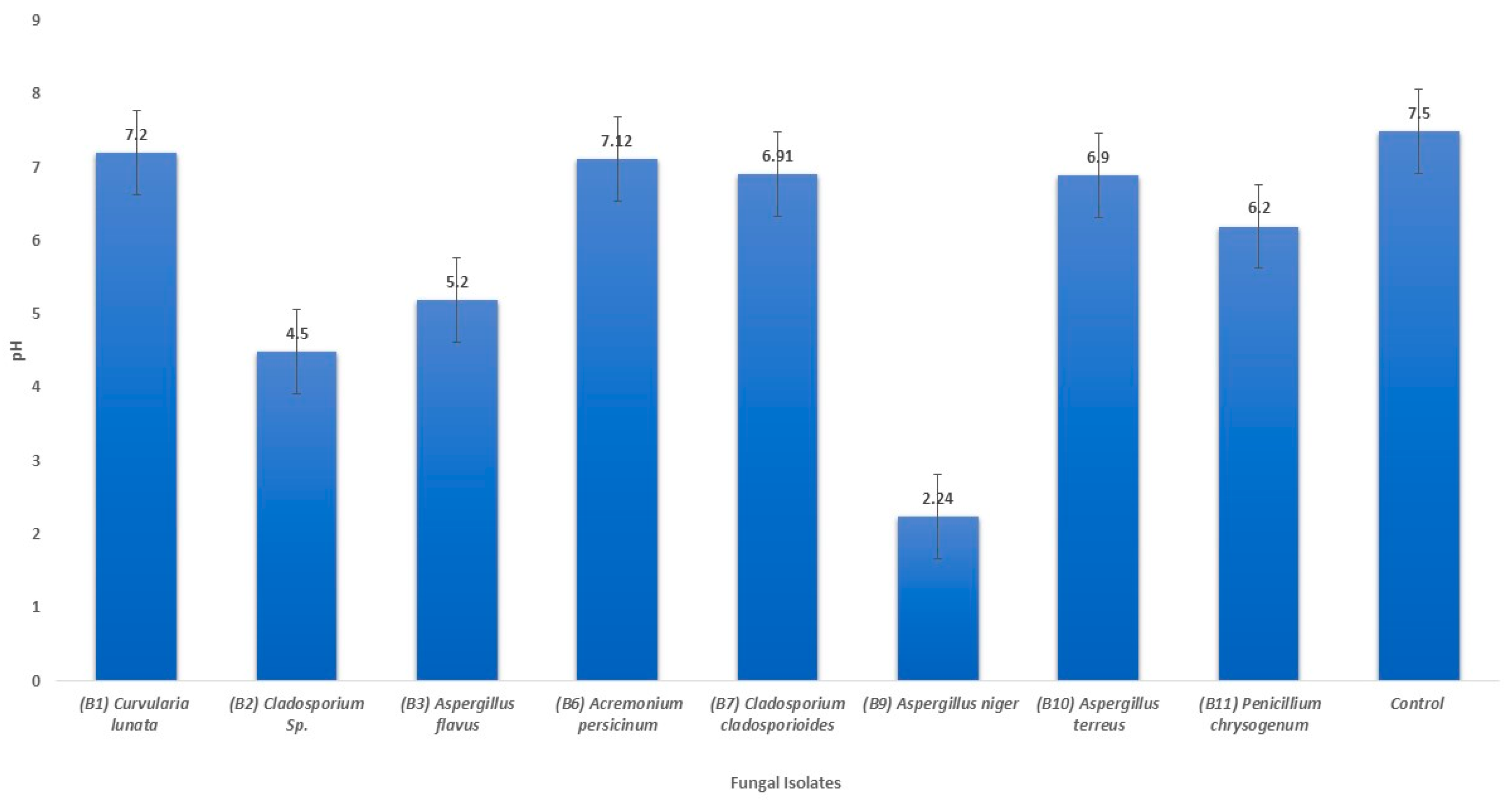
3.3. Changes in pH of the Liquid Medium
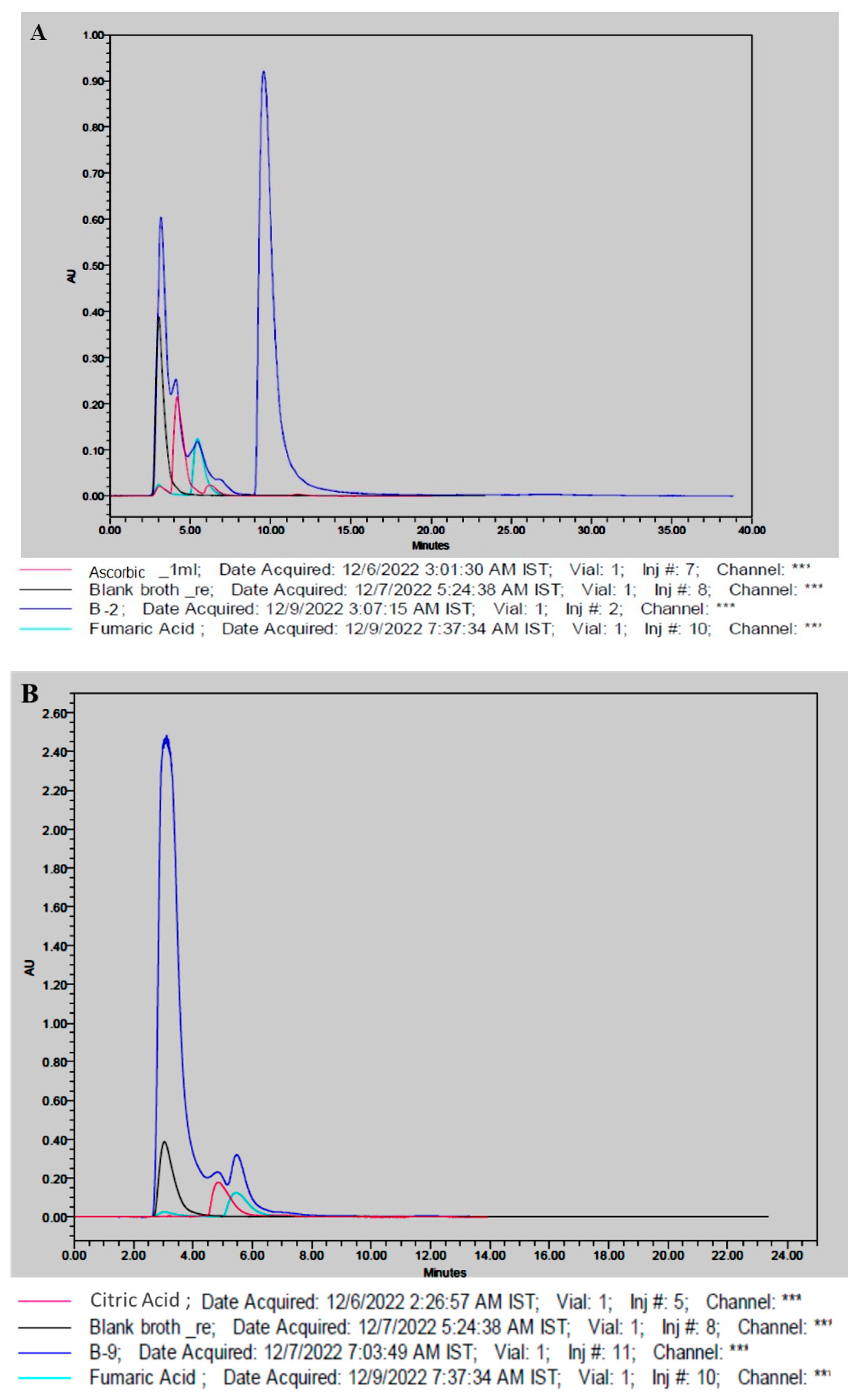
3.4. Production of Organic Acids
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ghany, T.M.A.; Omar, A.; Elwkeel, F.M.; Al Abboud, M.A.; Alawlaqi, M. Fungal deterioration of limestone false-door monument. Heliyon 2019, 5, e02673. [Google Scholar] [CrossRef] [PubMed]
- Siedel, H.; Siegesmund, S. Characterization of stone deterioration on buildings. In Stone in Architecture: Properties, Durability; Springer: Berlin/Heidelberg, Germany, 2014; pp. 349–414. [Google Scholar]
- Hawksworth, D.L.; Lücking, R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 2017, 5, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Sterflinger, K. Fungi as geologic agents. Geomicrobiol. J. 2000, 17, 97–124. [Google Scholar] [CrossRef]
- Gadd, G.M. Geomicrobiology of the built environment. Nat. Microbiol. 2017, 2, 16275. [Google Scholar] [CrossRef] [PubMed]
- Ljaljević-Grbić, M.V.; Stupar, M.; Savković, D.; Knežević, A.Z.; Dimkić, I.Z.; Kosel, J.J.; Tavzes, M.; Unković, N.D. From on-site to in-lab: Microscopic observation of fungal proliferation on 17th century mural paintings. Zb. Matice Srp. Za Prir. Nauk. 2022, 7–14. [Google Scholar] [CrossRef]
- Gadd, G.M. Fungi, rocks, and minerals. Elem. Int. Mag. Mineral. Geochem. Petrol. 2017, 13, 171–176. [Google Scholar] [CrossRef]
- Kosel, J.; Ropret, P. Overview of fungal isolates on heritage collections of photographic materials and their biological potency. J. Cult. Heritage 2021, 48, 277–291. [Google Scholar] [CrossRef]
- Gadd, G.M. Geomycology: Biogeochemical transformations of rocks, minerals, metals and radionuclides by fungi, bioweathering and bioremediation. Mycol. Res. 2007, 111, 3–49. [Google Scholar] [CrossRef]
- Burford, E.P.; Fomina, M.; Gadd, G.M. Fungal involvement in bioweathering and biotransformation of rocks and minerals. Miner. Mag. 2003, 67, 1127–1155. [Google Scholar] [CrossRef]
- Gadd, G.M. Fungi in Biogeochemical Cycles; Cambridge University Press: Cambridge, UK, 2006. [Google Scholar]
- Popović, S.; Unković, N.; Janakiev, T.; Knežević, A.; Dimkić, I.; Kosel, J.; Tavzes, Č.; Subakov-Simić, G.; Ljaljević-Grbić, M. Biofilm constituents as deteriogens of ancient Roman monument Mitrej above Rožanec (Slovenia). In Proceedings of the FEMS Conference on Microbiology in association with Serbian Society of Microbiology, Belgrade, Serbia, 30 June–2 July 2022. [Google Scholar]
- Scheerer, S.; Ortega-Morales, O.; Gaylarde, C. Microbial deterioration of stone monuments—An updated overview. Adv. Appl. Microbiol. 2009, 66, 97–139. [Google Scholar]
- Rosado, T.; Mirão, J.; Gil, M.; Candeias, A.; Caldeira, A.T.; Rogerio-Candelera, M.A. Role of Microorganisms in Mural Paintings Decay; CRC Press; Balkema Taylor & Francis Group: London, UK, 2014; pp. 217–222. [Google Scholar]
- Piñar, G.; Garcia-Valles, M.; Gimeno-Torrente, D.; Fernandez-Turiel, J.L.; Ettenauer, J.; Sterflinger, K. Microscopic, chemical, and molecular-biological investigation of the decayed medieval stained window glasses of two Catalonian churches. Int. Biodeterior. Biodegrad. 2013, 84, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Sterflinger, K.; Prillinger, H. Molecular taxonomy and biodiversity of rock fungal communities in an urban environment (Vienna, Austria). Antonie Van Leeuwenhoek 2001, 80, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Bindschedler, S.; Cailleau, G.; Verrecchia, E. Role of fungi in the biomineralization of calcite. Minerals 2016, 6, 41. [Google Scholar] [CrossRef]
- Savković, Ž.; Unković, N.; Stupar, M.; Franković, M.; Jovanović, M.; Erić, S.; Šarić, K.; Stanković, S.; Dimkić, I.; Vukojević, J.; et al. Diversity and biodeteriorative potential of fungal dwellers on ancient stone stela. Int. Biodeterior. Biodegrad. 2016, 115, 212–223. [Google Scholar] [CrossRef]
- Trovão, J.; Gil, F.; Catarino, L.; Soares, F.; Tiago, I.; Portugal, A. Analysis of fungal deterioration phenomena in the first Portuguese King tomb using a multi-analytical approach. Int. Biodeterior. Biodegrad. 2020, 149, 104933. [Google Scholar] [CrossRef]
- Mihajlovski, A.; Seyer, D.; Benamara, H.; Bousta, F.; Di Martino, P. An overview of techniques for the characterization and quantification of microbial colonization on stone monuments. Ann. Microbiol. 2015, 65, 1243–1255. [Google Scholar] [CrossRef]
- Brancaccio, P.; Selin, H. Cave Architecture of India. In Encyclopaedia of the History of Science, Technology, and Medicine in Non-Western Cultures; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–9. [Google Scholar]
- Agrawal, S.; Nandeibam, J.; Devi, I. Danger of exposure to keratinophilic fungi and other dermatophytes in recreational place in the northeast region of India. Aerobiologia 2021, 37, 755–766. [Google Scholar] [CrossRef]
- Pitt, J.I. The Genus Penicillium and Its Teleomorphic States Eupenicillium and Talaromyces; Academic Press Inc. Ltd.: London, UK, 1979. [Google Scholar]
- Samson, R.A.; Visagie, C.M.; Houbraken, J.; Hong, S.B.; Hubka, V.; Klaassen, C.H.; Perrone, G.; Seifert, K.A.; Susca, A.; Tanney, J.B.; et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 2014, 78, 141–173. [Google Scholar] [CrossRef]
- Samson, R.; Yilmaz, N.; Houbraken, J.; Spierenburg, H.; Seifert, K.; Peterson, S.; Varga, J.; Frisvad, J. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud. Mycol. 2011, 70, 159–183. [Google Scholar] [CrossRef]
- Visagie, C.; Houbraken, J.; Frisvad, J.; Hong, S.-B.; Klaassen, C.; Perrone, G.; Seifert, K.; Varga, J.; Yaguchi, T.; Samson, R. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef]
- Yilmaz, N.; Visagie, C.; Houbraken, J.; Frisvad, J.; Samson, R. Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 2014, 78, 175–341. [Google Scholar] [CrossRef] [PubMed]
- Woudenberg, J.; Groenewald, J.Z.; Binder, M.; Crous, P.W. Alternaria redefined. Stud. Mycol. 2013, 75, 171–212. [Google Scholar] [CrossRef] [PubMed]
- Bensch, K.; Braun, U.; Groenewald, J.; Crous, P. The genus Cladosporium. Stud. Mycol. 2012, 72, 1–401. [Google Scholar] [CrossRef] [PubMed]
- Madrid, H.; Groenewald, J.Z.; Binder, M.; Crous, P.W. Novel Curvularia species from clinical specimens. Persoonia-Mol. Phylogeny Evol. Fungi 2014, 33, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Summerbell, R.C.; Gueidan, C.; Schroers, H.-J.; de Hoog, G.S.; Starink, M.; Rosete, Y.A.; Guarro, J.; Scott, J.A. Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Stud. Mycol. 2011, 68, 139–162. [Google Scholar] [CrossRef] [PubMed]
- Glenn, A.E.; Bacon, C.W.; Price, R.; Hanlin, R.T. Molecular phylogeny of Acremonium and its taxonomic implications. Mycologia 1996, 88, 369. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Kumar, S.; Tamura, K.; Nei, M. MEGA: Molecular evolutionary genetics analysis software for microcomputers. Bioinformatics 1994, 10, 189–191. [Google Scholar] [CrossRef]
- Ma, W.; Wu, F.; Tian, T.; He, D.; Zhang, Q.; Gu, J.-D.; Duan, Y.; Ma, D.; Wang, W.; Feng, H. Fungal diversity and its contribution to the biodeterioration of mural paintings in two 1700-year-old tombs of China. Int. Biodeterior. Biodegrad. 2020, 152, 104972. [Google Scholar] [CrossRef]
- Han, Y.; Du, J.; Li, J.; Li, M. Quantification of the organic acids in hawthorn wine: A comparison of two HPLC methods. Molecules 2019, 24, 2150. [Google Scholar] [CrossRef]
- Hughes, J.; Howind, T. Science and Art: A Future for Stone. In Proceedings of the 13th International Congress on the Deterioration and Conservation of Stone, Glasgow, UK, 6–10 September 2016; University of the West of Scotland: Scotland, UK, 2016; Volume 2. [Google Scholar]
- Gaylarde, C.; Baptista-Neto, J.A.; Ogawa, A.; Kowalski, M.; Celikkol-Aydin, S.; Beech, I. Epilithic and endolithic microorganisms and deterioration on stone church facades subject to urban pollution in a sub-tropical climate. Biofouling 2017, 33, 113–127. [Google Scholar] [CrossRef]
- Gaylarde, C.C.; Baptista-Neto, J.A. Microbiologically induced aesthetic and structural changes to dimension stone. NPJ Mater. Degrad. 2021, 5, 33. [Google Scholar] [CrossRef]
- Strzelczyk, A.B. Observations on aesthetic and structural changes induced in Polish historic objects by microorganisms. Int. Biodeterior. Biodegrad. 2003, 53, 151–156. [Google Scholar] [CrossRef]
- Gaylarde, P.; Gaylarde, C. Deterioration of siliceous stone monuments in Latin America: Microorganisms and mechanisms. Corros. Rev. 2004, 22, 395–416. [Google Scholar] [CrossRef]
- Ortega-Morales, B.O.; Narváez-Zapata, J.; Reyes-Estebanez, M.; Quintana, P.; Rosa-García, S.d.C.D.l.; Bullen, H.; Gómez-Cornelio, S.; Chan-Bacab, M.J. Bioweathering potential of cultivable fungi associated with semi-arid surface microhabitats of Mayan buildings. Front. Microbiol. 2016, 7, 201. [Google Scholar] [CrossRef] [PubMed]
- Mustoe, G.E. Biogenic Weathering: Solubilization of iron from minerals by epilithic freshwater algae and cyanobacteria. Microorganisms 2018, 6, 8. [Google Scholar] [CrossRef]
- Barrionuevo, M.R.E.; Englert, G.E.; Gaylarde, C.C. Physical and microbiological analysis of sandstone deterioration in the Argentine Jesuit missions. Geomicrobiol. J. 2016, 33, 671–676. [Google Scholar] [CrossRef]
- Leventhal, G.E.; Ackermann, M.; Schiessl, K.T. Why microbes secrete molecules to modify their environment: The case of iron-chelating siderophores. J. R. Soc. Interface 2019, 16, 20180674. [Google Scholar] [CrossRef]
- Priyadarshini, E.; Priyadarshini, S.S.; Pradhan, N. Heavy metal resistance in algae and its application for metal nanoparticle synthesis. Appl. Microbiol. Biotechnol. 2019, 103, 3297–3316. [Google Scholar] [CrossRef]
- Pathak, A.; Kothari, R.; Vinoba, M.; Habibi, N.; Tyagi, V. Fungal bioleaching of metals from refinery spent catalysts: A critical review of current research, challenges, and future directions. J. Environ. Manag. 2021, 280, 111789. [Google Scholar] [CrossRef]
- Kramer, J.; Özkaya, Ö.; Kümmerli, R. Bacterial siderophores in community and host interactions. Nat. Rev. Genet. 2020, 18, 152–163. [Google Scholar] [CrossRef]
- Winkelmann, G. Ecology of siderophores with special reference to the fungi. BioMetals 2007, 20, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Văcar, C.L.; Mircea, C.; Pârvu, M.; Podar, D. Diversity and Metabolic Activity of Fungi Causing Biodeterioration of Canvas Paintings. J. Fungi 2022, 8, 589. [Google Scholar] [CrossRef] [PubMed]
- Pangallo, D.; Kraková, L.; Chovanová, K.; Šimonovičová, A.; De Leo, F.; Urzì, C. Analysis and comparison of the microflora isolated from fresco surface and from surrounding air environment through molecular and biodegradative assays. World J. Microbiol. Biotechnol. 2012, 28, 2015–2027. [Google Scholar] [CrossRef] [PubMed]
- Honeyborne, D.B. Weathering and Decay of Masonry. In Conservation of Building and Decorative Stone; Routledge: New York, NY, USA, 2007; pp. 153–178. [Google Scholar]
- Vasanthakumar, A.; DeAraujo, A.; Mazurek, J.; Schilling, M.; Mitchell, R. Microbiological survey for analysis of the brown spots on the walls of the tomb of King Tutankhamun. Int. Biodeterior. Biodegrad. 2013, 79, 56–63. [Google Scholar] [CrossRef]
- Jimenez-Quero, A.; Pollet, E.; Zhao, M.; Marchioni, E.; Averous, L.; Phalip, V. Itaconic and fumaric acid production from biomass hydrolysates by Aspergillus strains. J. Microbiol. Biotechnol. 2016, 26, 1557–1565. [Google Scholar] [CrossRef]
- Yang, L.; Lübeck, M.; Lübeck, P.S. Aspergillus as a versatile cell factory for organic acid production. Fungal Biol. Rev. 2017, 31, 33–49. [Google Scholar] [CrossRef]
- Gutarowska, B.; Czyżowska, A. The ability of filamentous fungi to produce acids on indoor building materials. Ann. Microbiol. 2009, 59, 807–813. [Google Scholar] [CrossRef]

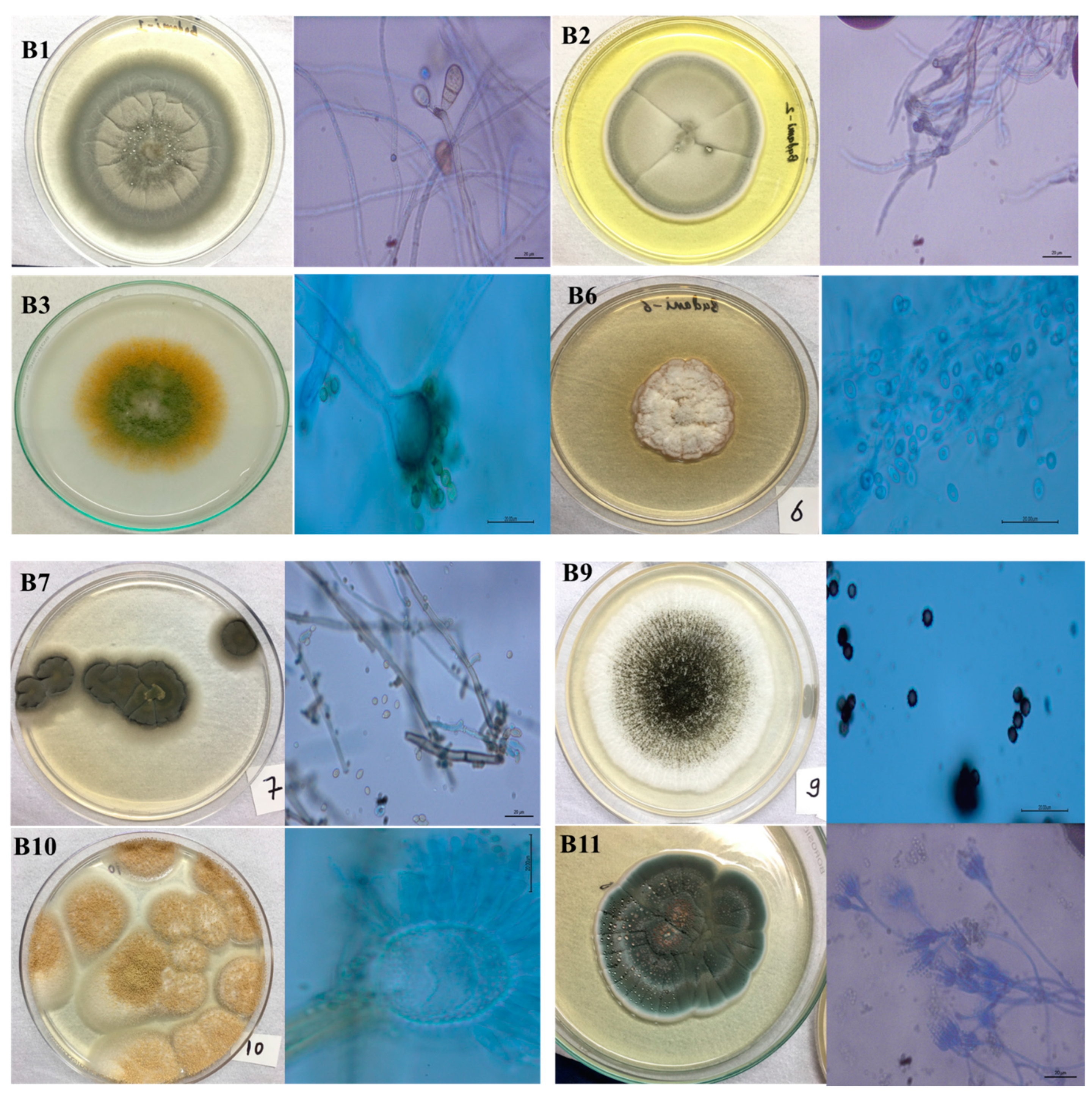

| Strain | Morphological Identification | Molecular Identification and BLAST Similarity | Identity (%) | Gen Bank Accession Number |
|---|---|---|---|---|
| B1 | Curvularia sp.; | Curvularia lunata; (MT163336.1) | 100% | OQ874797.1 |
| B2 | Cladosporium sp.; | Cladosporium sp., (MN634540.1) | 73% | OQ874798.1 |
| B3 | Aspergillus sp.; | Aspergillus flavus; (MK418229.1) | 100% | OQ874799.1 |
| B6 | Acremonium sp.; | Acremonium persicinum; (JQ599382.1) | 99% | OQ874800.1 |
| B7 | Cladosporium sp.; | Cladosporium cladosporioides; (MN634579.1) | 99% | OQ874801.1 |
| B9 | Aspergillus sp.; | Aspergillus niger; (OQ653131.1) | 100% | OQ874802.1 |
| B10 | Aspergillus sp.; | Aspergillus terreus; (OP758079.1) | 100% | OQ874803.1 |
| B11 | Penicillium sp.; | Penicillium chrysogenum; (KX266831.1) | 100% | OQ874804.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agrawal, S.; Khumlianlal, J.; Devi, S.I. Uncovering the Fungal Diversity and Biodeterioration Phenomenon on Archaeological Carvings of the Badami Cave Temples: A Microcosm Study. Life 2024, 14, 28. https://doi.org/10.3390/life14010028
Agrawal S, Khumlianlal J, Devi SI. Uncovering the Fungal Diversity and Biodeterioration Phenomenon on Archaeological Carvings of the Badami Cave Temples: A Microcosm Study. Life. 2024; 14(1):28. https://doi.org/10.3390/life14010028
Chicago/Turabian StyleAgrawal, Shivankar, Joshua Khumlianlal, and Sarangthem Indira Devi. 2024. "Uncovering the Fungal Diversity and Biodeterioration Phenomenon on Archaeological Carvings of the Badami Cave Temples: A Microcosm Study" Life 14, no. 1: 28. https://doi.org/10.3390/life14010028
APA StyleAgrawal, S., Khumlianlal, J., & Devi, S. I. (2024). Uncovering the Fungal Diversity and Biodeterioration Phenomenon on Archaeological Carvings of the Badami Cave Temples: A Microcosm Study. Life, 14(1), 28. https://doi.org/10.3390/life14010028








