1. Introduction
According to recent studies, the DNA G-quadruplexes (G4s), one of the most widely investigated non B-form DNA structures, are considered to be an integral part of complex regulatory systems in both normal and pathological cells. G4s are four-stranded nucleic acid secondary structures formed from guanine-rich sequences by the stacking of two or more G-tetrads, planar arrangements of four guanine residues stabilized by Hoogsteen hydrogen bonding and coordination to a central cation. They can adopt many different topologies depending on how the primary DNA structure folds into quadruplex arrangements. In recent years, new tools have been developed to introduce some improvements in the prediction of the G4-forming DNA sequences (G4 motifs) throughout the genomes [
1]. They radically differ from the standard and most commonly used Quadparser algorithm G
nL
1-7G
nL
1-7G
nL
1-7G
n, where G
n corresponds to G-tracts with n consecutive guanines (n ≥ 3), and L is a semi-arbitrary 1-7-nucleotide sequence corresponding to quadruplex loops; this tool is based on a simple folding rule representing four runs of guanosines separated by relatively short loops. Bioinformatics analysis has shown that G4 motifs are widely represented in the genomes of all organisms, although they are predominantly enriched in eukaryotic genomes. Thus, 95 families of G4 motifs have been identified in the human genome [
2]. They often cluster at key regulatory elements and provide a significant potential repertoire for the formation of short-lived G4 structures that influence essential biological processes such as DNA replication, telomere maintenance, gene expression, recombination, and DNA repair, as well as genetic and epigenetic integrity [
3]. Thus, G4s may contribute to replication-dependent genome instability due to replication fork arrest [
4,
5,
6] and increase the frequency of mutations, deletions, and recombination events [
7,
8]. G4s represent a strong block for replication and transcription, not only by themselves, but also due to oxidative lesions, which are especially susceptible to DNA regions containing a large number of guanine bases [
9,
10]. It is well known that DNA oxidation occurs mainly at guanines, since they have the lowest redox potential among all native nucleobases [
11].
Given that G4 structures alter DNA accessibility to damaging agents and affect the efficiency and accuracy of DNA repair pathways [
12], they are subjected to natural selection and may be the driving force behind genome evolution [
13]. It can be assumed that it is these noncanonical structures that switch the activity of genes that ensure the development of
Vertebratas during the transition from gill to pulmonary respiration. At the same time, evolutionary conservation of G4 motifs located in certain gene regions should be observed if related G4 structures are biologically relevant.
Here, we used bioinformatics tools to analyze the presence and variability of G4-forming sequences in the promoter regions of the
TERT gene for 158 mammalian species. The human TERT protein (hTERT) is the catalytic subunit of telomerase, an enzyme complex that is primarily responsible for maintaining telomere length and replication potency of stem cells [
14]; its activity is thought to be vital for the immortalization of cancer cells [
15]. The core region of the
hTERT promoter (approximately −180 ÷ +1 from the transcription start site (TSS)) contains a 68 nt G-rich site with twelve consecutive G-tracts, which have been shown to fold into three stacked parallel-stranded G4s [
16]. Given the key role of the TERT enzyme and the obvious importance of telomere biology in the life of multicellular animals, it can be assumed that the
TERT genes are the subject of evolution, reflecting the adaptation of organisms to various living conditions.
The TERT protein contains three canonical domains: (1) a telomerase essential N-terminal domain, (2) a telomerase RNA-binding domain, and (3) a reverse transcriptase domain [
17]. Such a structural organization makes it possible to search for putative genes encoding TERT, to determine their promoter regions, and to perform bioinformatics analysis using publicly available genomic data sets and specially adapted software for the joint alignment of stable blocks in the promoter regions of
TERT genes across different orders of mammals.
The main goal of our study was to test whether the G4 motifs of the TERT promoters in the mammalian class are evolutionarily conserved and therefore biologically significant. We searched for G4 motifs separately in the coding and noncoding strands of the TERT promoter regions for mammalian species belonging to different orders. This was carried out to determine whether potential G4 structures are involved in the regulation of gene expression at the transcriptional level (if G4 motifs are located in the non-coding strand) or whether they are an element of possible translational regulation if G4 motifs are located in the coding strand and, subsequently, in the RNA transcript. We also analyzed the type and frequency of nucleotide substitutions causing the genome mutations in conserved regions of TERT G4 motifs.
2. Results
2.1. Selection of Mammalian TERT Promoters
Initially, we downloaded data on
TERT genes from 158 mammalian species available in NCBI Orthologs at
https://www.ncbi.nlm.nih.gov/ (accessed on 13 August 2022) For all the
TERT genes studied, we found a putative transcription start site (TSS) and downloaded the 1000 bp upstream region. We call it the “promoter region” because it contains the core
TERT promoter, a proximal promoter part containing recognition sites for many transcription factors and a more distant hypermethylation oncological region implicated in cancer progression. After a revision aimed at removing promoter regions with unreliable sequencing results and erroneously determined coordinates, 141
TERT genes and their promoter regions remained.
2.2. Search Tools for G4 Motifs
To detect G4 motifs in
TERT promoter regions, we tested three tools with different algorithms: G4Hunter, pqsfinder, and QGRS mapper. Our choice of tool was based on the ability to compare matches to identify conserved G4 motifs. Pqsfinder [
18] was developed to search for G4 motifs that allow for imperfect quadruplex formation with potential single bulges and mismatches in the quadruplex core and with a wider range of G-tract lengths and loop sizes. G4Hunter [
19] takes into account G-richness and G-skewness (G/C) asymmetry between complementary DNA strands of a given sequence and uses simple encoding and sliding window statistics that can account for various types of quadruplex defects. As a result, it provides a score (propensity to form G4 structure). It is not clear how to compare such regions in different promoters. QGRS Mapper [
20] provides a pattern search: G
3+ L
1-30 G
3+ L
1-30 G
3+ L
1-30 G
3+.
Since the search results of three different G4 predictors varied significantly, we chose the QGRS mapper as the main tool. Using the QGRS mapper, we can lose a number of G4 motifs that possibly form defective G4 structures. On the other hand, we have the ability to easily compare matches in G-tracts and to detect substitutions within loops of conserved G4 motifs. In addition, the pqsfinder and G4Hunter programs were rejected because they identify G4 motifs with dinucleotide G-tracts that do not provide the formation of a thermodynamically stable G4 structures. Additional constraints adopted in our study: (i) the maximal total length of the G4 motif does not exceed 45 nucleotide units; (ii) each G-tract must fit into the sequence window defined by the starting position of the first G-tract; (iii) each subsequent G-tract must lie behind the 3′-end of the previous one (without overlap). All of these options can be freely configured using the QGRS mapper package.
2.3. Distribution of G4 Motifs in TERT Promoters across the Mammalian Class
The search for G4 motifs was carried out in the
TERT promoter regions of 141 mammalian species belonging to 20 orders, 5 of which are
Primates (primates),
Artiodactyla (artiodactyls),
Carnivora (predators),
Chiroptera (bats), and
Rodentia (rodents) and which contain more than 10 species. All other orders were considered together. A list of mammalian species used in this study is shown in
Table S1.
Figure S2 demonstrates taxonomictrees for five orders of mammals.
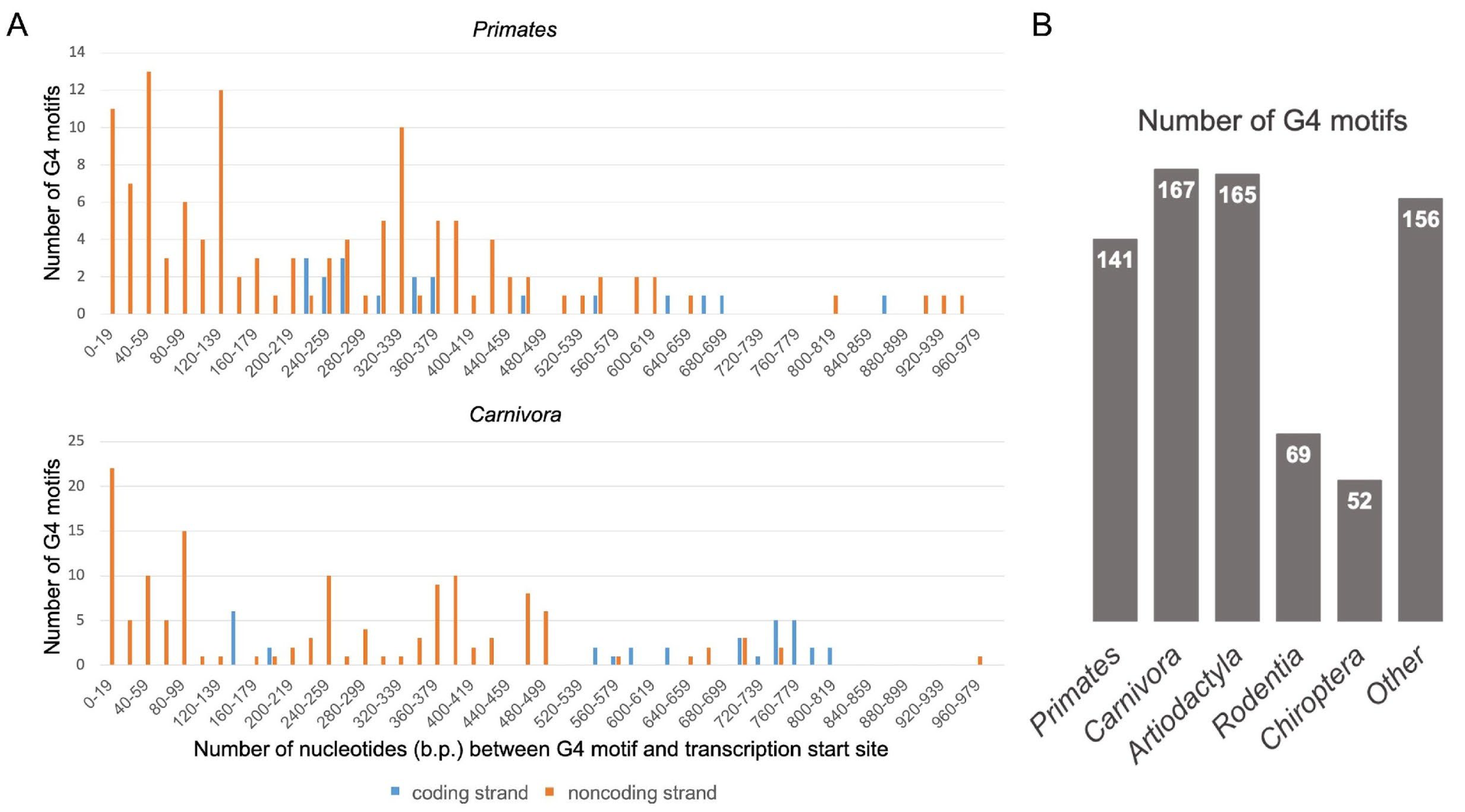
At the first stage, we compared histograms of distances (the number of nucleotide units) between a certain G4 motif and TSS on the coding and noncoding strands of the 1000 bp TERT promoter regions for all studied species of mammals. The NCBI gene database used in the work indicates which of the TERT gene strands contains the coding sequence and which is complementary to it (i.e., is noncoding or template).
Figure 1A shows examples of histograms obtained for
Primates and
Carnivora (similar histograms for other orders of mammals are shown in
Figure S1); the number of found G4 motifs for all studied orders is listed in
Figure 1B. Numerical data on the localization of G4 motifs in the
TERT promoter zones 0–180, 181–500 and 501–1000 bp from TSS are presented in
Table 1.
Only 6 out of 141 mammalian species tested do not contain G4 motifs. The absence of G4 motifs is observed in four rodent and one bat species that are cancer resistant. Diversity in the G4 motif distribution in different orders of mammals is shown in
Table 1. In general, the total number of G4 motifs on the noncoding strand of
TERT promoters is significantly higher than on the coding strand, this effect being more pronounced for the proximal parts of the promoter than for the distal one. In addition, just the core and proximal parts of the
TERT promoters (~0–400 bp) are enriched in G4 motifs compared to the distal part.
We also estimated the average number of all G4 motifs per species for orders of the following parameters were determined in the block: mammals (
Figure S3) and revealed that it strongly depends on the order type.
2.4. The Search for Conserved G4 Motifs in the TERT Promoter Regions of Five Mammalian Orders
Although the TERT protein is widely ubiquitous across mammals, not much is known about whether the G4 motifs of TERT promoters are conserved in the mammalian kingdom through their evolutionary history.
Note that a multiple alignment of any set of G4 motifs with the accepted restrictions has at least twelve identical columns corresponding to four G-tracts with three consecutive guanosine residues. To avoid false positive predictions of conserved G4 motifs, we used a step-by-step procedure. First, we found blocks of reliable alignments in the promoters of each of the five orders of mammals using the Nucleotide PanGenome (NPG) explorer program (
https://github.com/npge/npge, accessed on 13 August 2022). In this work, NPG-explorer input promoters instead of genomes and G4 motif coordinates instead of gene coordinates. The output was a set of alignments consisting of fragments of several (not necessarily all) promoters with more than 80% identical columns and at least 100 columns in length. Using this tool, we performed a cross-species comparison of all found G4 motifs, containing no more than 45 bp, in both coding and noncoding strands of
TERT promoter regions. The numbers of aligned blocks and
TERT promoter fragments in each block are shown in
Table S2. Clusters of the G4s motifs with intersecting coordinates within the block alignment were found in the blocks. The procedure was carried out separately for the species of each mammalian order. Groups of conserved G4 motifs were identified after visual examination of all clusters.
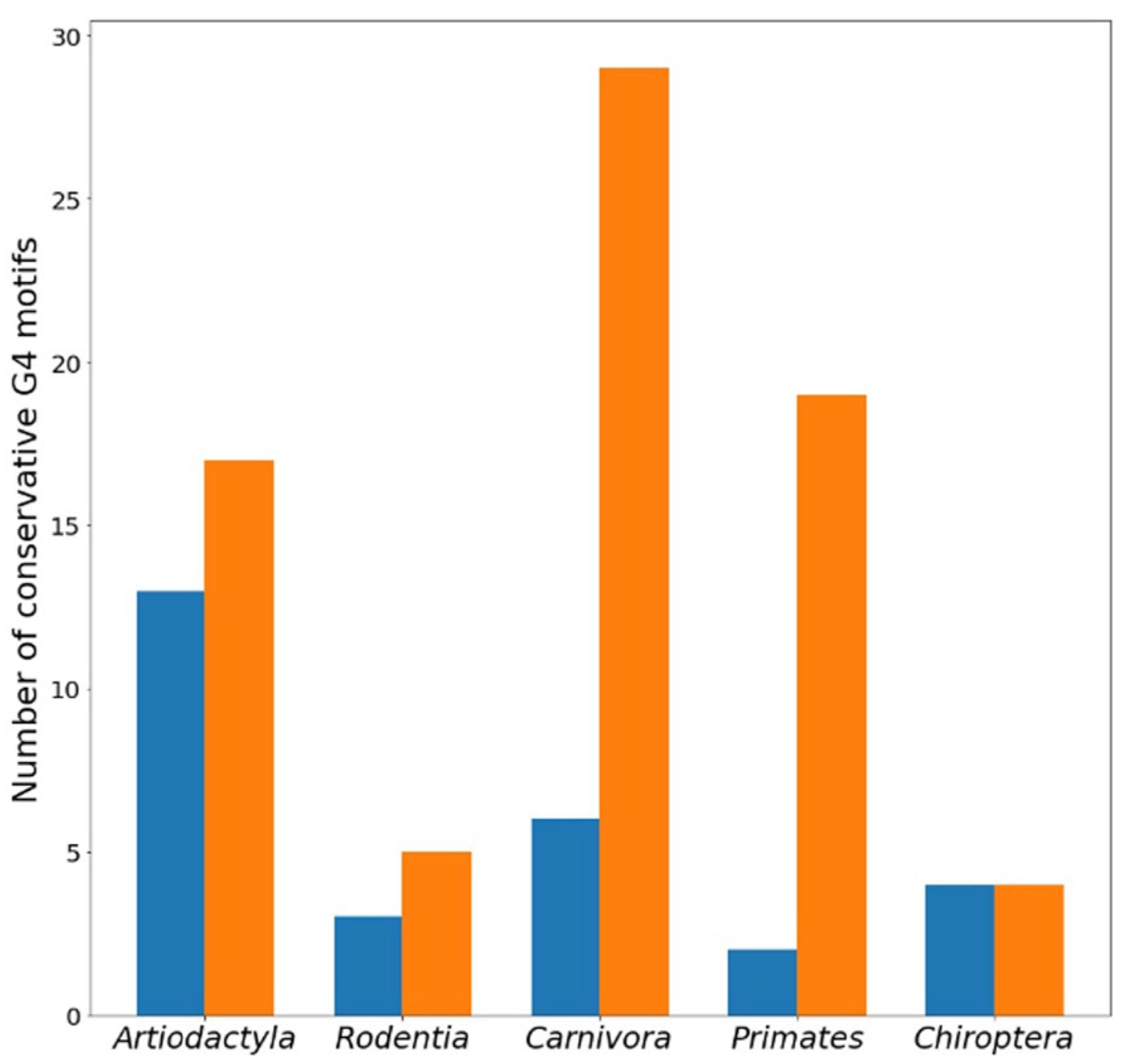
The numbers of conserved G4 motifs located in different
TERT promoter zones in coding and noncoding strands for five mammalian orders are presented in
Table 2. Comparison of the data in
Table 1 and
Table 2 shows that the proportion of conserved G4 motifs in relation to all found ones across the compared set of mammalian species depends on the type of order, varying from 18/22% (coding/noncoding strands) for
Carnivora to 13/11% for
Rodentia. As can be seen, conserved G4 motif loci lie primarily on the noncoding strand of
TERT promoter regions (
Figure 2), although strand preference is mammalian order specific and correlates with strand-dependent arrangement of G4 motifs (
Table 1). Most (79 out of 102) conserved G4 motifs are located in the core and proximal promoter zones.
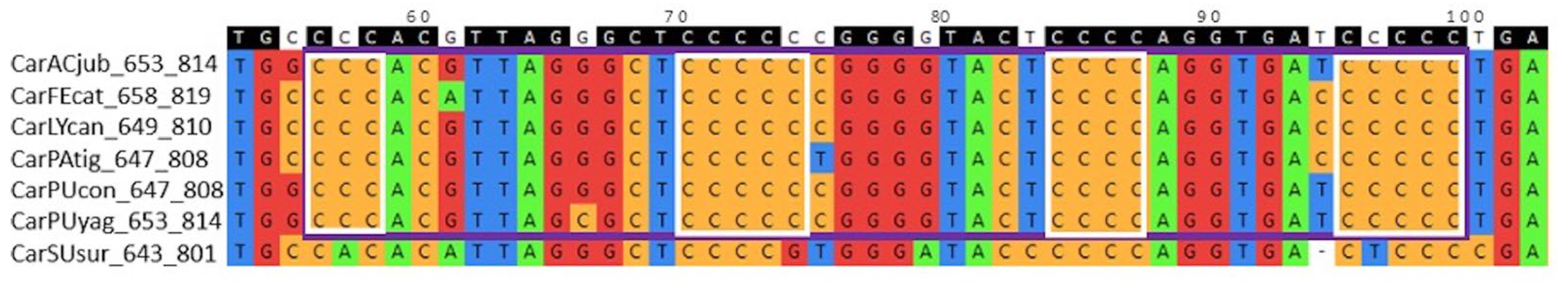
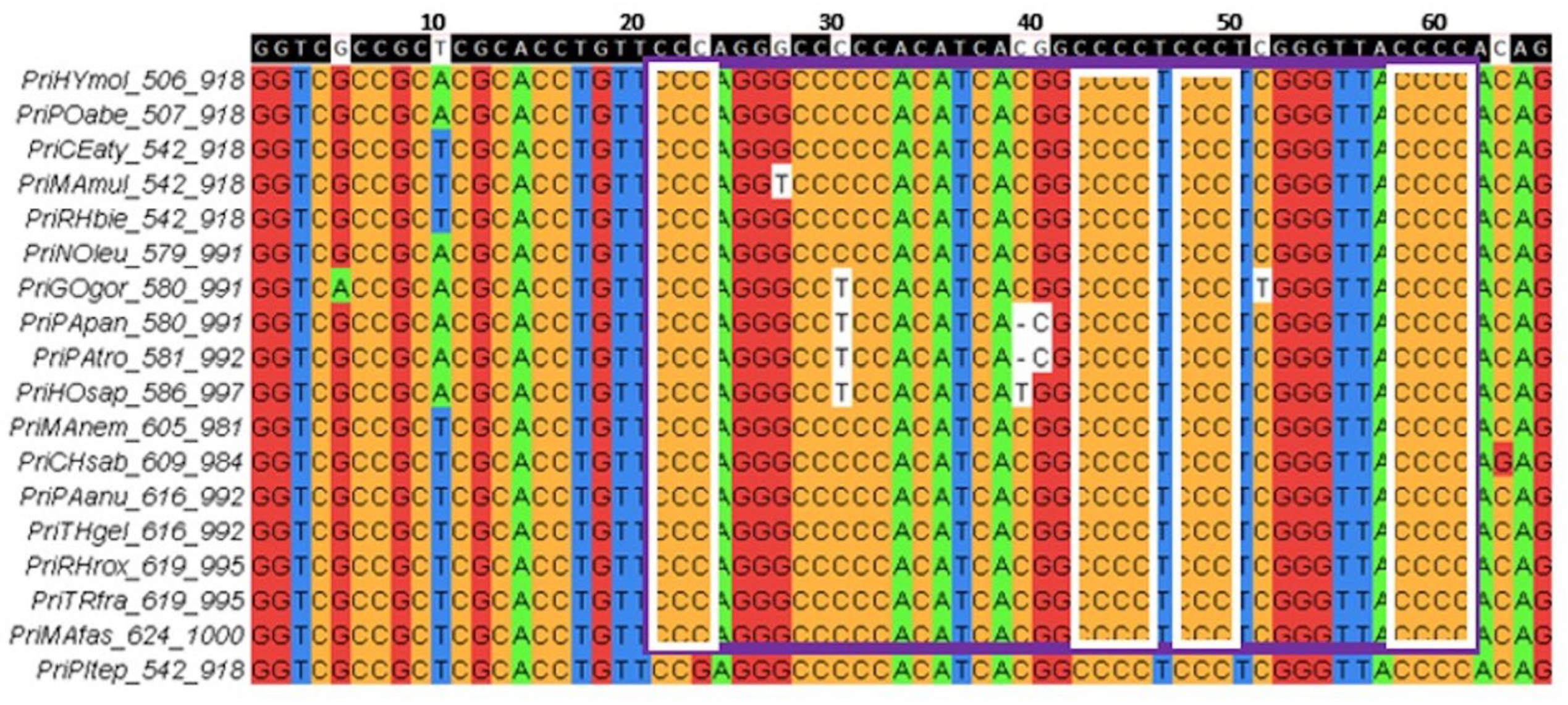
An example of a group of conserved G4 motifs in the block alignment of
TERT promoters for seven species of the order
Carnivora is shown in
Figure 3.
We then compared the coordinates of the conserved G4 motifs of the
TERT promoter regions with the binding sites of several known transcription factors: Sp1, Ets, c-Myc, etc. It was shown that G4 motifs from 23 primate species overlap with the first and second binding sites of Sp1 and with Etsrecognition site immediately before the TSS (
Figure S4). The recognition sites of both transcription factors are conserved in the TERT promoters of 23 primate species studied. The Ets site islocated in the loop of conserved G4 motifs. We intend to continue studying this phenomenon in other orders of mammals.
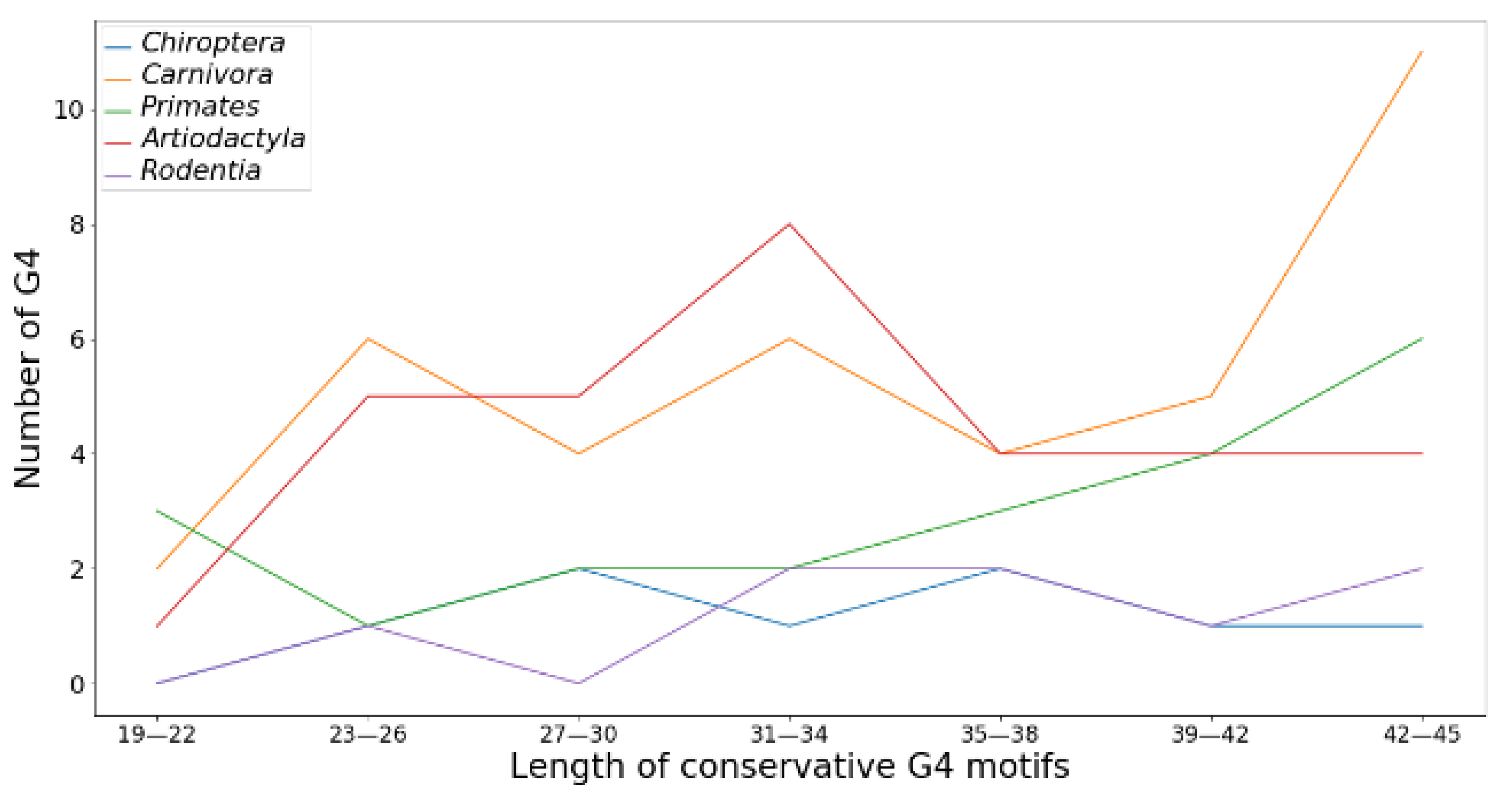
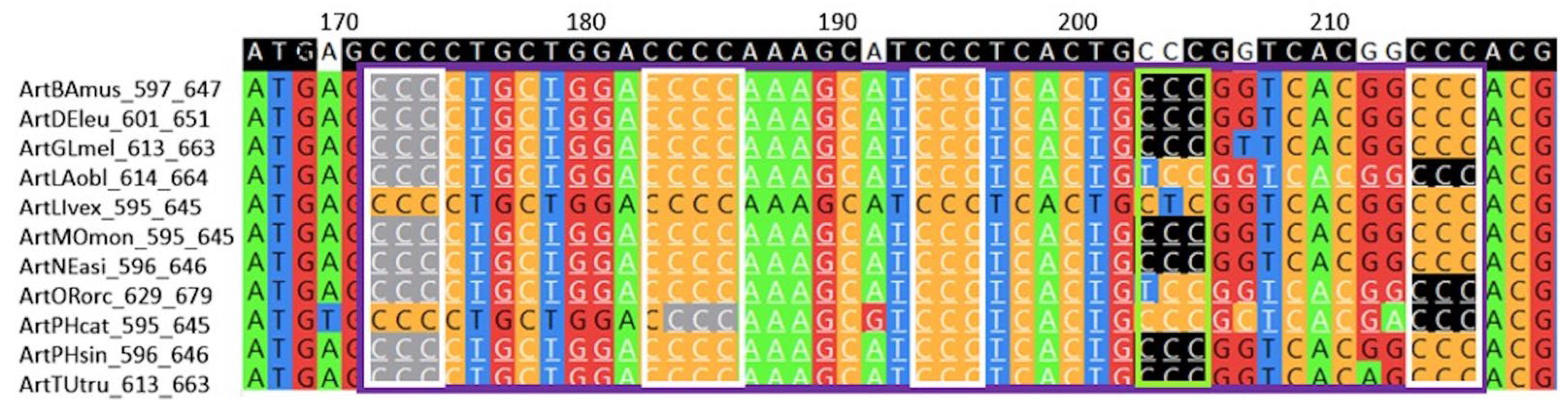
The length of the conserved G4 depends on the type of mammalian order (
Figure 4). As can be seen from the obtained data, species of the order
Carnivora have an increased number of conserved G4 motifs in
TERT promoters 40 or more nucleotides in length, while 31–34 nt conserved G4 motifs are most represented in
Artiodactyla. In mammals of the order
Rodentia and
Chiroptera, the distribution of conserved G4 motifs along the length is more or less uniform, without pronounced maxima and minima.
2.5. Single-Nucleotide Variants in Groups of Conserved G4 Motifs in the TERT Promoter Regions of Five Mammalian Orders
Since G4 DNA contributes to regulating multiple essential cellular processes, its sequence motifs should evolve under natural selection. For estimating the diversity in G4 motifs of the TERT promoters from different orders of mammals, we also used a block alignment approach based on the NPG-explorer program. Nucleotide substitutions can be reliably identified only within groups of conservative G4 motifs. Comparisons of a unique predicted G4 motif within a block alignment with homologous fragments of a different species within a block that do not contain the predicted G4 motifs or with predicted G4 motifs that only partially overlap with the G4 motif in question are less reliable.
Point mutations throughout the coding strand of G4 motifs located in
TERT promoters for 18 species of the order
Primates are shown in
Figure 5 as an example.
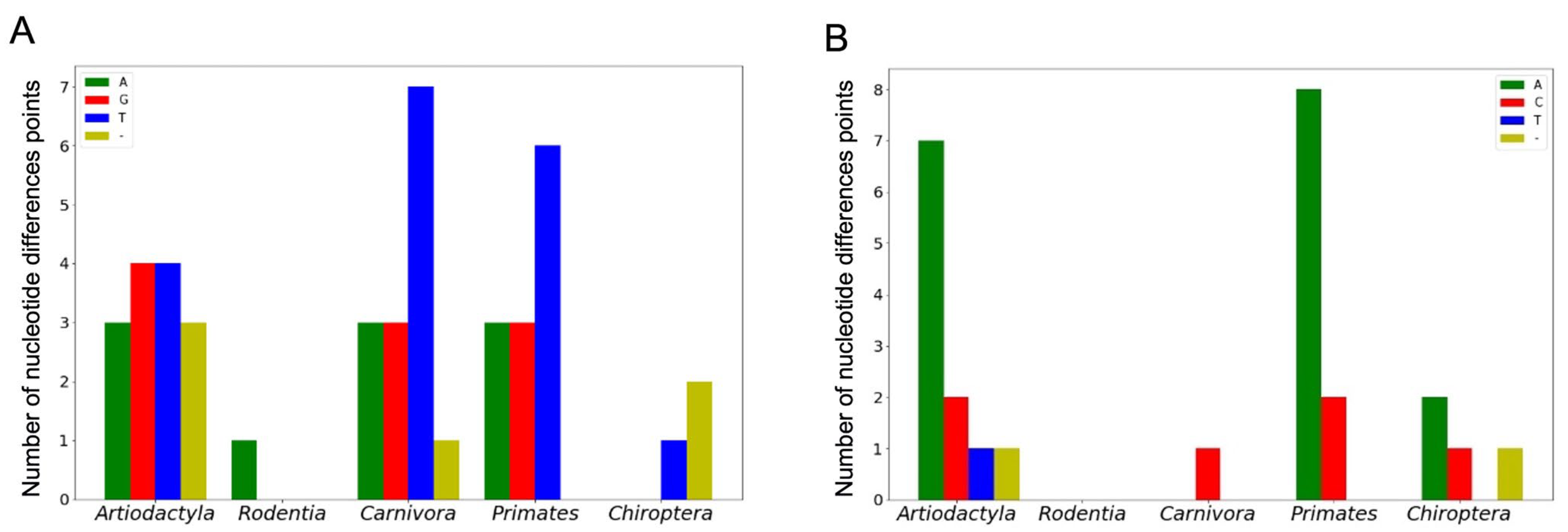
We separately considered nucleotide substitutions in G4 structure-disruptive positions (occurring in G-tracts) and positions corresponding to G4 loop-forming sequences. Since genome nucleotide substitutions leading to point mutations are usually indicated on the coding strand, the structurally disruptive positions correspond to C-tracts if the G4 motif lies on the noncoding strand and to G-tracts if the G4-forming sequence is located in the coding one.
Figure 6A,B shows the number of nucleotide difference points in C- and G-tracts, respectively, depending on the type of mammalian order.
As can be seen, the number and variability of nucleotide difference positions in both C- and G-tracts are taxon-dependent, and C↔T/G↔A substitutions are the most frequent, although a wider set of single-nucleotide variants occurred when G4 motifs are located on the noncoding strand (
Figure 6A).
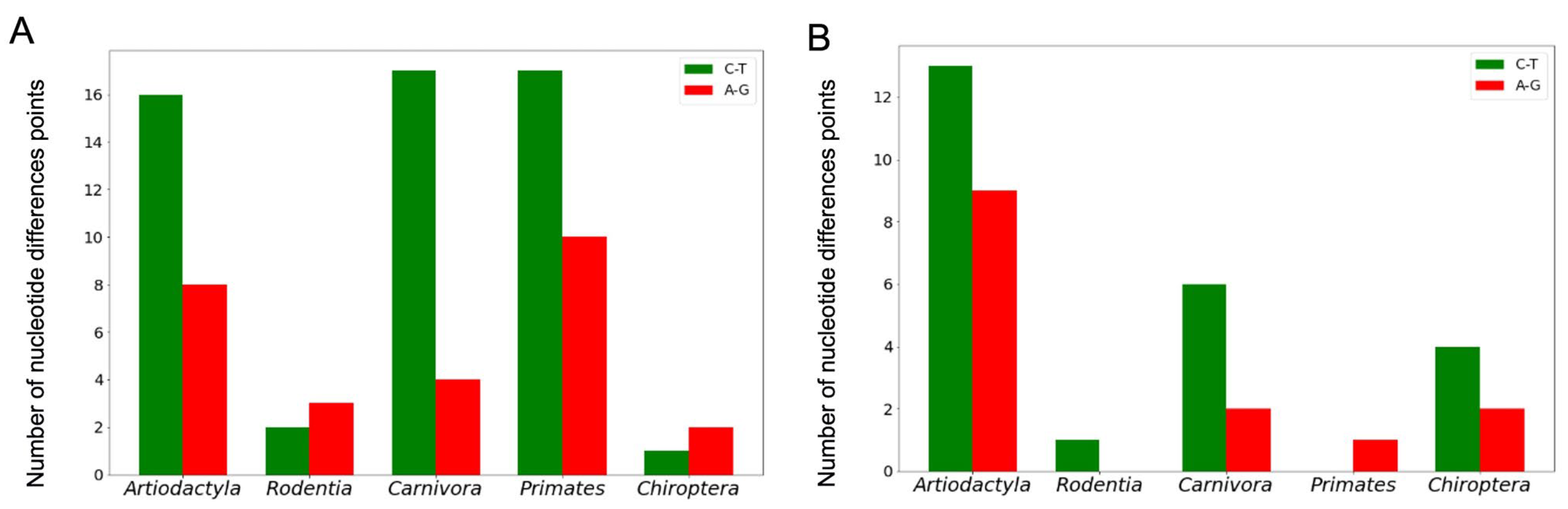
The number of nucleotide difference points and the type of nucleotide substitutions throughout quadruplex loop-forming sequences in conserved G4 motifs were also evaluated in five orders of mammals.
Table S3 summarizes these data for the coding and noncoding strands of the
TERT promoter regions. Based on these data, it can be concluded that the A↔G and C↔T substitutions are the most frequent. Their distribution in the loop-forming sequences of the conserved
TERT promoter G4 motifs among different mammalian orders is shown in
Figure 7.
Next, for each group of conserved G4 motifs, we determined the density of nucleotide substitutions as the ratio of substitution number in G4 loop-forming sequences to the total loop length and compared it with the density of nucleotide substitutions of the control sequence. As a control, we used the
TERT promoter region equal in length to the total length of the loops either before the initial G4 motif or after the final G4 motif, which did not intersect with any G4 motifs either in the same or in any other blocks. Comparison of the calculated densities of nucleotide substitution for five orders of mammals revealed a higher density of single-nucleotide variants in G4 motifs compared to regions lacking these motifs in each of the studied mammalian order (
Table 3).
To evaluate the statistical significance of this statement, we used paired Wilcoxon signed rank test. As a rule, the established difference between the compared parameters is statistically significant when the
p value ≤ 0.05 [
21]. According to the data presented in
Table 3, after the application of the Holm–Bonferroni correction, only the order of
Primates meets this criterion.
3. Discussion
The trigger for a detailed bioinformatics analysis of whether the G4 motifs of the
TERT promoters are evolutionarily conserved across the orders of mammals was the data on G4-driven genomic instability in the
hTERT promoter region associated with the development of oncological diseases. It has been recently shown by chemical probing and spectroscopic methods that 96 nt DNAs modeling the G-rich strand of the
hTERT promoter and its variants with G > A point substitutions corresponding to somatic driver mutations fold into three stacked parallel G4s with a local destabilization at the substitution site [
16]. Mutations in the
hTERT promoter play a critical role in the immortality of cancer cells [
15] and increase clinical diagnostic potential in thyroid cancer [
22], melanoma [
23], glioblastoma [
24], and bladder cancer [
25]. One of the reasons for the selective advantage of malignant cells is the recruitment of a transcription factor (multimeric GA-binding protein, GABP), whose interaction with the mutated promoter provides a mechanism to overcome the downregulation of
TERT expression and replicative senescence [
26,
27].
Previously, functional genetic studies have identified several cancer-specific point mutations within the
hTERT core promoter region [
28]. However, not much was known about the role of G4 structures in mutagenesis and how conserved G4 motifs are between mammalian species and orders.
In our work, we used bioinformatics analysis to determine the distribution of G4 motifs in
TERT promoters, as well as the stability of their composition and sequence across mammalian species of different orders, since the risk of developing cancer is better described in mammals [
29]. Some species were removed because their sequences of interest contained more than 100 unread nucleotides or they were not aligned with their closest relatives by the blast2seq service. The search for G4 motifs was performed in the
TERT promoter regions of 141 mammalian species using the QGRS mapper software program; the size of the analyzed regions was taken as equal to 1000 bp from TSS. The diversity in the distribution of the
TERT G4 motifs in different orders of mammals (
Table 1) is an additional argument in favor of the evolution of
TERT-mediated regulation. Despite the ubiquity of G4 motifs in promoters of eukaryotic oncogenes [
30], their presence is not universal. G4 motifs were not found in six mammalian species (
Table 1), including four rodent species and one species of bats that are cancer resistant [
31]. This conclusion is consistent with the proposed involvement of G4 structures in the malignant transformation of eukaryotic cells. In other cases, G4 motifs were found to be predominantly located in the region proximal to the TSS (up to 400 bp) and are over-represented on the noncoding strand of the
TERT promoters, although the choice of strand depends on the order of mammals (
Figure 1 and
Figure S1,
Table 1). The higher density of G4 motifs on the noncoding versus the coding strand suggests that G4s regulate gene expression at the transcriptional rather than the translational level. The latter is possible only if the G4 motifs are located in the RNA transcript [
32].
To search for conserved G4 motifs in
TERT promoter regions of different mammalian species, we applied for the first time a block alignment approach using the NPG-explorer (the number of aligned blocks and
TERT promoter fragments in each block is indicated in
Table S2) and showed that
TERT promoter sequences capable of folding into G4 structures are evolutionarily conserved throughout the class
Mammalia (
Figure 2 and
Figure 3,
Table 2) and are therefore biologically relevant. The proportion of conserved G4 motifs in relation to all found G4 motifs across the compared set of mammalian species depended on the type of mammalian order and varied from 11 to 22% (
Table 1 and
Table 2). The conserved G4 motif loci lie mainly on the noncoding strand of
TERT promoter regions in the core and proximal promoter zones (
Figure 2 and
Figure 4). Thus, the formation of G4 may prevent binding of the RNA polymerase complex or transcription factors to the promoter.
In recent years, the deleterious effects of G4s on genome integrity and their potential role in regulating the DNA repair machinery have become the subject of intense research [
12]. Recent findings suggest that G4 structures, in collaboration with various specialized proteins, lead to genetic instability, alter the accessibility of DNA to damaging agents, and affect the efficiency and accuracy of DNA repair pathways. In a series of our studies, we have experimentally shown that G4 formation can have a negative impact on DNA mismatch repair that leads to a high mutation rate in G/C-rich regions of oncogene promoters [
16,
33]. In this study, we found the most frequent nucleotide substitutions (C↔T and G↔A) at positions disrupting G4 structure (occurring in G-tracts) and positions corresponding to G4 loop-forming sequences (
Figure 6 and
Figure 7,
Table S3) and showed that a statistically significant higher frequency of nucleotide substitutions in the conserved G4 motifs of the
TERT promoters compared to the surrounding regions was only confirmed for the order Primates (
Table 3). These data support the hypothesis that the high mutation frequency in the G/C-rich promoters of human oncogenes may be associated with quadruplex-controlled changes in the function of repair proteins.
5. Conclusions
The computational predictions indicate that G4 motifs are predominant in human oncogene promoters compared to the rest of the genome, strongly suggesting a role of G4 structures in cancer progression. In this study, we identified G4 motifs in the TERT promoter regions of 141 mammalian species and compared their location, degree of conservation and mutability potential using bioinformatics analysis. In order to address this, we used a combination of known and newly developed approaches. For searching for G4 motifs, we applied three tools: pqsfinder, G4Hunter and QGRS mapper; the latter was chosen as the main one. To identify conserved G4 motifs in the TERT promoter regions of five mammalian orders: Primates (primates), Artiodactyla (artiodactyls), Carnivora (predators), Chiroptera (bats), and Rodentia (rodents), we used for the first time a block alignment approach based on the NPG-explorer. Thus far, nucleotide pangenome software has predicted evolutionary relationship at the nucleotide level between two or more entire genomes. Here, the study focused on comparing functionally important genome regions across a large number of mammalian species. Under the selected similarity criteria (identity of block 80% or more, their length is not less than 100 columns), blocks of significant alignments containing G4 motifs were found in the TERT promoters of each of the five mammalian orders. According to the data obtained, the proportion of conserved G4 motifs in relation to all found G4 motifs in the compared set of mammalian species depended on the type of mammalian order and varied from 11 to 22%. Conserved G4 motifs were over-represented on the noncoding strand of TERT promotes, and their number, length, and localization varied across mammalian orders. The level of diversity in the conserved G4 motifs was also assessed using a block alignment approach: a statistically significant higher frequency of nucleotide substitutions in the conserved G4 motifs compared to the surrounding regions was confirmed only for the order Primates. This finding supports our hypothesis that G4 structures formed in oncogene promoters may act as mutagenic factors leading to genome instability in malignant cells due to interference with the DNA repair machinery.