Virtual Screening of a Marine Natural Product Database for In Silico Identification of a Potential Acetylcholinesterase Inhibitor
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset of Marine Natural Compounds
2.2. Structure-Based Pharmacophore Modelling and Virtual Screening
2.3. Structure Preparation
2.4. Prediction of Drug-Likeness Features
2.5. Molecular Docking
2.6. Toxicity Estimation
2.7. Molecular Dynamics Simulations
2.8. Density Functional Theory
3. Results and Discussion
3.1. Structure-Based Pharmacophore Modelling
3.2. ADME Studies
3.3. Molecular Docking of Compounds from the CMNPD Database
3.4. Toxicity Estimation
3.5. Molecular Dynamics Simulations
3.6. DFT Calculations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Available online: https://www.who.int/data/gho/data/themes/global-dementia-observatory-gdo (accessed on 22 March 2023).
- NIA. Available online: https://www.nia.nih.gov/health/what-are-signs-alzheimers-disease (accessed on 1 January 2020).
- Serrano-Pozo, A.; Frosch, M.P.; Masliah, E.; Hyman, B.T. Neuropathological Alterations in Alzheimer Disease. Cold Spring Harb. Perspect. Med. 2011, 1, a006189. [Google Scholar] [CrossRef] [PubMed]
- Grill, J.D.; Cummings, J.L. Current Therapeutic Targets for the Treatment of Alzheimer’s Disease. Expert Rev. Neurother. 2010, 10, 711–728. [Google Scholar] [CrossRef] [PubMed]
- FDA. Available online: https://www.fda.gov/news-events/press-announcements/fda-grants-accelerated-approval-alzheimers-disease (accessed on 23 March 2023).
- FDA. Available online: https://www.fda.gov/news-events/press-announcements/fda-grants-accelerated-approval-alzheimers-disease-treatment;Treatment-alzheimers-disease (accessed on 23 March 2023).
- Barthold, D.; Joyce, G.; Ferido, P.; Drabo, E.F.; Marcum, Z.A.; Gray, S.L.; Zissimopoulos, J.; Akushevich, I. Pharmaceutical Treatment for Alzheimer’s Disease and Related Dementias: Utilization and Disparities. J. Alzheimer’s Dis. 2020, 76, 579. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xu, S.; Liu, L.; Feng, R.; Gong, Y.; Zhao, X.; Li, J.; Cai, J.; Feng, N.; Wang, L.; et al. Multifunctional Compound AD-35 Improves Cognitive Impairment and Attenuates the Production of TNF-α and IL-1β in an Aβ25-35-Induced Rat Model of Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 56, 1403–1417. [Google Scholar] [CrossRef] [PubMed]
- Yiannopoulou, K.G.; Anastasiou, A.I.; Zachariou, V.; Pelidou, S.H. Reasons for Failed Trials of Disease-Modifying Treatments for Alzheimer Disease and Their Contribution in Recent Research. Biomedicines 2019, 7, 97. [Google Scholar] [CrossRef] [PubMed]
- Talesa, V.N. Acetylcholinesterase in Alzheimer’s Disease. Mech. Ageing Dev. 2001, 122, 1961–1969. [Google Scholar] [CrossRef]
- Bartolini, M.; Bertucci, C.; Cavrini, V.; Andrisano, V. β-Amyloid Aggregation Induced by Human Acetylcholinesterase: Inhibition Studies. Biochem. Pharmacol. 2003, 65, 407–416. [Google Scholar] [CrossRef]
- García-Ayllón, M.S.; Small, D.H.; Avila, J.; Sáez-Valero, J. Revisiting the Role of Acetylcholinesterase in Alzheimer’s Disease: Cross-Talk with β-Tau and p-Amyloid. Front. Mol. Neurosci. 2011, 4, 22. [Google Scholar] [CrossRef]
- Castro, A.; Martinez, A. Targeting Beta-Amyloid Pathogenesis Through Acetylcholinesterase Inhibitors. Curr. Pharm. Des. 2006, 12, 4377–4387. [Google Scholar] [CrossRef]
- Hansen, R.A.; Gartlehner, G.; Webb, A.P.; Morgan, L.C.; Moore, C.G.; Jonas, D.E. Efficacy and Safety of Donepezil, Galantamine, and Rivastigmine for the Treatment of Alzheimer’s Disease: A Systematic Review and Meta-Analysis. Clin. Interv. Aging 2008, 3, 211. [Google Scholar] [PubMed]
- Sharma, K. Cholinesterase Inhibitors as Alzheimer’s Therapeutics (Review). Mol. Med. Rep. 2019, 20, 1479–1487. [Google Scholar] [CrossRef]
- Vijayakumar, S.; Manogar, P.; Prabhu, S.; Sanjeevkumar Singh, R.A. Novel Ligand-Based Docking; Molecular Dynamic Simulations; and Absorption, Distribution, Metabolism, and Excretion Approach to Analyzing Potential Acetylcholinesterase Inhibitors for Alzheimer’s Disease. J. Pharm. Anal. 2018, 8, 413. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Mai, X.; Wu, X.; Hu, X.; Luo, X.; Zhang, G. Exploring the Inhibition of Quercetin on Acetylcholinesterase by Multispectroscopic and In Silico Approaches and Evaluation of Its Neuroprotective Effects on PC12 Cells. Molecules 2022, 27, 7971. [Google Scholar] [CrossRef] [PubMed]
- Lyu, C.; Chen, T.; Qiang, B.; Liu, N.; Wang, H.; Zhang, L.; Liu, Z. CMNPD: A Comprehensive Marine Natural Products Database towards Facilitating Drug Discovery from the Ocean. Nucleic Acids Res. 2021, 49, D509–D515. [Google Scholar] [CrossRef]
- Sunseri, J.; Koes, D.R. Pharmit: Interactive Exploration of Chemical Space. Nucleic Acids Res. 2016, 44, W442–W448. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated Data and New Features for Efficient Prediction of Protein Targets of Small Molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Drwal, M.N.; Banerjee, P.; Dunkel, M.; Wettig, M.R.; Preissner, R. ProTox: A Web Server for the in Silico Prediction of Rodent Oral Toxicity. Nucleic Acids Res. 2014, 42, W53. [Google Scholar] [CrossRef]
- Shaikh, G.M.; Murahari, M.; Thakur, S.; Kumar, M.S.; YC, M. Studies on Ligand-Based Pharmacophore Modeling Approach in Identifying Potent Future EGFR Inhibitors. J. Mol. Graph. Model. 2022, 112, 108114. [Google Scholar] [CrossRef]
- Pathania, S.; Singh, P.K. Analyzing FDA-Approved Drugs for Compliance of Pharmacokinetic Principles: Should There Be a Critical Screening Parameter in Drug Designing Protocols? Expert Opin. Drug Metab. Toxicol. 2021, 17, 351–354. [Google Scholar] [CrossRef]
- Breznik, M.; Ge, Y.; Bluck, J.P.; Briem, H.; Hahn, D.F.; Christ, C.D.; Mortier, J.; Mobley, D.L.; Meier, K. Prioritizing Small Sets of Molecules for Synthesis through In-Silico Tools: A Comparison of Common Ranking Methods. ChemMedChem 2023, 18, e202200425. [Google Scholar] [CrossRef] [PubMed]
- Warren, G.L.; Andrews, C.W.; Capelli, A.M.; Clarke, B.; LaLonde, J.; Lambert, M.H.; Lindvall, M.; Nevins, N.; Semus, S.F.; Senger, S.; et al. A Critical Assessment of Docking Programs and Scoring Functions. J. Med. Chem. 2006, 49, 5912–5931. [Google Scholar] [CrossRef] [PubMed]
- Son, M.; Park, C.; Rampogu, S.; Zeb, A.; Lee, K.W. Discovery of Novel Acetylcholinesterase Inhibitors as Potential Candidates for the Treatment of Alzheimer’s Disease. Int. J. Mol. Sci. 2019, 20, 1000. [Google Scholar] [CrossRef]
- Nogara, P.A.; Saraiva, R.D.A.; Caeran Bueno, D.; Lissner, L.J.; Lenz Dalla Corte, C.; Braga, M.M.; Rosemberg, D.B.; Rocha, J.B.T. Virtual Screening of Acetylcholinesterase Inhibitors Using the Lipinski’s Rule of Five and ZINC Databank. BioMed Res. Int. 2015, 2015, 870389. [Google Scholar] [CrossRef] [PubMed]
- Junaid, M.; Islam, N.; Hossain, M.K.; Ullah, M.O.; Halim, M.A. Metal Based Donepezil Analogues Designed to Inhibit Human Acetylcholinesterase for Alzheimer’s Disease. PLoS ONE 2019, 14, e0211935. [Google Scholar] [CrossRef]
- Biabani, M.A.F.; Laatsch, H.; Helmke, E.; Weyland, H. Delta-Indomycinone: A New Member of Pluramycin Class of Antibiotics Isolated from Marine Streptomyces sp. J. Antibiot. 1997, 50, 874–877. [Google Scholar] [CrossRef]
- Tietze, L.F.; Singidi, R.R.; Gericke, K.M. Total Synthesis of the Proposed Structure of the Anthrapyran Metabolite δ-Indomycinone. Chem. A Eur. J. 2007, 13, 9939–9947. [Google Scholar] [CrossRef]
- Ellman, G.L.; Courtney, K.D.; Andres, V.; Featherstone, R.M. A New and Rapid Colorimetric Determination of Acetylcholinesterase Activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Pouplana, S.; Espargaro, A.; Galdeano, C.; Viayna, E.; Sola, I.; Ventura, S.; Munoz-Torrero, D.; Sabate, R. Thioflavin-S Staining of Bacterial Inclusion Bodies for the Fast, Simple, and Inexpensive Screening of Amyloid Aggregation Inhibitors. Curr. Med. Chem. 2014, 21, 1152–1159. [Google Scholar] [CrossRef]
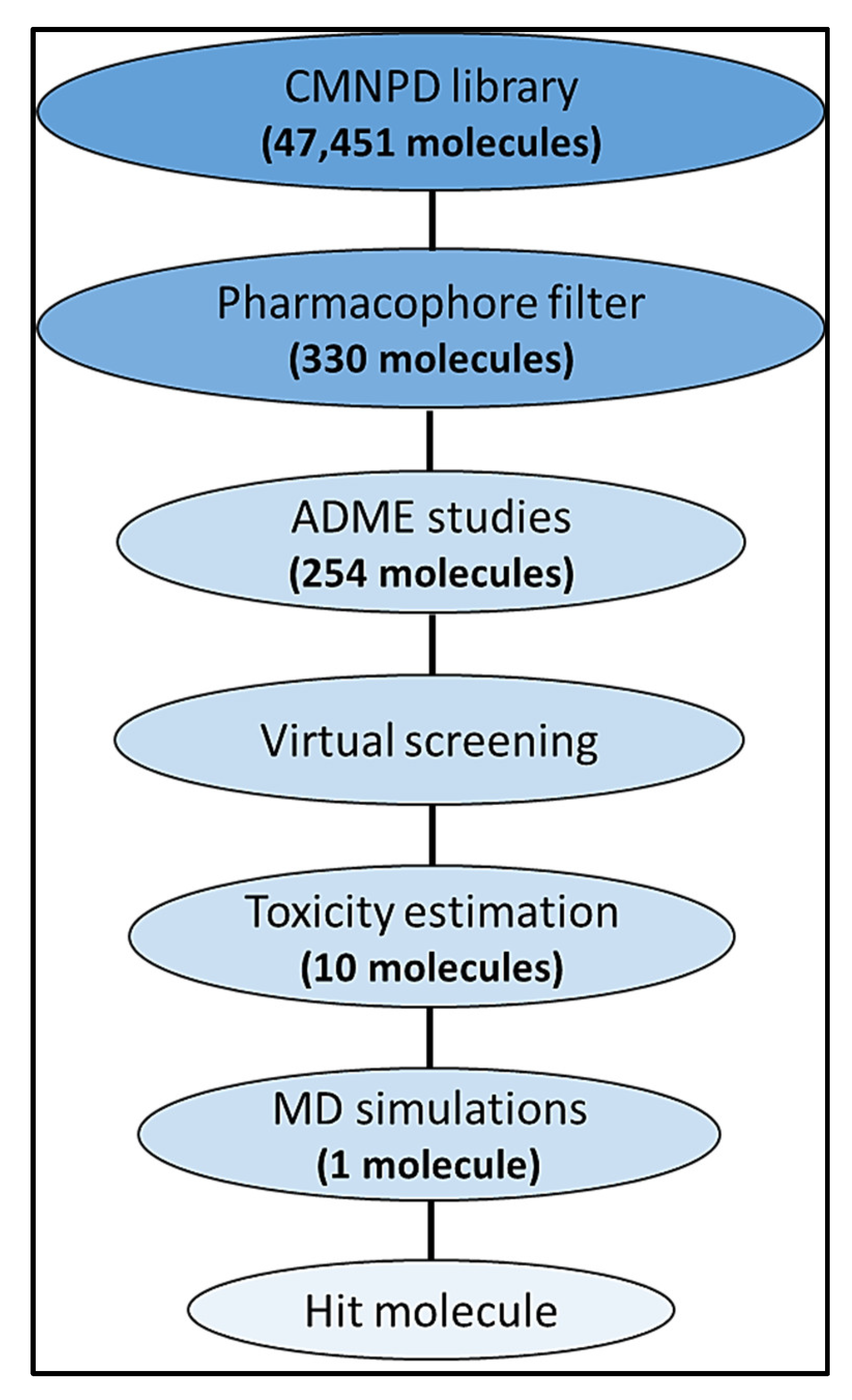

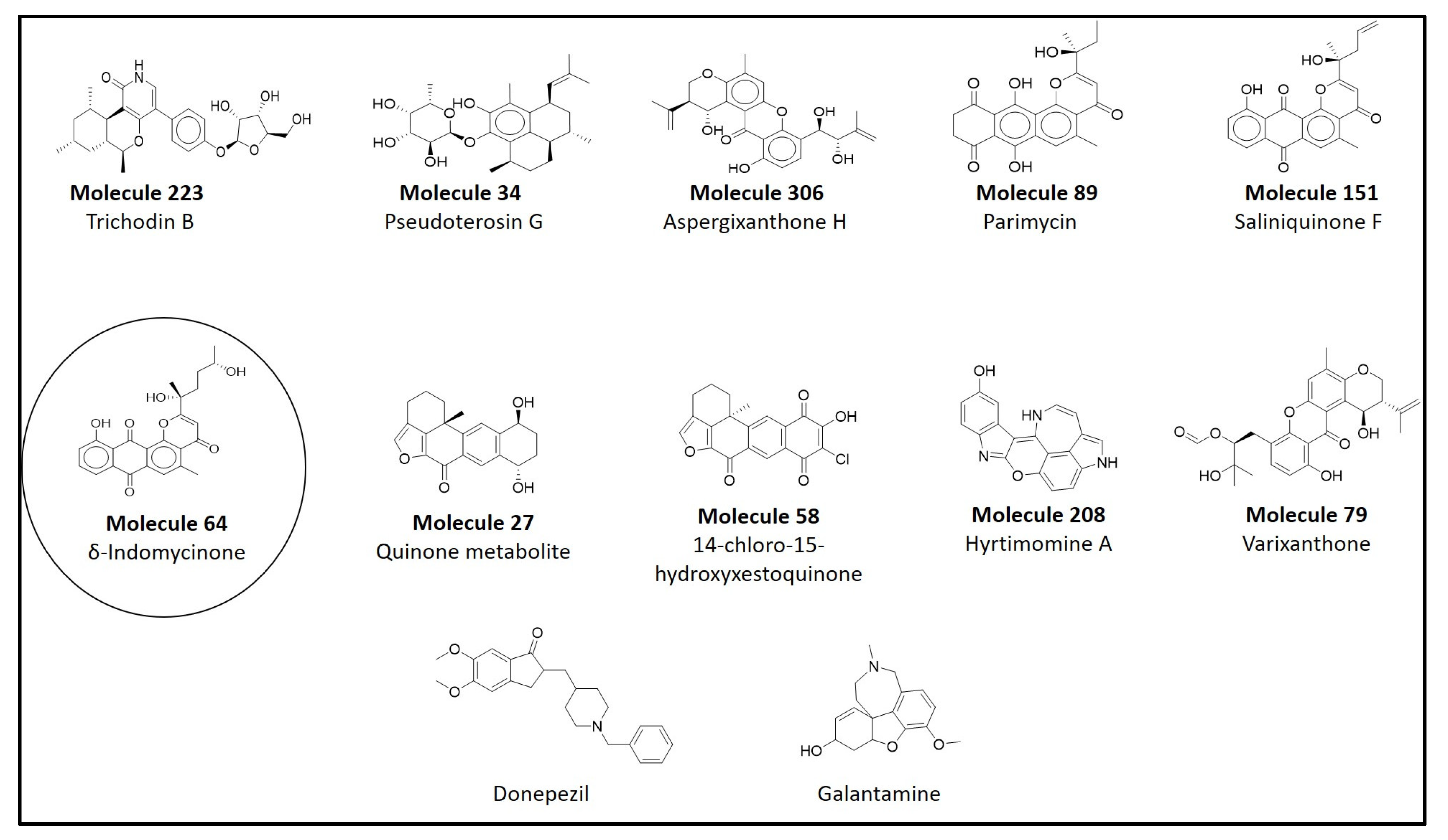

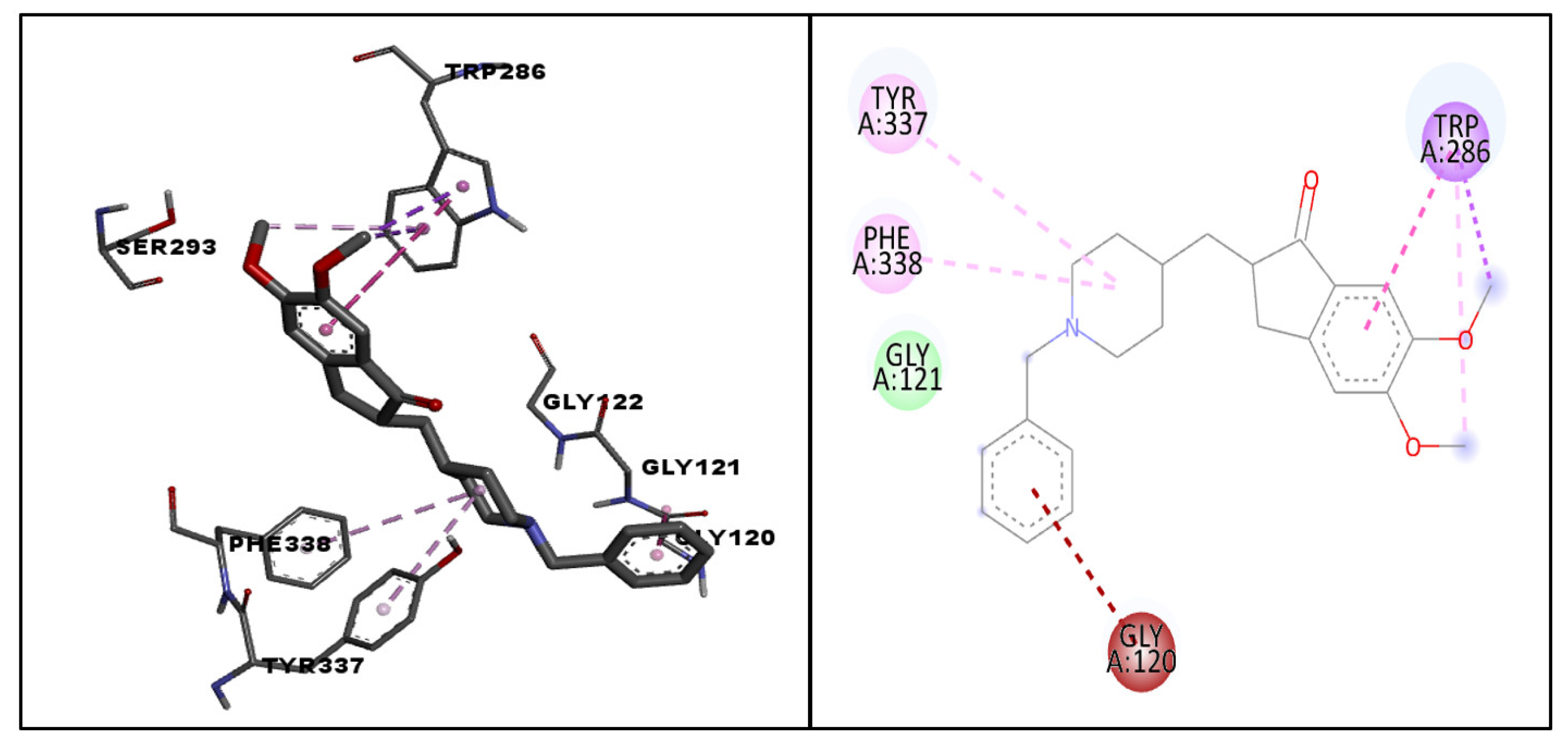

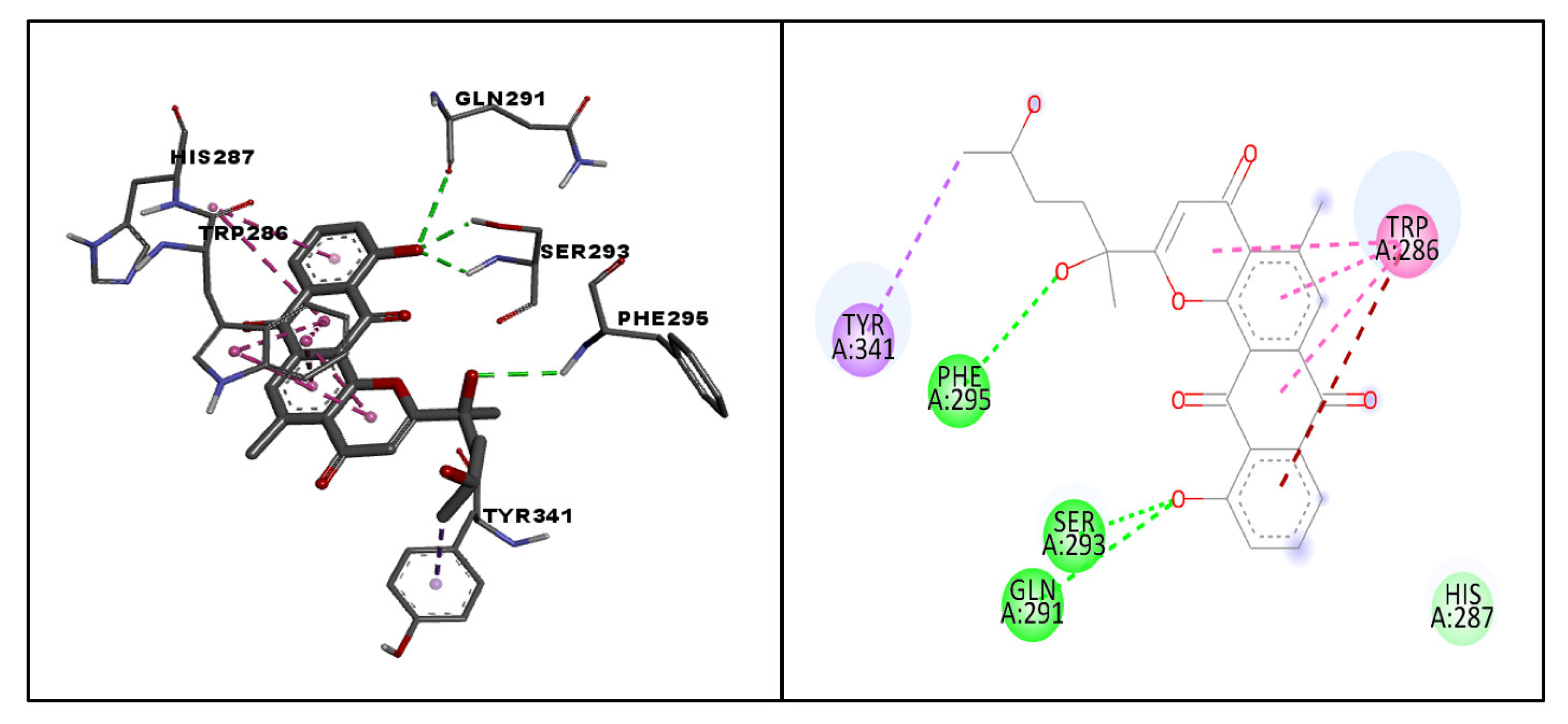
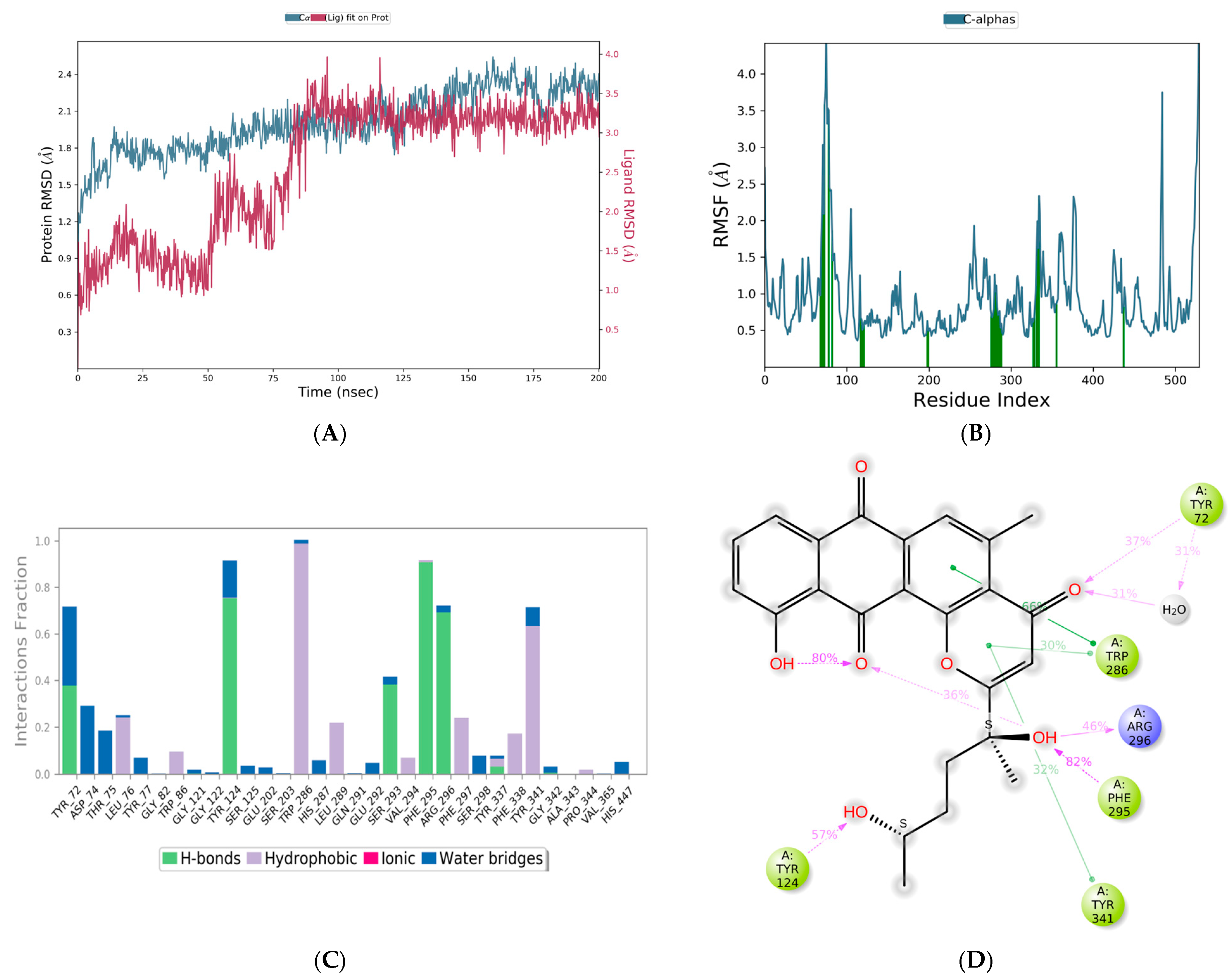
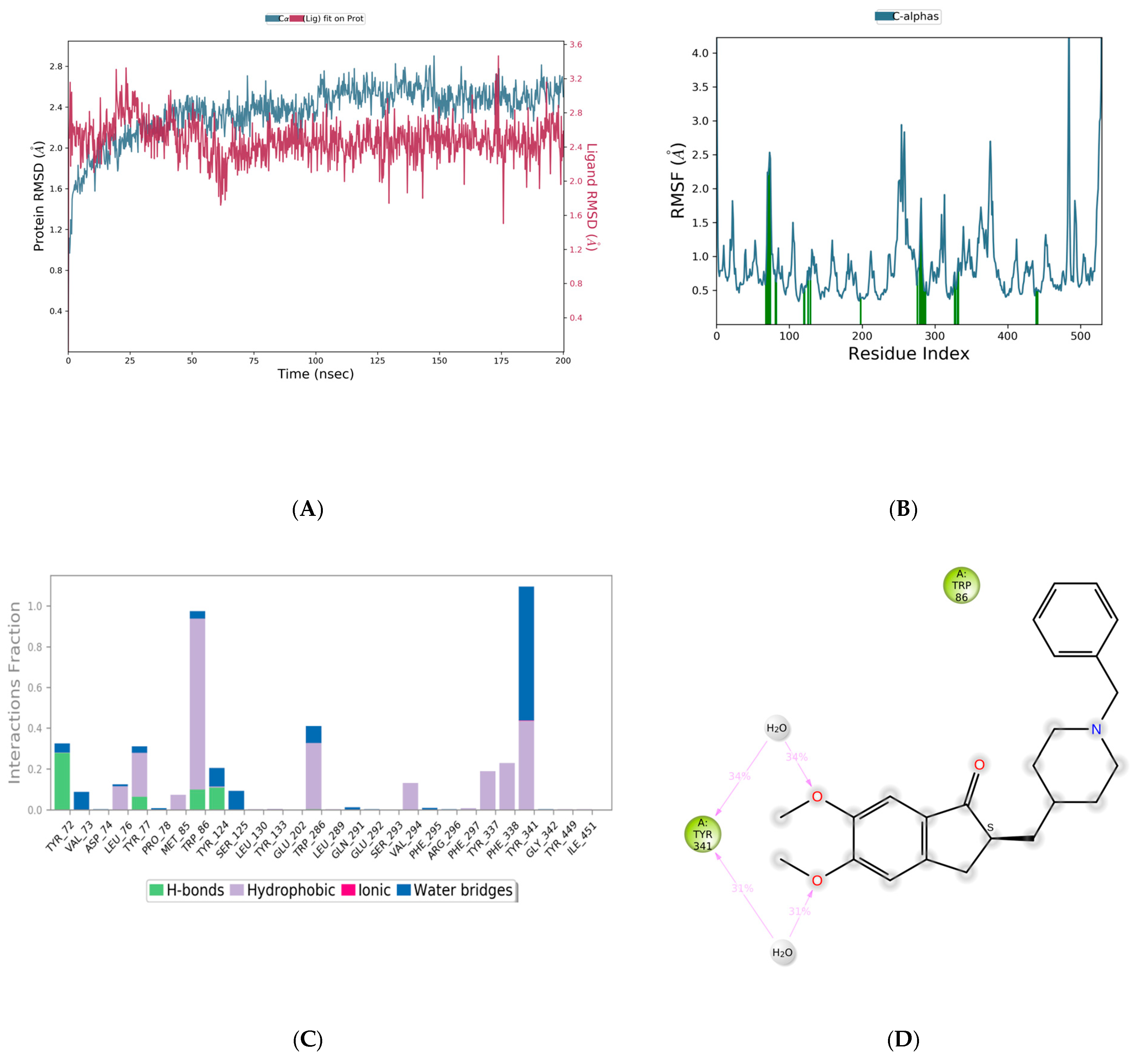
| Molecule | CMNPD ID | Docking Score | Interacting Amino Acid Residues | Interactions Involved |
|---|---|---|---|---|
| 223 | CMNPD24838 | −12.6 | GLY122, GLY121 | Conventional hydrogen bonding |
| VAL294, GLU202, GLY448 | Carbon hydrogen bonding | |||
| PHE295 | Donor–donor interaction | |||
| TRP86, TYR337 | Pi–donor hydrogen bond | |||
| TRP286 | Pi–sigma interaction | |||
| TRP286 | Pi–pi stacked interaction | |||
| TYR341, TYR124, TYR337 | Pi–pi T-shaped interaction | |||
| TRP286 | Pi–alkyl interaction | |||
| 34 | CMNPD4433 | −11.7 | TRP86, SER125 | Conventional hydrogen bonding |
| TYR124 | Pi–pi T-shaped interaction | |||
| TRP286 | Pi–pi stacked interaction | |||
| TRP286 | Pi–sigma interaction | |||
| TRP86 | Pi–donor hydrogen bonding | |||
| TYR72, PHE297, TYR341, PHE338, TRP286 | Pi–alkyl interaction | |||
| 306 | CMNPD30440 | −11.4 | GLY122, GLY121 | Conventional hydrogen bonding |
| HIS447 | Carbon hydrogen bonding | |||
| TRP286 | Pi–sigma interaction | |||
| TYR124 | Pi–lone pair interaction | |||
| TRP286 | Pi–pi stacked interaction | |||
| TYR124, TYR337, TRP86 | Pi–pi T-shaped interaction | |||
| TYR72, PHE297, PHE338 | Pi–alkyl interaction | |||
| 89 | CMNPD13187 | −11.3 | PHE295, SER293 | Conventional hydrogen bonding |
| TRP286, TYR341 | Pi–pi stacked interaction | |||
| 151 | CMNPD19682 | −11.2 | TYR341, GLN291, SER293 | Conventional hydrogen bonding |
| TRP286 | Pi–pi stacked interaction | |||
| TRP286 | Amide pi-stacked interaction | |||
| PHE338, TYR337 | Pi–alkyl interaction | |||
| HIS287 | Van der Waal’s interaction | |||
| 64 | CMNPD8741 | −11.1 | PHE295, SER293, GLN291 | Conventional hydrogen bonding |
| TRP286 | Pi–pi stacked interaction | |||
| TRP286 | Amide pi-stacked interaction | |||
| HIS287 | Van der Waal’s interaction | |||
| TYR341 | Pi–sigma interaction | |||
| 27 | CMNPD3303 | −11.0 | TRP286 | Pi–pi stacked interaction, pi–alkyl interaction |
| TYR341 | Pi–donor hydrogen bonding | |||
| 58 | CMNPD7644 | −10.9 | PHE295 | Conventional hydrogen bonding |
| VAL294 | Carbon hydrogen bonding | |||
| TRP286 | Pi–pi stacked interaction, pi–alkyl interaction | |||
| TYR341 | Pi–pi stacked interaction | |||
| 208 | CMNPD23795 | −10.8 | PHE295 | Conventional hydrogen bonding |
| TRP286 | Pi–pi stacked interaction | |||
| TYR341 | Pi–donor hydrogen bonding | |||
| 79 | CMNPD12415 | −10.7 | PHE295, SER293 | Conventional hydrogen bonding |
| TYR341, TRP286 | Pi–pi stacked interaction | |||
| TRP286 | Pi–donor hydrogen bonding | |||
| TYR337 | Pi–alkyl interaction | |||
| TYR341 | Pi–sigma interaction | |||
| PHE297 | Pi–pi T-shaped interaction | |||
| LEU289 | Alkyl interaction | |||
| Standard | Donepezil | −10.4 | GLY121 | Van der Waal’s interaction |
| TRP286 | Pi–sigma interaction | |||
| TRP286 | Pi–pi stacked interaction | |||
| GLY120 | Amide pi-stacked interaction | |||
| TYR337, PHE338, TRP286 | Pi–alkyl interaction | |||
| Standard | Galantamine | −9.1 | GLU202 | Conventional hydrogen bonding |
| TYR337 | Carbon hydrogen bonding | |||
| TRP86 | Pi–sigma interaction | |||
| GLY121 | Amide pi-stacked interaction | |||
| TRP286, PHE338, PHE297, PHE295, GLY122, HIS447 | Pi–alkyl interaction | |||
| GLY122 | Van der Waal’s interaction |
| Molecule | Compound ID | Docking Score (kcal/mol) | Molecular Formula | Predicted Toxic Dose (mg/kg) | Toxicity Class |
|---|---|---|---|---|---|
| 223 | CMNPD24838 | −12.6 | C26H33NO7 | 3000 | 5 |
| 34 | CMNPD4433 | −11.7 | C26H38O6 | 3000 | 5 |
| 306 | CMNPD30440 | −11.4 | C25H26O7 | 832 | 4 |
| 89 | CMNPD13187 | −11.3 | C22H20O7 | 2000 | 4 |
| 151 | CMNPD19682 | −11.2 | C23H18O6 | 450 | 4 |
| 64 | CMNPD8741 | −11.1 | C24H22O7 | 4000 | 5 |
| 27 | CMNPD3303 | −11.0 | C20H20O4 | 2400 | 5 |
| 58 | CMNPD7644 | −10.9 | C20H13ClO5 | 2000 | 4 |
| 208 | CMNPD23795 | −10.8 | C19H11N3O2 | 1600 | 4 |
| 79 | CMNPD12415 | −10.7 | C26H28O8 | 832 | 4 |
| Standard | Donepezil | −10.4 | C24H29NO3 | 505 | 4 |
| Standard | Galantamine | −9.1 | C17H21NO3 | 85 | 3 |
| Compound Name | HOMO | EHOMO (ev) | LUMO | ELUMO (ev) | Energy Gap (Δev) |
|---|---|---|---|---|---|
| Molecule 64 |  | −7.4600 | 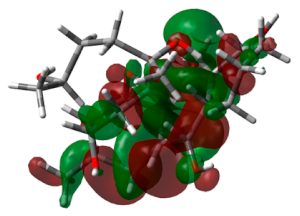 | −4.6123 | 2.8476 |
| Standard |  | −0.2621 | 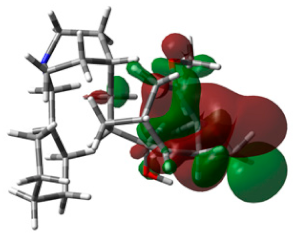 | −5.5059 | 1.6280 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gade, A.C.; Murahari, M.; Pavadai, P.; Kumar, M.S. Virtual Screening of a Marine Natural Product Database for In Silico Identification of a Potential Acetylcholinesterase Inhibitor. Life 2023, 13, 1298. https://doi.org/10.3390/life13061298
Gade AC, Murahari M, Pavadai P, Kumar MS. Virtual Screening of a Marine Natural Product Database for In Silico Identification of a Potential Acetylcholinesterase Inhibitor. Life. 2023; 13(6):1298. https://doi.org/10.3390/life13061298
Chicago/Turabian StyleGade, Anushree Chandrashekhar, Manikanta Murahari, Parasuraman Pavadai, and Maushmi Shailesh Kumar. 2023. "Virtual Screening of a Marine Natural Product Database for In Silico Identification of a Potential Acetylcholinesterase Inhibitor" Life 13, no. 6: 1298. https://doi.org/10.3390/life13061298
APA StyleGade, A. C., Murahari, M., Pavadai, P., & Kumar, M. S. (2023). Virtual Screening of a Marine Natural Product Database for In Silico Identification of a Potential Acetylcholinesterase Inhibitor. Life, 13(6), 1298. https://doi.org/10.3390/life13061298







