Unlocking Cowpea’s Defense Responses: Conserved Transcriptional Signatures in the Battle against CABMV and CPSMV Viruses
Abstract
1. Introduction
- Is there a cowpea conserved transcriptional response (up- or down-regulated) to the CABMV and CPSMV mechanical inoculations at early analyzed time points (1 or 16 hpi treatments)?
- If the conservation exists, what are the critical biological processes, molecular functions, and metabolic and signaling pathways associated with the conserved response of up-regulated transcripts for a 1 or 16 hpi treatment?
- Are the up-regulated co-expressed genes widely present among Viridiplantae species?
- Based on the scrutinized data, can a tentative model of the cowpea transcriptional conserved response be constructed?
2. Materials and Methods
2.1. Plant Material, Experimental Design, and Virus Mechanical Inoculation Strategy
2.2. Total RNA Processing and RNA-Seq Library Synthesis
2.3. RNA-Seq Data Assembly and Differential Expression Analysis
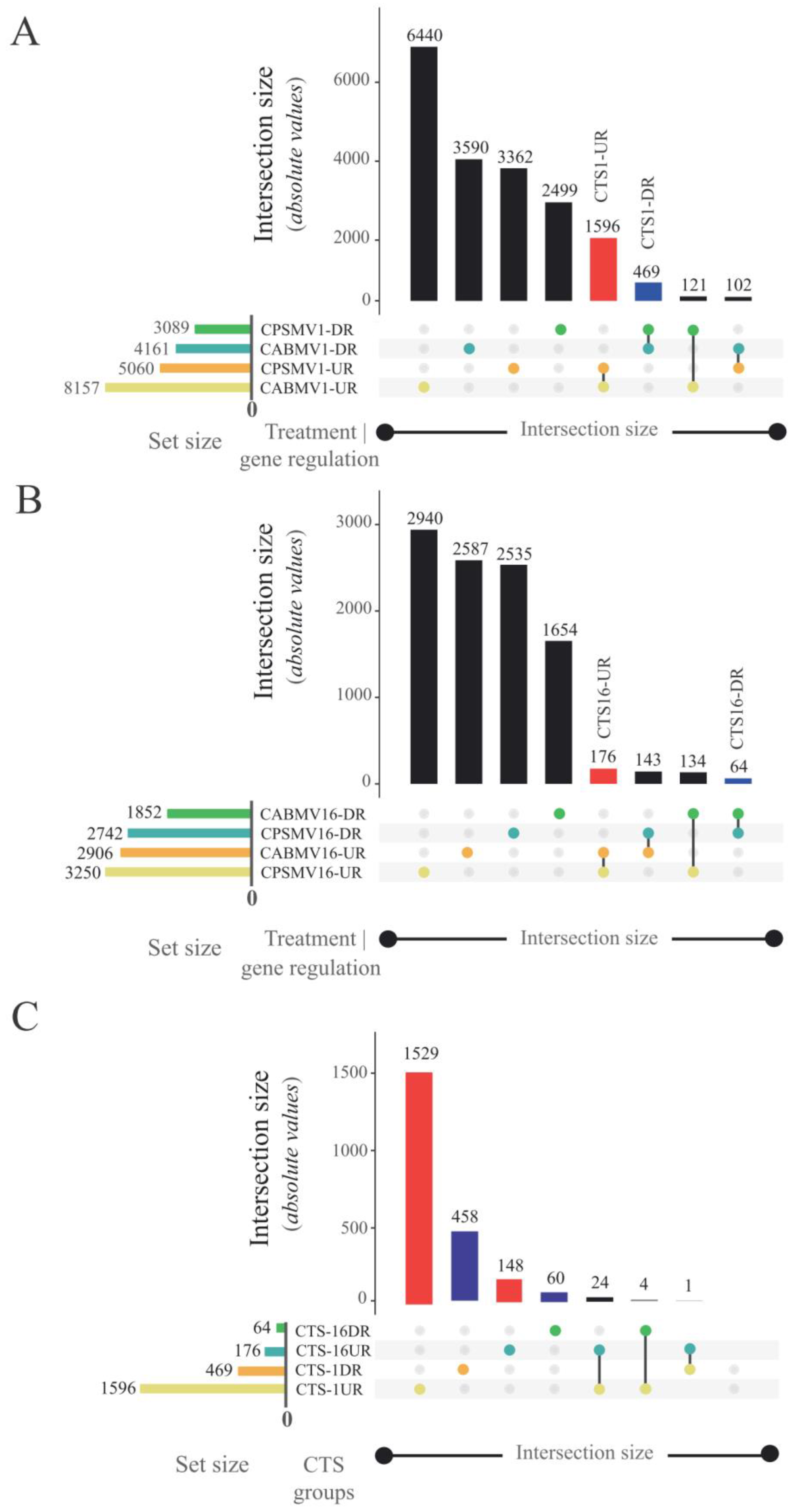
2.4. Conserved Transcriptional Signatures (CTS) Identification
- CTS-1UR (up-regulated transcripts in response to CABMV and CPSMV mechanical inoculations at one hpi treatments);
- CTS-1DR (down-regulated transcripts in response to CABMV and CPSMV mechanical inoculations at one hpi treatments);
- CTS-16UR (up-regulated transcripts in response to CABMV and CPSMV mechanical inoculations at 16 hpi treatments);
- CTS-16DR (down-regulated transcripts in response to CABMV and CPSMV mechanical inoculations at 16 hpi treatments).
2.5. Biological Processes and Molecular Functions Enrichment Analysis
2.6. MapMan Analysis of CTS Up-Regulated Groups
2.7. CTS Mining for Jasmonic Acid and Ethylene Biosynthesis Pathways
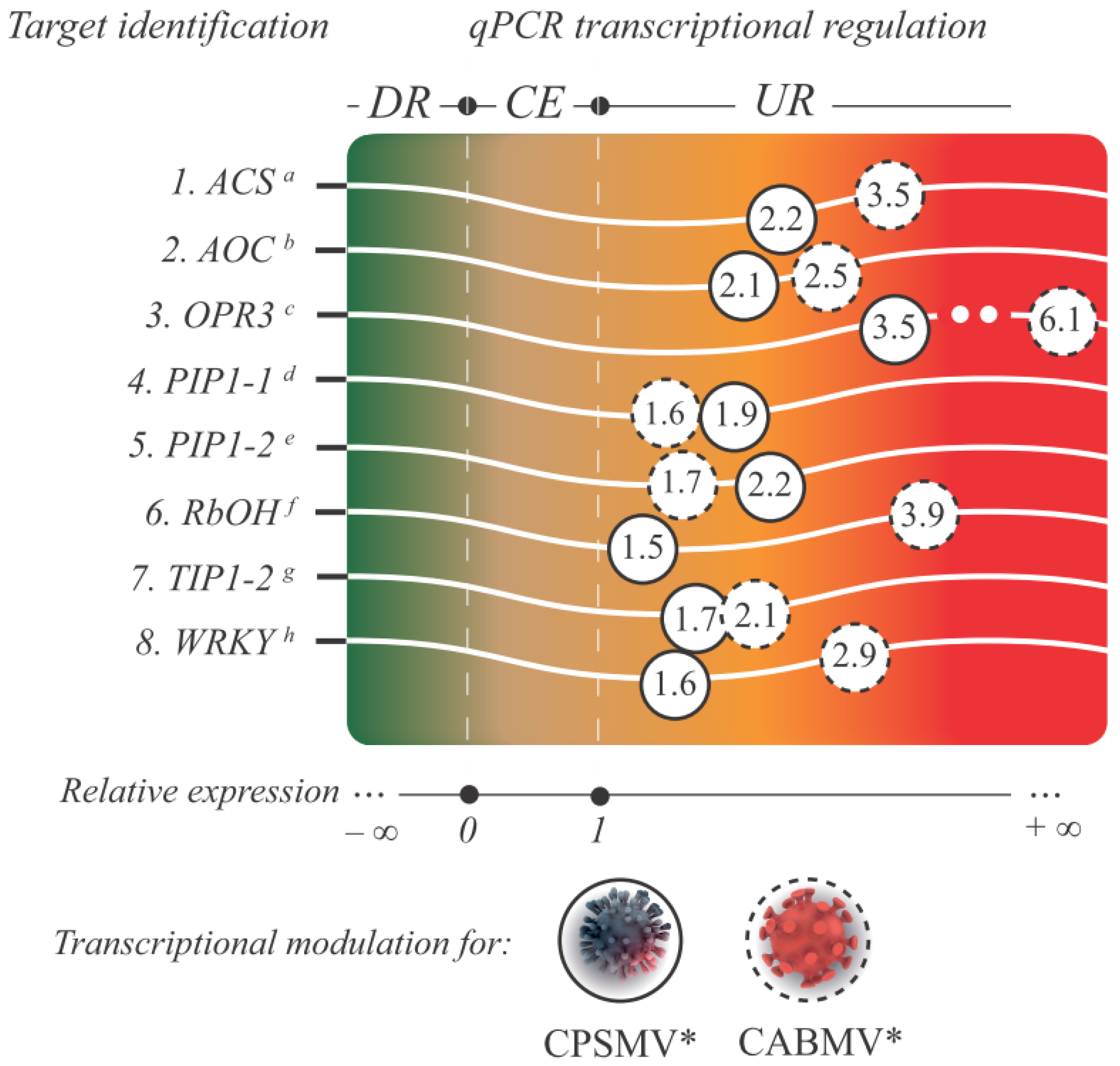
2.8. qPCR: Setup, cDNA Synthesis, Primers Efficiency Analysis, and Relative Expression
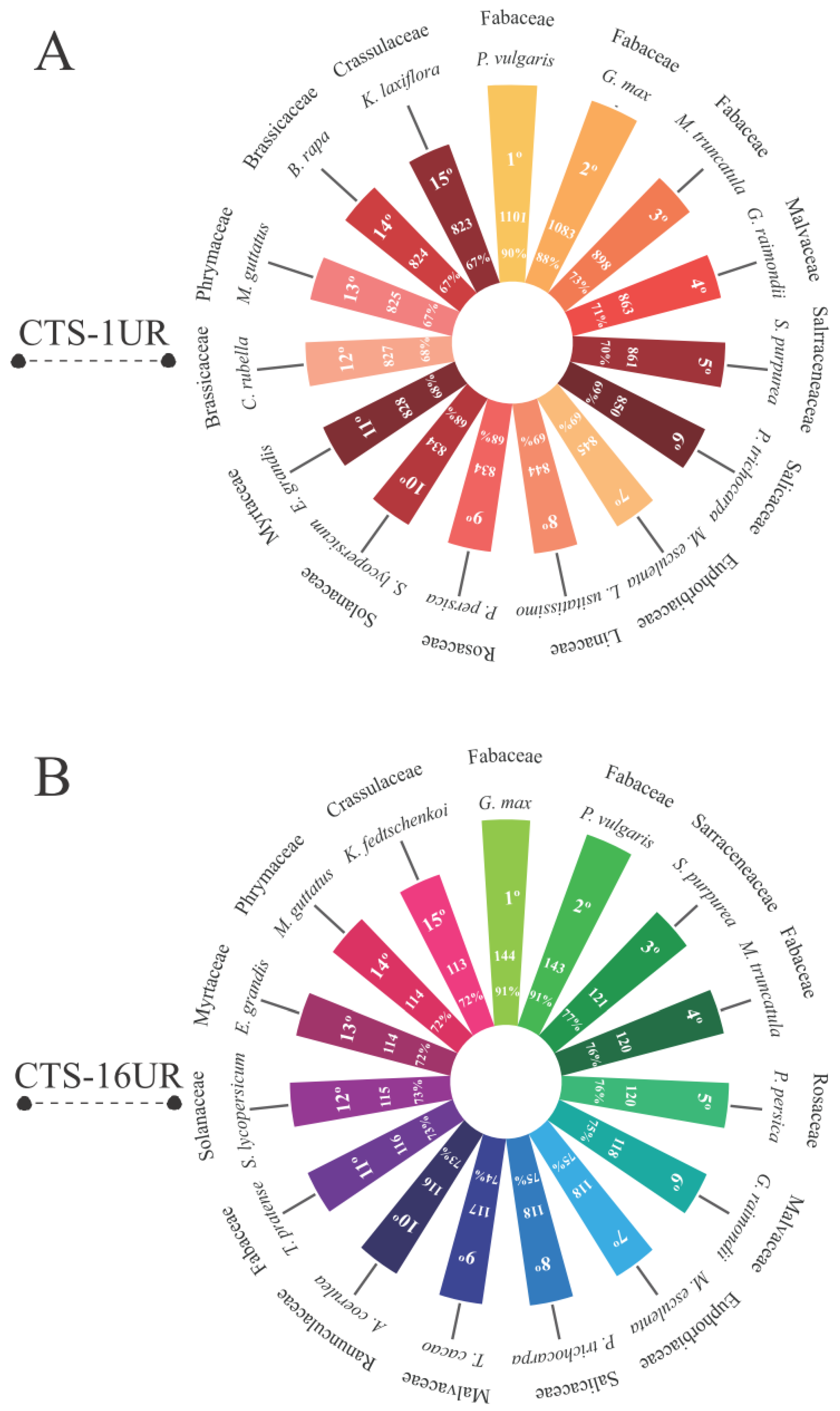
2.9. Orthology of CTS-Coding Loci in Viridiplantae Species
3. Results
3.1. Is There a Cowpea Conserved Transcriptional Response (Up-Regulated or Down-Regulated) to the CABMV and CPSMV Mechanical Inoculations at Early Analyzed Time Points (1 or 16 Hpi Treatments)?
3.2. What Are the Primary Biological Processes, Molecular Functions, and Metabolic and Signaling Pathways Associated with the Conserved Response?
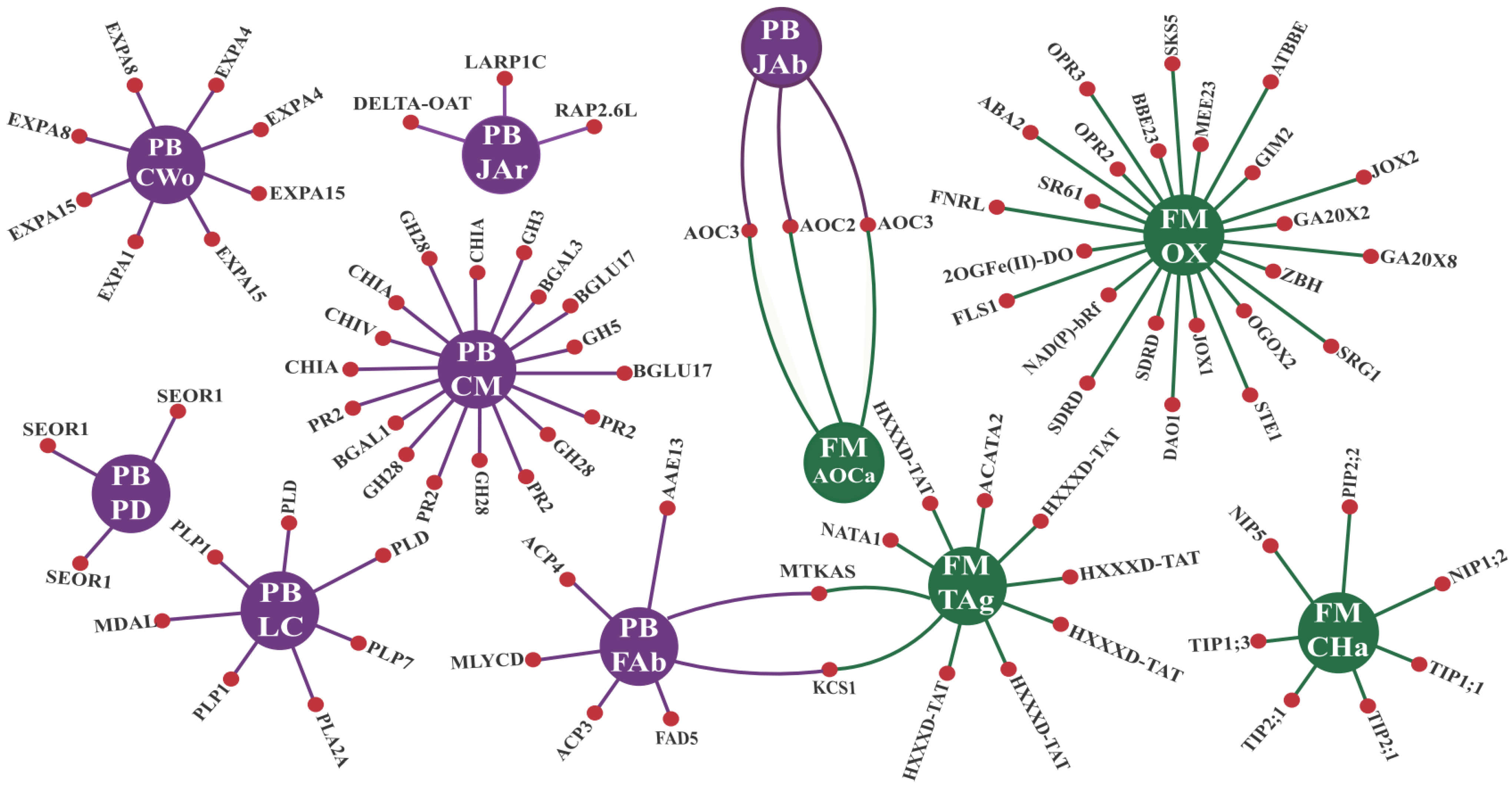
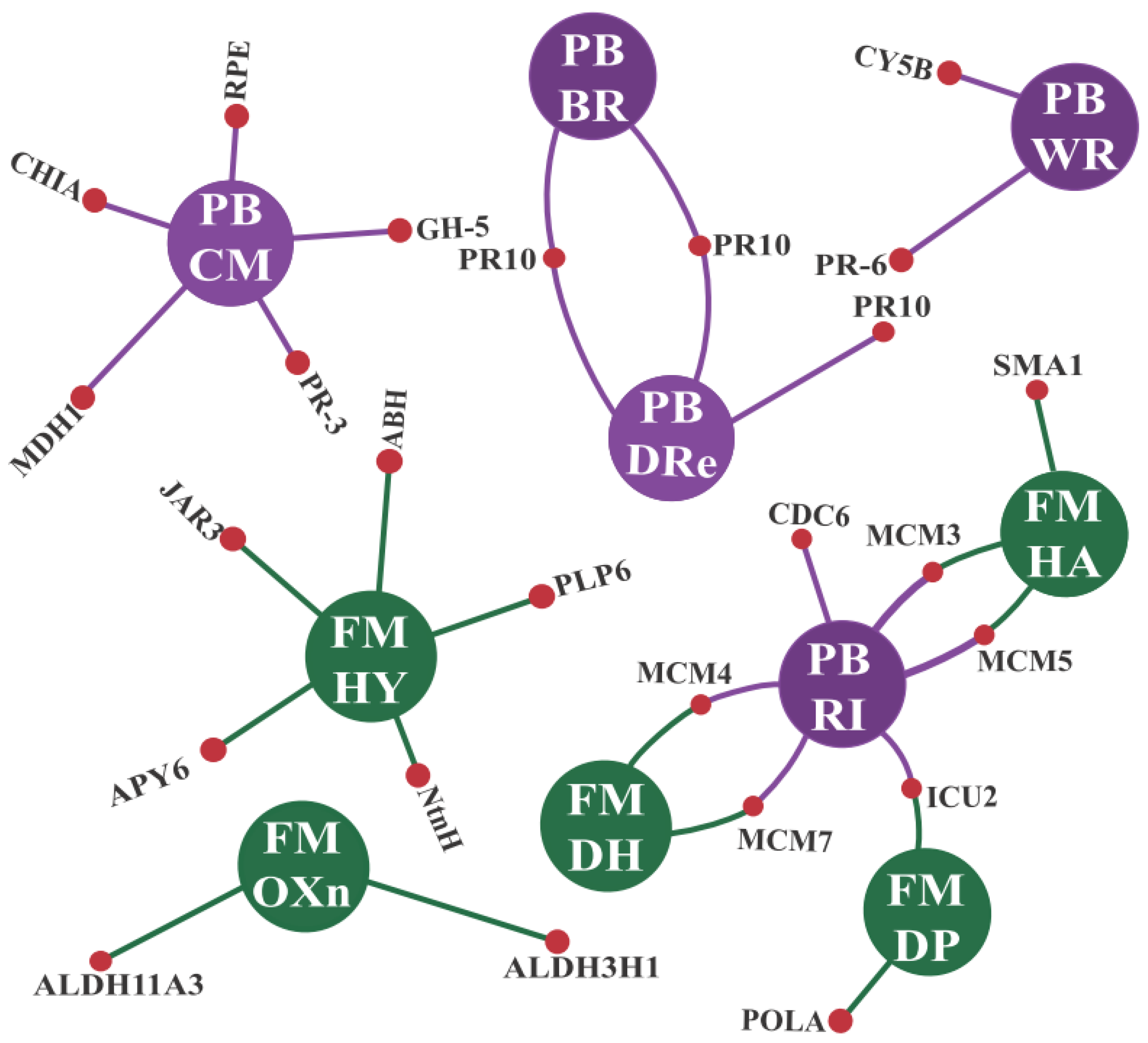
3.2.1. GO Enrichments Analysis for Up-Regulated CTS Groups: A Focus on Biological Processes and Molecular Functions
3.2.2. CTS and Their Intricate Metabolic and Signaling Pathways
- a.
- Enzymes responsive to oxidative stress or involved in hormonal metabolic processes
- b.
- Signaling proteins and transcription factors
- c.
- R genes and PR proteins
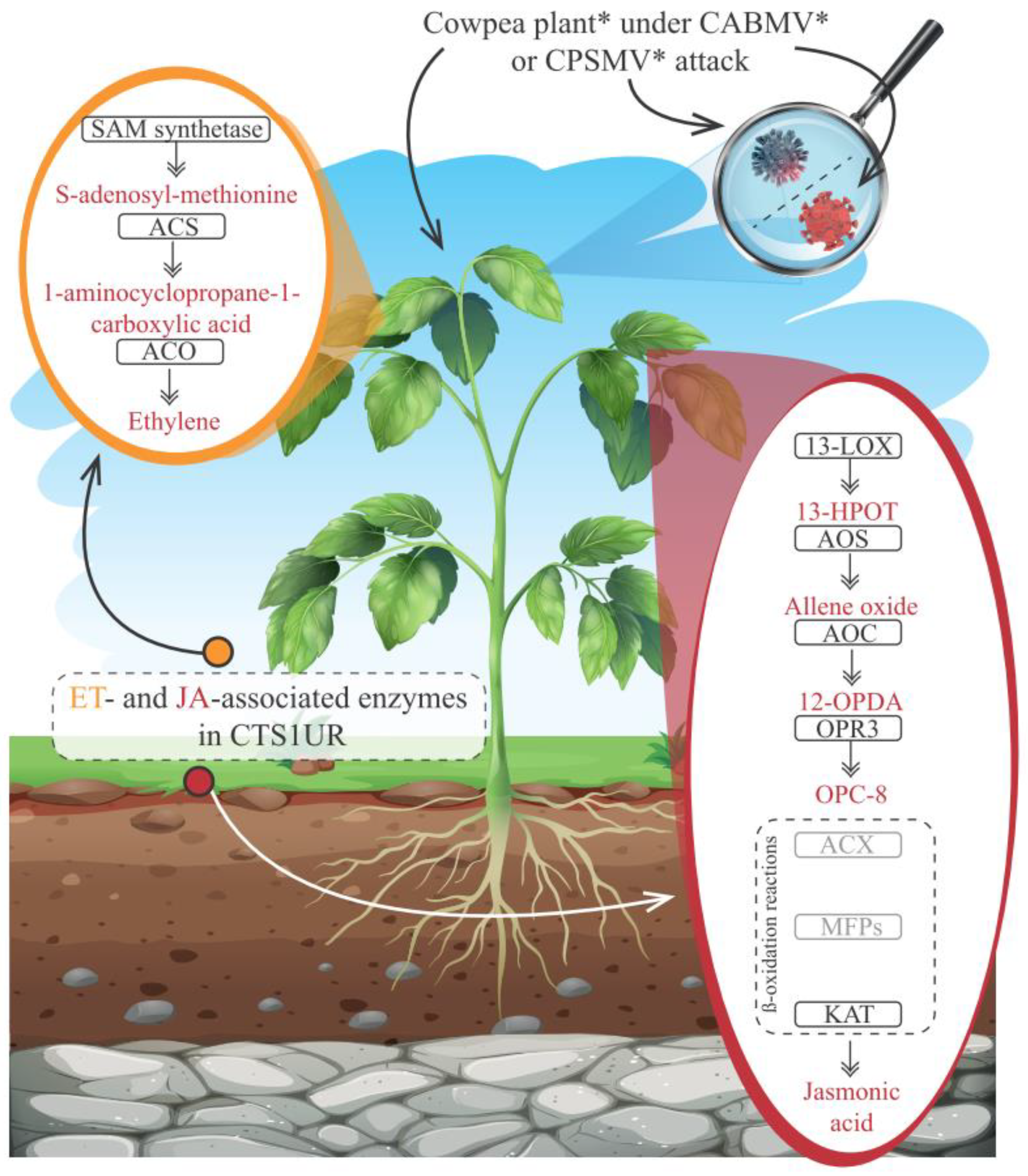
3.2.3. CTS and Phytohormones: An Emphasis on the JA and ET Biosynthesis Pathways
3.2.4. qPCR Validation of the CTS Expression
3.3. Are the Cowpea CTS-Encoding Genes Widely Distributed among Viridiplantae Species?
3.4. Based on the Scrutinized Data, Can a Tentative Model of the Cowpea Transcriptional Conserved Response Be Constructed?
4. Discussion
4.1. General Aspects of the Cowpea Conserved Response to CABMV and CPSMV Mechanical Inoculations
4.2. Enriched Biological Processes and Molecular Functions Unveiled Some Cowpea Molecular Weapons to Combat CABMV and CPSMV
4.3. JA and ET Emerged as Prominent Players in the Cowpea Conserved Response to CPSMV and CABMV Mechanical Inoculations
4.4. The Crucial Role of Redox Balance, Other Phytohormones, Signaling Pathways, R Genes, and Some Transcription Factors Groups
4.5. Conservation Encompasses Not Only the Target Genes Transcription but Also the Preservation of CTS-Anchoring Gene Loci in Viridiplantae
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boukar, O.; Fatokun, C.A.; Huynh, B.-L.; Roberts, P.A.; Close, T.J. Genomic Tools in Cowpea Breeding Programs: Status and Perspectives. Front. Plant Sci. 2016, 7, 757. [Google Scholar] [CrossRef] [PubMed]
- Mekonnen, T.W.; Gerrano, A.S.; Mbuma, N.W.; Labuschagne, M.T. Breeding of Vegetable Cowpea for Nutrition and Climate Resilience in Sub-Saharan Africa: Progress, Opportunities, and Challenges. Plants 2022, 11, 1583. [Google Scholar] [CrossRef] [PubMed]
- Booker, H.M.; Umaharan, P.; McDavid, C.R. Effect of Cowpea Severe Mosaic Virus on Crop Growth Characteristics and Yield of Cowpea. Plant Dis. 2005, 89, 515–520. [Google Scholar] [CrossRef]
- Neya, B.J.; Zida, P.E.; Sereme, D.; Lund, O.S.; Traore, O. Evaluation of Yield Losses Caused by Cowpea Aphid-Borne Mosaic Virus (CABMV) in 21 Cowpea (Vigna unguiculata (L.) Walp.) Varieties in Burkina Faso. Pak. J. Biol. Sci. 2015, 18, 304–313. [Google Scholar] [CrossRef][Green Version]
- Costa, C.L.; Lin, M.T.; Kitajima, E.W.; Santos, A.A.; Mesquita, R.C.M.; Freire, F.R.F. Cerotoma Arcuata (Oliv.) Um Crisomelídeo Vetor Do Mosaico Da Vigna No Brasil. Cerotoma Arcuata (Oliv.) Um Cris. Vetor Do Mosaico Da Vigna No Bras. 1978, 3, 81–82. [Google Scholar]
- Pirone, T.P. Viral Genes and Gene Products That Determine Insect Transmissibility. Semin. Virol. 1991, 2, 81–87. [Google Scholar]
- Gray, S.M. Plant Virus Proteins Involved in Natural Vector Transmission. Trends Microbiol. 1996, 4, 259–264. [Google Scholar] [CrossRef]
- Bastos, E.A. A Cultura de Feijão-Caupi No Brasil; Jorimá Marques Ferreira—Empresa Brasileira de Pesquisa Agropecuária-Embrapa Embrapa Meio-Norte: Teresina, PI, Brazil, 2016; Volume 2, pp. 1–66. [Google Scholar]
- Baebler, Š.; Krečič-Stres, H.; Rotter, A.; Kogovšek, P.; Cankar, K.; Kok, E.J.; Gruden, K.; Kovač, M.; Žel, J.; Pompe-Novak, M.; et al. PVYNTN Elicits a Diverse Gene Expression Response in Different Potato Genotypes in the First 12 h after Inoculation. Mol. Plant Pathol. 2009, 10, 263–275. [Google Scholar] [CrossRef]
- Widyasari, K.; Alazem, M.; Kim, K.-H. Soybean Resistance to Soybean Mosaic Virus. Plants 2020, 9, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Miozzi, L.; Napoli, C.; Sardo, L.; Accotto, G.P. Transcriptomics of the Interaction between the Monopartite Phloem-Limited Geminivirus Tomato Yellow Leaf Curl Sardinia Virus and Solanum lycopersicum Highlights a Role for Plant Hormones, Autophagy and Plant Immune System Fine Tuning during Infection. PLoS ONE 2014, 9, e89951. [Google Scholar] [CrossRef]
- Rodrigo, G.; Carrera, J.; Ruiz-Ferrer, V.; Del Toro, F.J.; Llave, C.; Voinnet, O.; Elena, S.F. A Meta-Analysis Reveals the Commonalities and Differences in Arabidopsis Thaliana Response to Different Viral Pathogens. PLoS ONE 2012, 7, e40526. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Neto, J.R.C.; Silva, M.D.; Rodrigues, F.A.; Nepomuceno, A.L.; Pandolfi, V.; Lima Morais, D.A.; Kido, E.A.; Benko-Iseppon, A.M. Importance of Inositols and Their Derivatives in Cowpea under Root Dehydration: An Omics Perspective. Physiol. Plant. 2021, 172, 441–462. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Neto, J.R.C.; Borges, A.N.D.C.; Da Silva, M.D.; Morais, D.A.D.L.; Bezerra-Neto, J.P.; Bourque, G.; Kido, E.A.; Benko-Iseppon, A.M. The Cowpea Kinome: Genomic and Transcriptomic Analysis Under Biotic and Abiotic Stresses. Front. Plant Sci. 2021, 12, 667013. [Google Scholar] [CrossRef] [PubMed]
- Matos, M.K.D.S.; Benko-Iseppon, A.M.; Bezerra-Neto, J.P.; Ferreira-Neto, J.R.C.; Wang, Y.; Liu, H.; Pandolfi, V.; Amorim, L.L.B.; Willadino, L.; Do Vale Amorim, T.C.; et al. The WRKY Transcription Factor Family in Cowpea: Genomic Characterization and Transcriptomic Profiling under Root Dehydration. Gene 2022, 823, 146377. [Google Scholar] [CrossRef]
- Rocha, M.M.; Lima, J.A.A.; Filho, F.R.F.; Lima, V.C.V. Resistência de Caupi de Tegumento Branco a Algumas Estirpes de Comovirus, Potyvirus e Cucumovirus. In IV Reunião Nacional de Pesquisa de Caupi—EMBRAPA—CPAMN; EMBRAPA Meio-Norte: Teresina, PI, Brazil, 1996; pp. 100–101. [Google Scholar]
- Oliveira, C.R.R.D.; Freire Filho, F.R.; Nogueira, M.D.S.D.R.; Barros, G.B.; Eiras, M.; Ribeiro, V.Q.; Lopes, Â.C.D.A. Reação de Genótipos de Feijão-Caupi Revela Resistência Às Coinfecções Pelo Cucumber Mosaic Virus, Cowpea Aphid-Borne Mosaic Virus e Cowpea Severe Mosaic Virus. Bragantia 2012, 71, 59–66. [Google Scholar] [CrossRef]
- Cardoso, M.J.; Filho, F.R.F. BR 14-Mulato. In Nova Cultivar de Feijão Macassar Para o Estado Do Piauí; Empresa Brasileira de Pesquisa Agropecuária—EMBRAPA: Brasília, Brazil, 1990; Volume 4, pp. 1–4. [Google Scholar]
- Hull, R. Mechanical Inoculation of Plant Viruses. Curr. Protoc. Microbiol. 2009, 16.B.6.1, 16.B.6.4. [Google Scholar] [CrossRef]
- Barna, B.; Kiraly, L. Host–Pathogen Relations: Diseases Caused by Viruses, Subviral Organisms, and Phytoplasmas. In Plant Toxicology; Hock, B., Elstner, E.F., Eds.; Routledge: New York, NY, USA, 2004; Volume 4, pp. 519–553. [Google Scholar]
- Bourgey, M.; Dali, R.; Eveleigh, R.; Chen, K.C.; Letourneau, L.; Fillon, J.; Michaud, M.; Caron, M.; Sandoval, J.; Lefebvre, F.; et al. GenPipes: An Open-Source Framework for Distributed and Scalable Genomic Analyses. GigaScience 2019, 8, giz037. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Zúñiga-León, E.; Carrasco-Navarro, U.; Fierro, F. NeVOmics: An Enrichment Tool for Gene Ontology and Functional Network Analysis and Visualization of Data from OMICs Technologies. Genes 2018, 9, 569. [Google Scholar] [CrossRef]
- Delgado-Salinas, A.; Thulin, M.; Pasquet, R.; Weeden, N.; Lavin, M. Vigna (Leguminosae) Sensu Lato: The Names and Identities of the American Segregate Genera. Am. J. Bot. 2011, 98, 1694–1715. [Google Scholar] [CrossRef]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. Mapman: A User-Driven Tool to Display Genomics Data Sets onto Diagrams of Metabolic Pathways and Other Biological Processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Zarembinski, T.I.; Theologis, A. Ethylene Biosynthesis and Action: A Case of Conservation. Plant Mol. Biol. 1994, 26, 1579–1597. [Google Scholar] [CrossRef]
- Vandenbussche, F.; Vaseva, I.; Vissenberg, K.; Van Der Straeten, D. Ethylene in Vegetative Development: A Tale with a Riddle. New Phytol. 2012, 194, 895–909. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Kombrink, E. Jasmonates: Structural Requirements for Lipid-Derived Signals Active in Plant Stress Responses and Development. ACS Chem. Biol. 2010, 5, 63–77. [Google Scholar] [CrossRef]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, Metabolism, and Signaling by Proteins Activating and Repressing Transciption. EXBOT J. 2016, 68, erw443. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Sonnhammer, E.L.L.; Östlund, G. InParanoid 8: Orthology Analysis between 273 Proteomes, Mostly Eukaryotic. Nucleic Acids Res. 2015, 43, 234–239. [Google Scholar] [CrossRef]
- Casteel, C.L.; De Alwis, M.; Bak, A.; Dong, H.; Whitham, S.A.; Jander, G. Disruption of Ethylene Responses by Turnip Mosaic Virus Mediates Suppression of Plant Defense against the Green Peach Aphid Vector. Plant Physiol. 2015, 169, 209–218. [Google Scholar] [CrossRef]
- Goyer, A.; Hamlin, L.; Crosslin, J.M.; Buchanan, A.; Chang, J.H. RNA-Seq Analysis of Resistant and Susceptible Potato Varieties during the Early Stages of Potato Virus Y Infection. BMC Genom. 2015, 16, 472. [Google Scholar] [CrossRef]
- Maurel, C.; Boursiac, Y.; Luu, D.-T.; Santoni, V.; Shahzad, Z.; Verdoucq, L. Aquaporins in Plants. Physiol. Rev. 2015, 95, 1321–1358. [Google Scholar] [CrossRef]
- Sade, D.; Sade, N.; Shriki, O.; Lerner, S.; Gebremedhin, A.; Karavani, A.; Brotman, Y.; Osorio, S.; Fernie, A.R.; Willmitzer, L.; et al. Water Balance, Hormone Homeostasis, and Sugar Signaling Are All Involved in Tomato Resistance to Tomato Yellow Leaf Curl Virus. Plant Physiol. 2014, 165, 1684–1697. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Wang, X.; Li, P.; Wang, H.; Ji, H.; Xie, J.; Qiu, Q.; Shen, D.; Dong, H. Plant Aquaporin AtPIP1;4 Links Apoplastic H2O2 Induction to Disease Immunity Pathways. Plant Physiol. 2016, 171, 1635–1650. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A.; Dangl, J.L. Functions of the Respiratory Burst Oxidase in Biotic Interactions, Abiotic Stress and Development. Curr. Opin. Plant Biol. 2005, 8, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Park, C.-J.; Kim, K.-J.; Shin, R.; Park, J.M.; Shin, Y.-C.; Paek, K.-H. Pathogenesis-Related Protein 10 Isolated from Hot Pepper Functions as a Ribonuclease in an Antiviral Pathway: Characterization of the Hot Pepper PR-10. Plant J. 2004, 37, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Su, Y.; Ling, H.; Ahmad, W.; Gao, S.; Guo, J.; Que, Y.; Xu, L. A Sugarcane Pathogenesis-Related Protein, ScPR10, Plays a Positive Role in Defense Responses under Sporisorium Scitamineum, SrMV, SA, and MeJA Stresses. Plant Cell Rep. 2017, 36, 1427–1440. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.H.; Gururani, M.A.; Chun, S.-C. Expression Analysis of Rice Pathogenesis-Related Proteins Involved in Stress Response and Endophytic Colonization Properties of Gfp-Tagged Bacillus Subtilis CB-R05. Appl. Biochem. Biotechnol. 2014, 174, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, P.; Rasool, S.; Gul, A.; Sheikh, S.A.; Akram, N.A.; Ashraf, M.; Kazi, A.M.; Gucel, S. Jasmonates: Multifunctional Roles in Stress Tolerance. Front. Plant Sci. 2016, 7, 813. [Google Scholar] [CrossRef]
- León, J.; Rojo, E.; Sánchez-Serrano, J.J. Wound Signalling in Plants. J. Exp. Bot. 2001, 52, 813. [Google Scholar] [CrossRef]
- Chauvin, A.; Caldelari, D.; Wolfender, J.; Farmer, E.E. Four 13-lipoxygenases Contribute to Rapid Jasmonate Synthesis in Wounded Arabidopsis Thaliana Leaves: A Role for Lipoxygenase 6 in Responses to Long-distance Wound Signals. New Phytol. 2013, 197, 566–575. [Google Scholar] [CrossRef]
- Glauser, G.; Dubugnon, L.; Mousavi, S.A.R.; Rudaz, S.; Wolfender, J.-L.; Farmer, E.E. Velocity Estimates for Signal Propagation Leading to Systemic Jasmonic Acid Accumulation in Wounded Arabidopsis. J. Biol. Chem. 2009, 284, 34506–34513. [Google Scholar] [CrossRef]
- Chakraborty, N.; Basak, J. Comparative Transcriptome Profiling of a Resistant vs. Susceptible Vigna Mungo Cultivar in Response to Mungbean Yellow Mosaic India Virus Infection Reveals New Insight into MYMIV Resistance. Curr. Plant Biol. 2018, 15, 8–24. [Google Scholar] [CrossRef]
- Heyman, J.; Canher, B.; Bisht, A.; Christiaens, F.; De Veylder, L. Emerging Role of the Plant ERF Transcription Factors in Coordinating Wound Defense Responses and Repair. J. Cell Sci. 2018, 15, 8–24. [Google Scholar] [CrossRef] [PubMed]
- Kende, H. Hormone Response Mutants. A Plethora of Surprises. Plant Physiol. 2001, 125, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Singh, R.P.; Tai, G.C.C. Molecular Characterization and Expression Analysis of 1-Aminocyclopropane-1-Carboxylate Oxidase Homologs from Potato under Abiotic and Biotic Stresses. Genome 2002, 45, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Dziurka, M.; Janeczko, A.; Juhász, C.; Gullner, G.; Oklestková, J.; Novák, O.; Saja, D.; Skoczowski, A.; Tóbiás, I.; Barna, B. Local and Systemic Hormonal Responses in Pepper Leaves during Compatible and Incompatible Pepper-Tobamovirus Interactions. Plant Physiol. Biochem. 2016, 109, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Clarke, S.F.; Burritt, D.J.; Jameson, P.E.; Guy, P.L. Influence of Plant Hormones on Virus Replication and Pathogenesis-Related Proteins In Phaseolus Vulgaris L. Infected with White Clover Mosaic Potexvirus. Physiol. Mol. Plant Pathol. 1998, 53, 195–207. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, S.H.; Palaniyandi, S.A.; Yang, S.H.; Suh, J.-W. Expression of Potato S-Adenosyl-l-Methionine Synthase (SbSAMS) Gene Altered Developmental Characteristics and Stress Responses in Transgenic Arabidopsis Plants. Plant Physiol. Biochem. 2015, 87, 84–91. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive Oxygen Species: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Whitham, S.A.; Yang, C.; Goodin, M.M. Global Impact: Elucidating Plant Responses to Viral Infection. MPMI 2006, 19, 1207–1215. [Google Scholar] [CrossRef]
- Durrant, W.E.; Dong, X. Systemic Acquired Resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Dong, X. Systemic Acquired Resistance: Turning Local Infection into Global Defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef]
- Gao, Q.-M.; Zhu, S.; Kachroo, P.; Kachroo, A. Signal Regulators of Systemic Acquired Resistance. Front. Plant Sci. 2015, 06, 228. [Google Scholar] [CrossRef] [PubMed]
- Alazem, M.; Lin, N.-S. Antiviral Roles of Abscisic Acid in Plants. Front. Plant Sci. 2017, 8, 1760. [Google Scholar] [CrossRef] [PubMed]
- Macho, A.P.; Zipfel, C. Plant PRRs and the Activation of Innate Immune Signaling. Mol. Cell 2014, 54, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Zorzatto, C.; Machado, J.P.B.; Lopes, K.V.G.; Nascimento, K.J.T.; Pereira, W.A.; Brustolini, O.J.B.; Reis, P.A.B.; Calil, I.P.; Deguchi, M.; Sachetto-Martins, G.; et al. NIK1-Mediated Translation Suppression Functions as a Plant Antiviral Immunity Mechanism. Nature 2015, 520, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Kundu, A.; Singh, P.K.; Dey, A.; Ganguli, S.; Pal, A. Complex Molecular Mechanisms Underlying MYMIV-Resistance in Vigna Mungo Revealed by Comparative Transcriptome Profiling. Sci. Rep. 2019, 9, 8858. [Google Scholar] [CrossRef]
- Qu, Y.; Sun, Y.; Yang, Z.; Ding, C. Calcium Ions Signaling: Targets for Attack and Utilization by Viruses. Front. Microbiol. 2022, 13, 889374. [Google Scholar] [CrossRef]
- Patel, A.; Dey, N.; Chaudhuri, S.; Pal, A. Molecular and Biochemical Characterization of a Vigna Mungo MAP Kinase Associated with Mungbean Yellow Mosaic India Virus Infection and Deciphering Its Role in Restricting the Virus Multiplication. Plant Sci. 2017, 262, 127–140. [Google Scholar] [CrossRef]
- Jin, H.; Liu, Y.; Yang, K.-Y.; Kim, C.Y.; Baker, B.; Zhang, S. Function of a Mitogen-Activated Protein Kinase Pathway in N Gene-Mediated Resistance in Tobacco: MAPK in N Gene-Mediated Resistance. Plant J. 2003, 33, 719–731. [Google Scholar] [CrossRef]
- Pan, J.; Zhang, M.; Kong, X.; Xing, X.; Liu, Y.; Zhou, Y.; Liu, Y.; Sun, L.; Li, D. ZmMPK17, a Novel Maize Group D MAP Kinase Gene, Is Involved in Multiple Stress Responses. Planta 2012, 235, 661–676. [Google Scholar] [CrossRef]
- Akhter, M.d.S.; Nakahara, K.S.; Masuta, C. Resistance Induction Based on the Understanding of Molecular Interactions between Plant Viruses and Host Plants. Virol. J. 2021, 18, 176. [Google Scholar] [CrossRef] [PubMed]
- Viswanath, K.K.; Kuo, S.-Y.; Tu, C.-W.; Hsu, Y.-H.; Huang, Y.-W.; Hu, C.-C. The Role of Plant Transcription Factors in the Fight against Plant Viruses. IJMS 2023, 24, 8433. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Cao, D.; Li, S.; Su, A.; Geng, J.; Grover, C.E.; Hu, S.; Hua, J. The Complete Mitochondrial Genome of Gossypium Hirsutum and Evolutionary Analysis of Higher Plant Mitochondrial Genomes. PLoS ONE 2013, 8, e69476. [Google Scholar] [CrossRef] [PubMed]
- Lonardi, S.; Muñoz-Amatriaín, M.; Liang, Q.; Shu, S.; Wanamaker, S.I.; Lo, S.; Tanskanen, J.; Schulman, A.H.; Zhu, T.; Luo, M.; et al. The Genome of Cowpea (Vigna unguiculata [L.] Walp.). Plant J. 2019, 98, 767–782. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borges-Martins, A.N.C.; Ferreira-Neto, J.R.C.; Silva, M.D.d.; Morais, D.A.d.L.; Pandolfi, V.; Silva, R.L.d.O.; Melo, A.L.T.M.d.; da Costa, A.F.; Benko-Iseppon, A.M. Unlocking Cowpea’s Defense Responses: Conserved Transcriptional Signatures in the Battle against CABMV and CPSMV Viruses. Life 2023, 13, 1747. https://doi.org/10.3390/life13081747
Borges-Martins ANC, Ferreira-Neto JRC, Silva MDd, Morais DAdL, Pandolfi V, Silva RLdO, Melo ALTMd, da Costa AF, Benko-Iseppon AM. Unlocking Cowpea’s Defense Responses: Conserved Transcriptional Signatures in the Battle against CABMV and CPSMV Viruses. Life. 2023; 13(8):1747. https://doi.org/10.3390/life13081747
Chicago/Turabian StyleBorges-Martins, Artemisa Nazaré Costa, José Ribamar Costa Ferreira-Neto, Manassés Daniel da Silva, David Anderson de Lima Morais, Valesca Pandolfi, Roberta Lane de Oliveira Silva, Ana Luiza Trajano Mangueira de Melo, Antônio Félix da Costa, and Ana Maria Benko-Iseppon. 2023. "Unlocking Cowpea’s Defense Responses: Conserved Transcriptional Signatures in the Battle against CABMV and CPSMV Viruses" Life 13, no. 8: 1747. https://doi.org/10.3390/life13081747
APA StyleBorges-Martins, A. N. C., Ferreira-Neto, J. R. C., Silva, M. D. d., Morais, D. A. d. L., Pandolfi, V., Silva, R. L. d. O., Melo, A. L. T. M. d., da Costa, A. F., & Benko-Iseppon, A. M. (2023). Unlocking Cowpea’s Defense Responses: Conserved Transcriptional Signatures in the Battle against CABMV and CPSMV Viruses. Life, 13(8), 1747. https://doi.org/10.3390/life13081747






