Genetic Basis of Acinetobacter sp. K1 Adaptation Mechanisms to Extreme Environmental Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin of the Bacterial Isolate Acinetobacter sp. K1 and Growth Conditions
2.2. DNA Extraction and Purification
2.3. The Whole-Genome Sequence Analysis
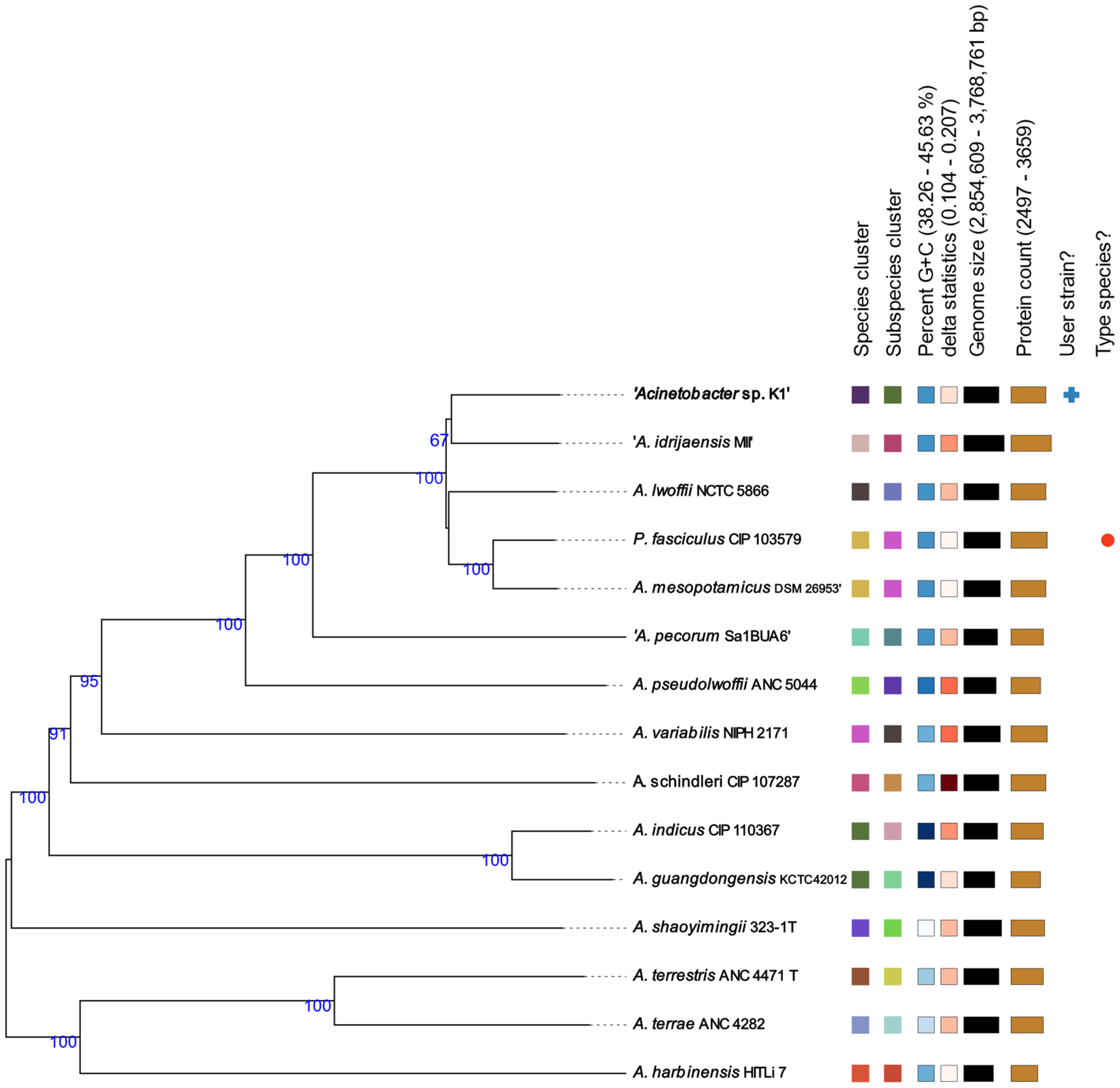
2.4. Taxogenomics
2.5. Identification of the Plasmid-Related Sequences and Localization of Determinants Important for the Adaptation Mechanisms of the K1 Isolate
3. Results
3.1. The Whole-Genome Sequence Analysis and Taxogenomics of the K1 Isolate
3.2. Identification of the Plasmid-Related Sequences in the K1 Genome
3.3. Genetic Determinants Important to Adaptation Mechanisms of the K1 Isolate
4. Discussion
4.1. Taxogenomic Placement of the K1 Isolate
4.2. Genetic Determinants Important for Adaptation of the K1 Strain to Extreme Environmental Conditions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Doughari, H.J.; Ndakidemi, P.A.; Human, I.S.; Benade, S. The Ecology, Biology and Pathogenesis of Acinetobacter spp.: An Overview. Microbes Environ. 2011, 26, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Veress, A.; Nagy, T.; Wilk, T.; Kömüves, J.; Olasz, F.; Kiss, J. Abundance of mobile genetic elements in an Acinetobacter lwoffii strain isolated from Transylvanian honey sample. Sci. Rep. 2020, 10, 2969. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.K.; Adams, F.G.; Brown, M.H. Diversity and function of capsular polysaccharide in Acinetobacter baumannii. Front. Microbiol. 2018, 9, 3301. [Google Scholar] [CrossRef]
- Robino, P.; Bert, E.; Tramuta, C.; Cerruti Sola, S.; Nebbia, P. Isolation of Acinetobacter lwoffii from a Lovebird (Agapornis roseicollis) with severe respiratory symptoms. Schweiz. Arch. Tierheilkd 2005, 147, 267–269. [Google Scholar] [CrossRef]
- Regalado, N.G.; Martin, G.; Antony, A.J. Acinetobacter lwoffii: Bacteremia associated with acute gastroenteritis. Travel Med. Infect. Dis. 2009, 7, 316–317. [Google Scholar] [CrossRef]
- Turton, J.F.; Shah, J.; Ozongwu, C.; Pike, R. Incidence of Acinetobacter species other than A. baumannii among clinical isolates of Acinetobacter: Evidence for emerging species. J. Clin. Microbiol. 2010, 48, 1445–1449. [Google Scholar] [CrossRef]
- Nakwan, N.; Wannaro, J.; Nakwan, N. Multidrug-resistant Acinetobacter lwoffii infection in neonatal intensive care units. Res. Rep. Neonatol. 2011, 1, 1–4. [Google Scholar] [CrossRef]
- Kulkarni, G.; Challa, J. The first Indian viridescent Acinetobacter lwoffii. Indian J. Med. Microbiol. 2021, 39, 130–132. [Google Scholar] [CrossRef]
- Kabala, C.; Galka, B.; Jezierski, P. Assessment and monitoring of soil and plant contamination with trace elements around Europe’s largest copper ore tailings impoundment. Sci. Total Environ. 2020, 738, 139918. [Google Scholar] [CrossRef]
- Uzarowicz, Ł.; Charzyński, P.; Greinert, A.; Hulisz, P.; Kabała, C.; Kusza, G.; Kwasowski, W.; Pędziwiatr, A. Studies of technogenic soils in Poland: Past, present, and future perspectives. Soil Sci. Ann. 2020, 71, 281–299. [Google Scholar] [CrossRef]
- Cimermanova, M.; Pristas, P.; Piknova, M. Biodiversity of Actinomycetes from Heavy Metal Contaminated Technosols. Microorganisms 2021, 9, 1635. [Google Scholar] [CrossRef] [PubMed]
- Abdel-El-Haleem, D. Minireview Acinetobacter: Environmental and biotechnological applications. Afr. J. Biotechnol. 2003, 2, 71–74. [Google Scholar] [CrossRef]
- Chojnacka, K. Biosorption and bioaccumulation—The prospects for practical applications. Environ. Int. 2010, 36, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Kopčáková, A.; Stramová, Z.; Kvasnová, S.; Godany, A.; Perhacova, Z.; Pristas, P. Need for database extension for reliable identification of bacteria from extreme environments using MALDI TOF mass spectrometry. Chem. Pap. 2014, 68, 1435–1442. [Google Scholar] [CrossRef]
- Rasulov, O.; Schwarz, M.; Horváth, A.; Zoirov, F.; Fayz, N. Analysis of soil contamination with heavy metals in (the three) highly contaminated industrial zones. SN Appl. Sci. 2020, 2, 2013. [Google Scholar] [CrossRef]
- Towner, K.J. Biology of Acinetobacter spp. In Acinetobacter, Microbiology, Epidemiology, Infections, Management; Bergogne-Bérézin, E., Joly-Guillou, M.-L., Towner, K.J., Eds.; CRC Press: Boca Raton, FL, USA, 1996; pp. 13–36. [Google Scholar]
- Šipošová, N.Š.; Liptáková, V.; Kvasnová, S.; Kosorínová, P.; Pristaš, P. Genetic diversity of Acinetobacter spp. adapted to heavy metal polluted environments. Nova Biotechnol. Chim. 2017, 16, 42–47. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Golosova, O.; Henderson, R.; Vaskin, Y.; Gabrielian, A.; Grekhov, G.; Nagarajan, V.; Oler, A.J.; Quiñones, M.; Hurt, D.; Fursov, M.; et al. Unipro UGENE NGS pipelines and components for variant calling, RNA-seq and ChIP-seq data analyses. PeerJ 2014, 2, e644. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.; Formsma, K.; Gerdes, S.; Glass, E.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic. Acids. Res. 2014, 42, 206–214. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-R, L.M.; Konstantinidis, K.T. Estimating coverage in metagenomic data sets and why it matters. ISME J. 2014, 8, 2349–2351. [Google Scholar] [CrossRef]
- Moore, R.M.; Harrison, A.O.; McAllister, S.M.; Polson, S.W.; Wommack, K.E. Iroki: Automatic customization and visualization of phylogenetic trees. PeerJ 2020, 8, e8584. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, 225–229. [Google Scholar] [CrossRef]
- Arredondo-Alonso, S.; Rogers, M.R.C.; Braat, J.C.; Verschuuren, T.D.; Top, J.; Corander, J.; Willems, R.J.L.; Schürch, A.C. mlplasmids: A user-friendly tool to predict plasmid- and chromosome-derived sequences for single species. Microb. Genom. 2018, 4, e000224. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Farris, J.S. Estimating phylogenetic trees from distance matrices. Am. Nat. 1972, 106, 645–667. Available online: https://www.jstor.org/stable/2459725 (accessed on 25 July 2023). [CrossRef]
- Holland, B.R.; Huber, K.T.; Dress, A.; Moulton, V. Delta plots: A tool for analyzing phylogenetic distance data. Mol. Biol. Evol. 2002, 19, 2051–2059. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, H.; Shin, D. The FeoA protein is necessary for the FeoB transporter to import ferrous iron. Biochem. Biophys. Res. Commun. 2012, 423, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.L.; Sudarsan, N.; Weinberg, Z.; Roth, A.; Stockbridge, R.B.; Breaker, R.R. Widespread Genetic Switches and Toxicity Resistance Proteins for Fluoride. Science 2012, 335, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Epstein, W. The KdpD sensor kinase of Escherichia coli responds to several distinct signals to turn on expression of the Kdp transport system. J. Bacteriol. 2016, 198, 212–220. [Google Scholar] [CrossRef]
- Levina, N.; Tötemeyer, S.; Stokes, N.R.; Louis, P.; Jones, M.A.; Booth, I.R. Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: Identification of genes required for MscS activity. EMBO J. 1999, 18, 1730–1737. [Google Scholar] [CrossRef]
- Khalid, M.; Hassani, S.; Abdollahi, M. Metal-induced oxidative stress: An evidence-based update of advantages and disadvantages. Curr. Opin. Toxicol. 2020, 20–21, 55–68. [Google Scholar] [CrossRef]
- Bouvet, P.J.M.; Grimont, P.A.D. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov. and Acinetobacter junii sp. nov. And emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Evol. Microbiol. 1986, 36, 228–240. [Google Scholar] [CrossRef]
- Campos-Guillén, J.; Pérez, J.C.; Medina, J.A.C.; Vera, C.M.; Rosas, L.M.S.; Gutiérrez, C.L.; Salinas, I.G.; Ramírez, M.R.H.; Alonso, G.S.; Hernández, A.C.; et al. Draft Genome Sequence of the Mercury-Resistant Bacterium Acinetobacter idrijaensis Strain MII, Isolated from a Mine-Impacted Area, Idrija, Slovenia. Genome Announc. 2014, 2, e01177-14. [Google Scholar] [CrossRef]
- Lapage, S.P.; Sneath, P.H.A.; Lessel, E.F.; Skerman, V.B.D.; Seelinger, H.P.R.; Clark, W.A. International Code of Nomenclature of Bacteria (1990 Revision); American Society for Microbiology: Washington, DC, USA, 1992; ISBN 1-55581-039-X. [Google Scholar]
- Nemec, A. Strain “Acinetobacter mesopotamicus” GC2 does not represent a novel species, but belongs to the species Acinetobacter lwoffii as revealed by whole-genome sequence-based analysis. Curr. Microbiol. 2021, 78, 369–370. [Google Scholar] [CrossRef]
- Acer, Ö.; Güven, K.; Poli, A.; Di Donato, P.; Leone, L.; Buono, L.; Güven, R.G.; Nicolaus, B.; Finore, I. Acinetobacter mesopotamicus sp. nov., Petroleum-degrading Bacterium, Isolated from Petroleum-Contaminated Soil in Diyarbakir, in the Southeast of Turkey. Curr. Microbiol. 2020, 77, 3192–3200. [Google Scholar] [CrossRef]
- Rajbanshi, A. Study on heavy metal resistant bacteria in Guheswori sewage treatment plant. Our Nat. 2008, 6, 52–57. [Google Scholar] [CrossRef]
- Raja, C.E.; Elvam, G.S.; Omine, K. Isolation, identification and characterization of heavy metal resistent bacteria from sewage. In International Joint Symposium on Geodisaster Prevention and Geoenvironment in Asia; JS: Fukuoka, Japan, 2009; pp. 205–211. [Google Scholar]
- El-Sayed, M.H. Multiple heavy metal and antibiotic resistance of Acinetobacter baumannii strain HAF—13 isolated from industrial effluents. Am. J. Microbiol. Res. 2016, 4, 26–36. [Google Scholar]
- Maslova, O.; Mindlin, S.; Beletsky, A.; Mardanov, A.; Petrova, M. Plasmids as Key Players in Acinetobacter Adaptation. Int. J. Mol. Sci. 2022, 23, 10893. [Google Scholar] [CrossRef]
- Moran, R.A.; Liu, H.; Doughty, E.L.; Hua, X.; Cummins, E.A.; Liveikis, T.; McNally, A.; Zhou, Z.; van Schaik, W.; Yu, Y. GR13-type plasmids in Acinetobacter potentiate the accumulation and horizontal transfer of diverse accessory genes. Microb. Genom. 2022, 8, mgen000840. [Google Scholar] [CrossRef]
- Brown, N.L.; Barrett, S.R.; Camakaris, J.; Lee, B.T.; Rouch, D.A. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol. Microbiol. 1995, 17, 1153–1166. [Google Scholar] [CrossRef]
- Williams, C.L.; Neu, H.M.; Alamneh, Y.A.; Reddinger, R.M.; Jacobs, A.C.; Singh, S.; Abu-Taleb, R.; Michel, S.L.J.; Zurawski, D.V.; Merrell, D.S. Characterization of Acinetobacter baumannii Copper Resistance Reveals a Role in Virulence. Front. Microbiol. 2020, 11, 16. [Google Scholar] [CrossRef]
- Mindlin, S.; Petrenko, A.; Petrova, M. Chromium resistance genetic element flanked by XerC/XerD recombination sites and its distribution in environmental and clinical Acinetobacter strains. FEMS Microbiol. Lett. 2018, 365, fny047. [Google Scholar] [CrossRef]
- Viti, C.; Marchi, E.; Decorosi, F.; Giovannetti, L. Molecular mechanisms of Cr(VI) resistance in bacteria and fungi. FEMS Microbiol. Rev. 2013, 38, 633–659. [Google Scholar] [CrossRef]
- Hussain, S.; Khan, M.; Sheikh, T.M.M.; Mumtaz, M.Z.; Chohan, T.A.; Shamim, S.; Liu, Y. Zinc Essentiality, Toxicity, and Its Bacterial Bioremediation: A Comprehensive Insight. Front. Microbiol. 2022, 13, 900740. [Google Scholar] [CrossRef]
- Gutiérrez-Barranquero, J.A.; de Vicente, A.; Carrión, V.J.; Sundin, G.W.; Cazorla, F.M. Recruitment and Rearrangement of Three Different Genetic Determinants into a Conjugative Plasmid Increase Copper Resistance in Pseudomonas syringae. Appl. Environ. Microbiol. 2013, 79, 1028–1033. [Google Scholar] [CrossRef]
- Ou, H.-Y.; Kuang, S.N.; He, X.; Molgora, B.M.; Ewing, P.J.; Deng, Z.; Osby, M.; Chen, W.; Xu, H.H. Complete genome sequence of hypervirulent and outbreak-associated Acinetobacter baumannii strain LAC-4: Epidemiology, resistance genetic determinants and potential virulence factors. Sci. Rep. 2015, 5, 8643. [Google Scholar] [CrossRef] [PubMed]
- Mindlin, S.; Petrenko, A.; Kurakov, A.; Beletsky, A.; Mardanov, A.; Petrova, M. Resistance of permafrost and modern Acinetobacter lwoffii strains to heavy metals and arsenic revealed by genome analysis. BioMed Res. Int. 2016, 2016, 3970831. [Google Scholar] [CrossRef] [PubMed]
- Suhadolnik, M.L.S.; Salgado, A.P.C.; Scholte, L.L.S.; Bleicher, L.; Costa, P.S.; Reis, M.P.; Dias, M.F.; Ávila, M.P.; Barbosa, F.A.R.; Chartone-Souza, E.; et al. Novel arsenic-transforming bacteria and the diversity of their arsenic-related genes and enzymes arising from arsenic-polluted freshwater sediment. Front. Microbiol. 2017, 7, 11231. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Zhu, J.; Rensing, C.; Liu, Y.; Gao, S.; Chen, W.; Huang, Q.; Liu, Y.-R. Recent advances in exploring the heavy metal(loid) resistant microbiome. Comput. Struct. Biotechnol. 2021, 19, 94–109. [Google Scholar] [CrossRef] [PubMed]
- Urano, H.; Yoshida, M.; Ogawa, A.; Yamamoto, K.; Ishihama, A.; Ogasawara, H. Cross-regulation between two common ancestral response regulators, HprR and CusR, in Escherichia coli. Microbiology 2017, 163, 243–252. [Google Scholar] [CrossRef]
- Urano, H.; Yamamoto, K.; Ogasawara, H.; Ishihama, A.; Umezawa, Y. Cooperative regulation of the common target genes between H2O2-sensing YedVW and Cu2+-sensing CusSR in Escherichia coli. Microbiology 2015, 161, 729–738. [Google Scholar] [CrossRef]
- Gudipaty, S.A.; Larsen, A.S.; Rensing, C.; McEvoy, M.M. Regulation of Cu(I)/Ag(I) efflux genes in Escherichia coli by the sensor kinase CusS. FEMS Microbiol. Lett. 2012, 130, 30–37. [Google Scholar] [CrossRef]
- Pristas, P.; Kvasnova, S.; Judova, J.; Perhacova, Z.; Vidova, B.; Sramkova, Z.; Godany, A. Non-ferrous metal industry waste disposal sites as a source of poly-extremotolerant bacteria. Nova Biotechnol. Chim. 2015, 14, 62–68. [Google Scholar] [CrossRef][Green Version]
- Yavankar, S.P.; Pardesi, K.R.; Chopade, B.A. Species distribution and physiological characterization of Acinetobacter genospecies from healthy human skin of tribal population in India. Indian J. Med. Microbiol. 2007, 25, 336–345. [Google Scholar] [CrossRef]
- Li, T.; Luo, D.; Ning, N.; Liu, X.; Chen, F.; Zhang, L.; Bao, C.; Li, Z.; Li, D.; Gu, H.; et al. Acinetobacter baumannii adaptation to the host pH microenvironment is mediated by allelic variation in a single residue of BauA protein. PNAS Nexus 2023, 2, pgad079. [Google Scholar] [CrossRef]
- Jung, J.; Park, W. Acinetobacter species as model microorganisms in environmental microbiology: Current state and perspectives. Appl. Microbiol. Biotechnol. 2015, 99, 2533–2548. [Google Scholar] [CrossRef]
- Padan, E.; Bibi, E.; Ito, M.; Krulwich, T.A. Alkaline pH homeostasis in bacteria: New insights. Biochim. Biophys. Acta Bioenerg. 2005, 1717, 67–88. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Ma, Z.; Gao, J.; Zhao, J.; Wei, L.; Liu, J.; Xu, N. Recent advances of pH homeostasis mechanisms in Corynebacterium glutamicum. World J. Microbiol. Biotechnol. 2019, 35, 192. [Google Scholar] [CrossRef] [PubMed]
- Krulwich, T.A.; Sachs, G.; Padan, E. Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 2011, 9, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Gunde-Cimerman, N.; Plemenitaš, A.; Oren, A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 2018, 42, 353–375. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, H.-M.; Yuan, J.-L.; Xu, L.; Sun, J.-Q. A comprehensive genomic analysis provides insights on the high environmental adaptability of Acinetobacter strains. Front. Microbiol. 2023, 14, 1177951. [Google Scholar] [CrossRef]
- Moe, P.C.; Blount, P.; Kung, C. Functional and structural conservation in the mechanosensitive channel MscL implicates elements crucial for mechanosensation. Mol. Microbiol. 1998, 28, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Pivetti, C.D.; Yen, M.-R.; Miller, S.; Busch, W.; Tseng, Y.-H.; Booth, I.R.; Saier, M.H. Two families of mechanosensitive channel proteins. Microbiol. Mol. Biol. Rev. 2003, 67, 66–85. [Google Scholar] [CrossRef]
- Naismith, J.H.; Booth, I.R. Bacterial mechanosensitive channels—MscS: Evolution’s solution to creating sensitivity in function. Annu. Rev. Biophys. 2012, 41, 157–177. [Google Scholar] [CrossRef]
- Kempf, B.; Bremer, E. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 1998, 170, 319–330. [Google Scholar] [CrossRef]
- Fasnacht, M.; Polacek, N. Oxidative stress in bacteria and the central dogma of molecular biology. Front. Mol. Biosci. 2021, 8, 671037. [Google Scholar] [CrossRef]
- Steimbrüch, B.A.; Sartorio, M.G.; Cortez, N.; Albanesi, D.; Lisa, M.-N.; Repizo, G.D. The distinctive roles played by the superoxide dismutases of the extremophile Acinetobacter sp. Ver3. Sci. Rep. 2022, 12, 4321. [Google Scholar] [CrossRef] [PubMed]
- Fridovich, I. Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 1995, 64, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Cussiol, J.R.R.; Alegria, T.G.P.; Szweda, L.I.; Netto, L.E.S. Ohr (organic hydroperoxide resistance protein) possesses a previously undescribed activity, lipoyl-dependent peroxidase. J. Biol. Chem. 2010, 285, 21943–21950. [Google Scholar] [CrossRef] [PubMed]
- Masip, L.; Veeravalli, K.; Georgiou, G. The many faces of glutathione in bacteria. Antioxid. Redox Signal. 2006, 8, 753–762. [Google Scholar] [CrossRef]
- Hu, Y.; Zheng, J.; Zhang, J. Natural transformation in Acinetobacter baumannii W068: A genetic analysis reveals the involvements of the CRP, XcpV, XcpW, TsaP, and TonB2. Front. Microbiol. 2022, 12, 738034. [Google Scholar] [CrossRef]
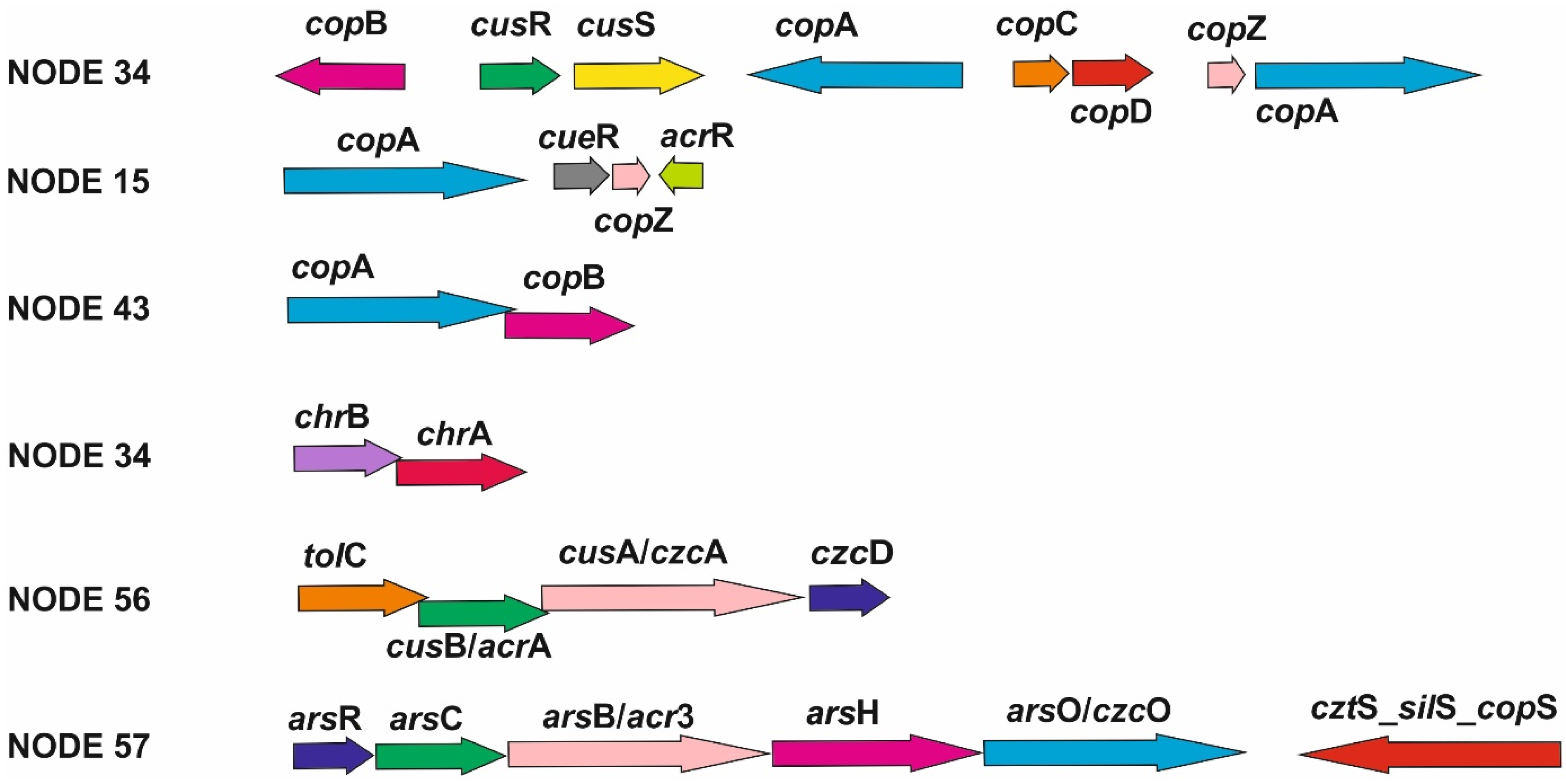

| Contig | Size (bp) | GenBank Accession No. | Replication | HGT | Resistance |
|---|---|---|---|---|---|
| NODE 4-2 | 34,994 | NZ_JALGQY010000005 | - | IS5_3 transposase superfamily | AcrA, AcrB; RND_mfp superfamily |
| NODE 13 | 78,976 | NZ_JALGQY010000014 | - | - | CzcD |
| NODE 17 | 62,178 | NZ_JALGQY010000018 | - | IS3, IS5, IS21, IS481, ISNCY transposase superfamilies; XerC integrase | TauE |
| NODE 29 | 44,217 | NZ_JALGQY010000029 | - | LGT_TIGR03299 phage/plasmid-like protein; COG5377 phage protein superfamily; AlpA superfamily; Inovirus_Gp2 superfamily; IS66 transposase family; XerC integrase | Fur; SmtA |
| NODE 38 | 27,781 | NZ_JALGQY010000038.1 | rep_pAB02_ORF2 | IS3, IS481, IS6 transposase families; XerC integrase | RcnR-FrmR-like_DUF156; MefA protein family; AcrR; GST_N_GTT1/GST_C superfamily |
| NODE 39 | 26,463 | NZ_JALGQY010000039 | - | IS5_2 transposase family | RND_mfp superfamily OtsA, OtsB |
| NODE 43 | 18,602 | NZ_JALGQY010000042.1 | RepM | IS481 and IS3 transposase families Relaxase; MobC | CopA, CopB |
| NODE 48 | 14,852 | NZ_JALGQY010000047 | - | IS3 transposase family | ArsR/SmtB family; TauE |
| NODE 55 | 8994 | NZ_JALGQY010000052.1 | Rep_3; RepM; rep_pAB02_ORF2; | MobA_MobL; TraA_Ti | TerC superfamily; SUL1 superfamily |
| NODE 57 | 8544 | NZ_JALGQY010000054 | - | IS5_3 transposase family | ArsH, ArsC, ArsO, ArsR, Acr3; cztS_silS_copS Pfam00520 |
| NODE 66 | 6322 | NZ_JALGQY010000055.1 | Rep_3; RepM; rep_pAB02_ORF2; | - | RamA |
| NODE 130 | 3934 | NZ_JALGQY010000063 | - | IS6, IS240 transposase family | CadR-PbrR Kup |
| NODE 211 | 3202 | NZ_JALGQY010000065 | - | Phage_antiter_Q | Beta-lactamase; Phenol_MetA_deg |
| NODE 375 | 2618 | NZ_JALGQY010000069 | - | Serine recombinase PinE recombinase | ABC_trans_N |
| NODE 1680 | 1426 | NZ_JALGQY010000076 | - | - | CrcB |
| NODE 1947 | 1329 | NZ_JALGQY010000077 | - | - | CzcD; MerR; CadR-PbrR; ZntA; ZitB |
| NODE 2568 | 1144 | NZ_JALGQY010000083 | - | MobA_MobL; TraA_Ti | TelA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrová, N.; Kisková, J.; Kolesárová, M.; Pristaš, P. Genetic Basis of Acinetobacter sp. K1 Adaptation Mechanisms to Extreme Environmental Conditions. Life 2023, 13, 1728. https://doi.org/10.3390/life13081728
Petrová N, Kisková J, Kolesárová M, Pristaš P. Genetic Basis of Acinetobacter sp. K1 Adaptation Mechanisms to Extreme Environmental Conditions. Life. 2023; 13(8):1728. https://doi.org/10.3390/life13081728
Chicago/Turabian StylePetrová, Nikola, Jana Kisková, Mariana Kolesárová, and Peter Pristaš. 2023. "Genetic Basis of Acinetobacter sp. K1 Adaptation Mechanisms to Extreme Environmental Conditions" Life 13, no. 8: 1728. https://doi.org/10.3390/life13081728
APA StylePetrová, N., Kisková, J., Kolesárová, M., & Pristaš, P. (2023). Genetic Basis of Acinetobacter sp. K1 Adaptation Mechanisms to Extreme Environmental Conditions. Life, 13(8), 1728. https://doi.org/10.3390/life13081728









