Circadian Genes MBOAT2/CDA/LPCAT2/B4GALT5 in the Metabolic Pathway Serve as New Biomarkers of PACA Prognosis and Immune Infiltration
Abstract
Simple Summary
Abstract
1. Introduction
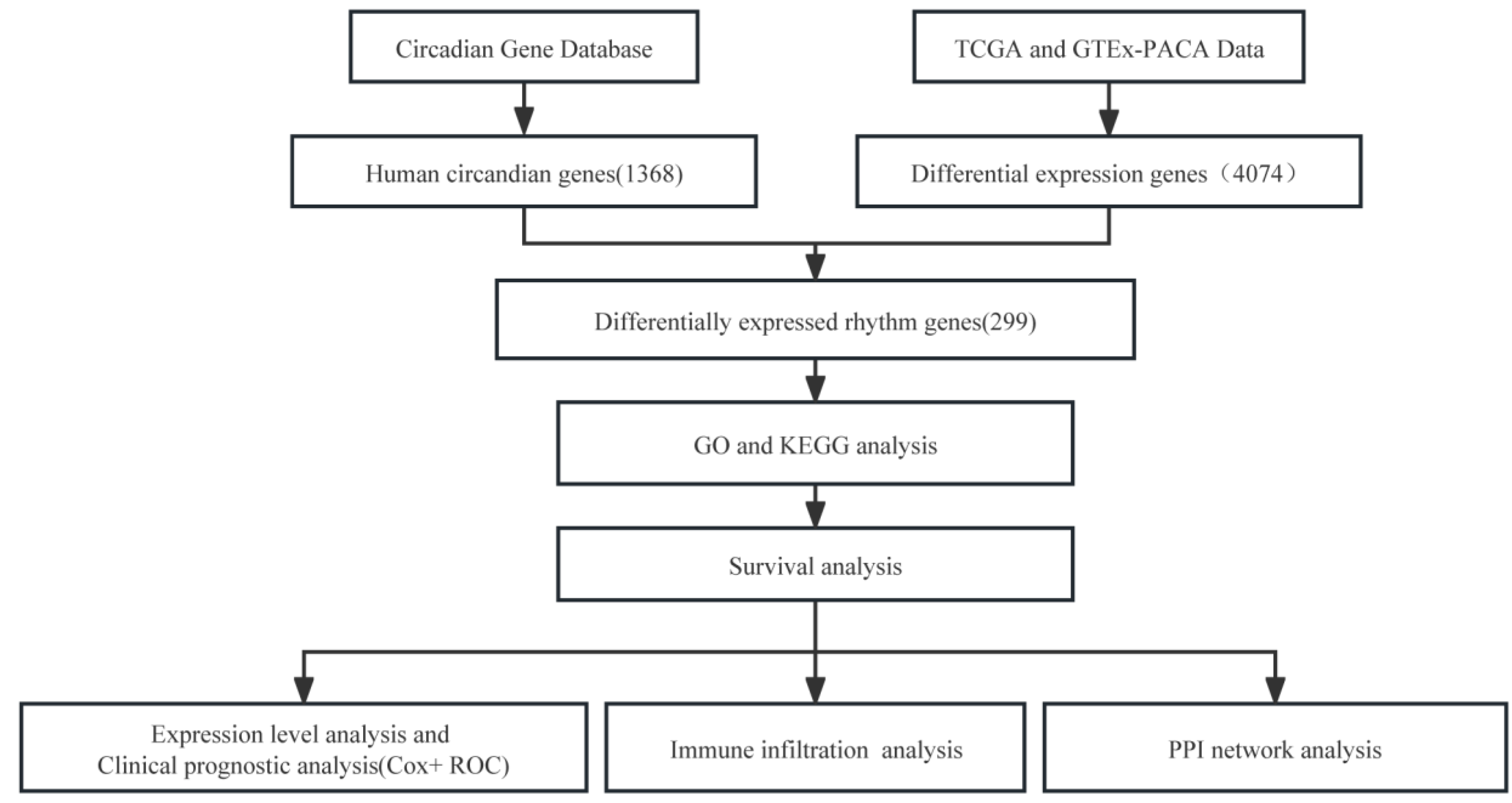
2. Materials and Methods
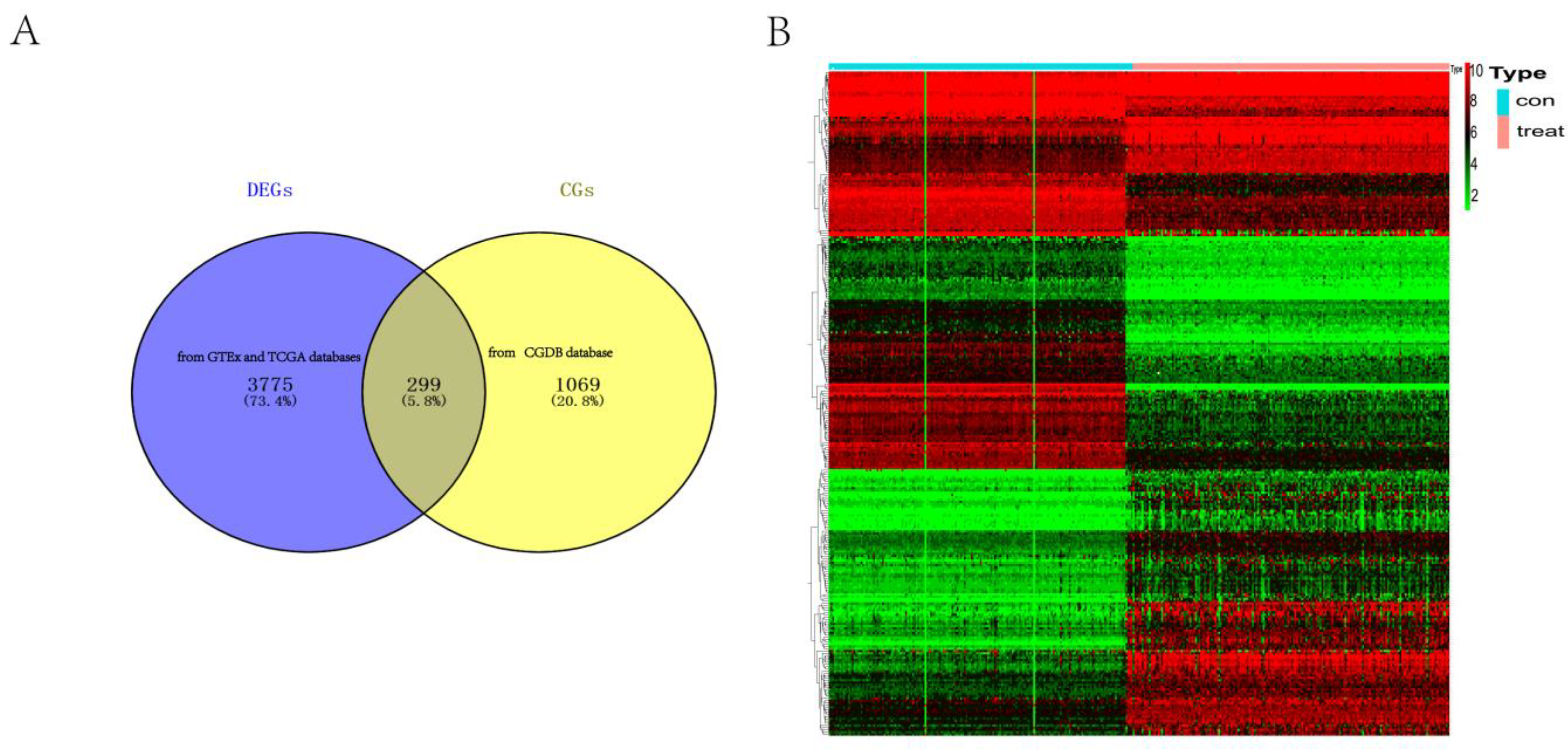
2.1. DERGs Identification
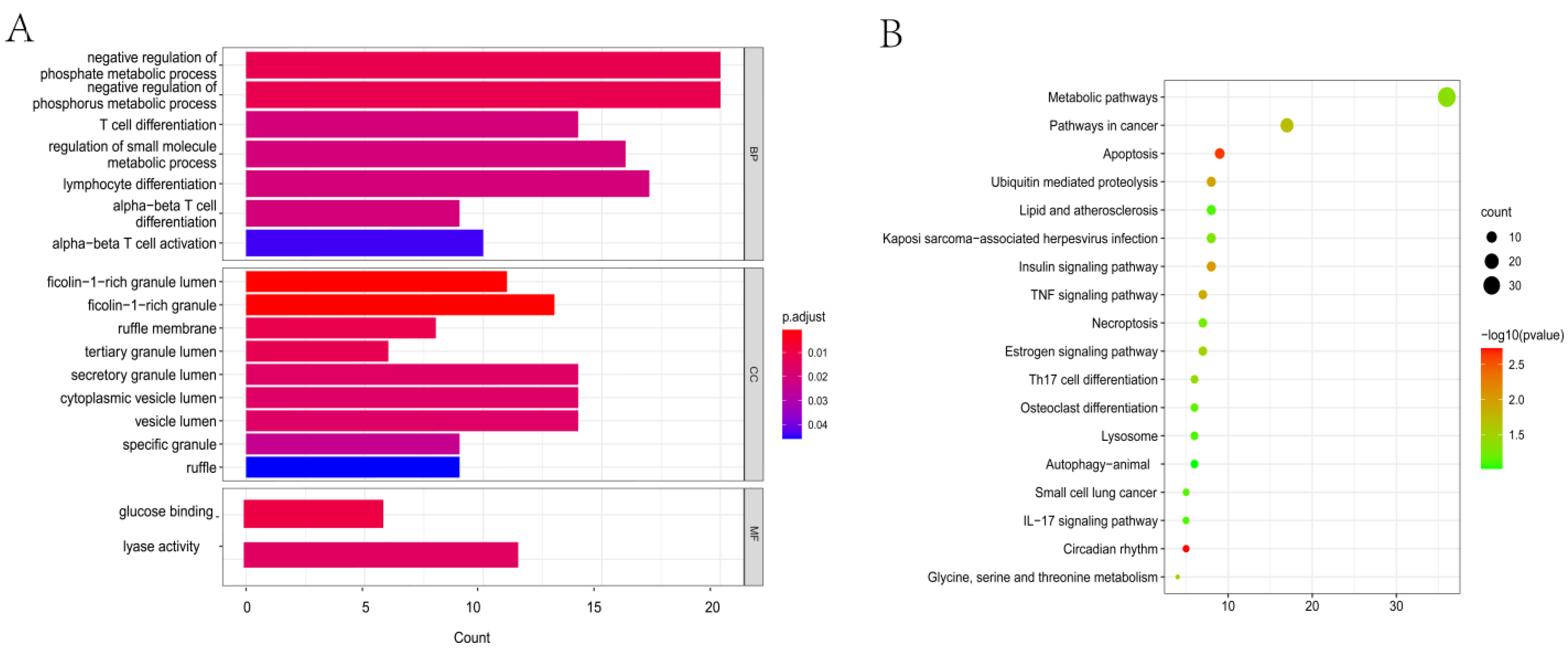
2.2. GO and KEGG Pathway Analyses
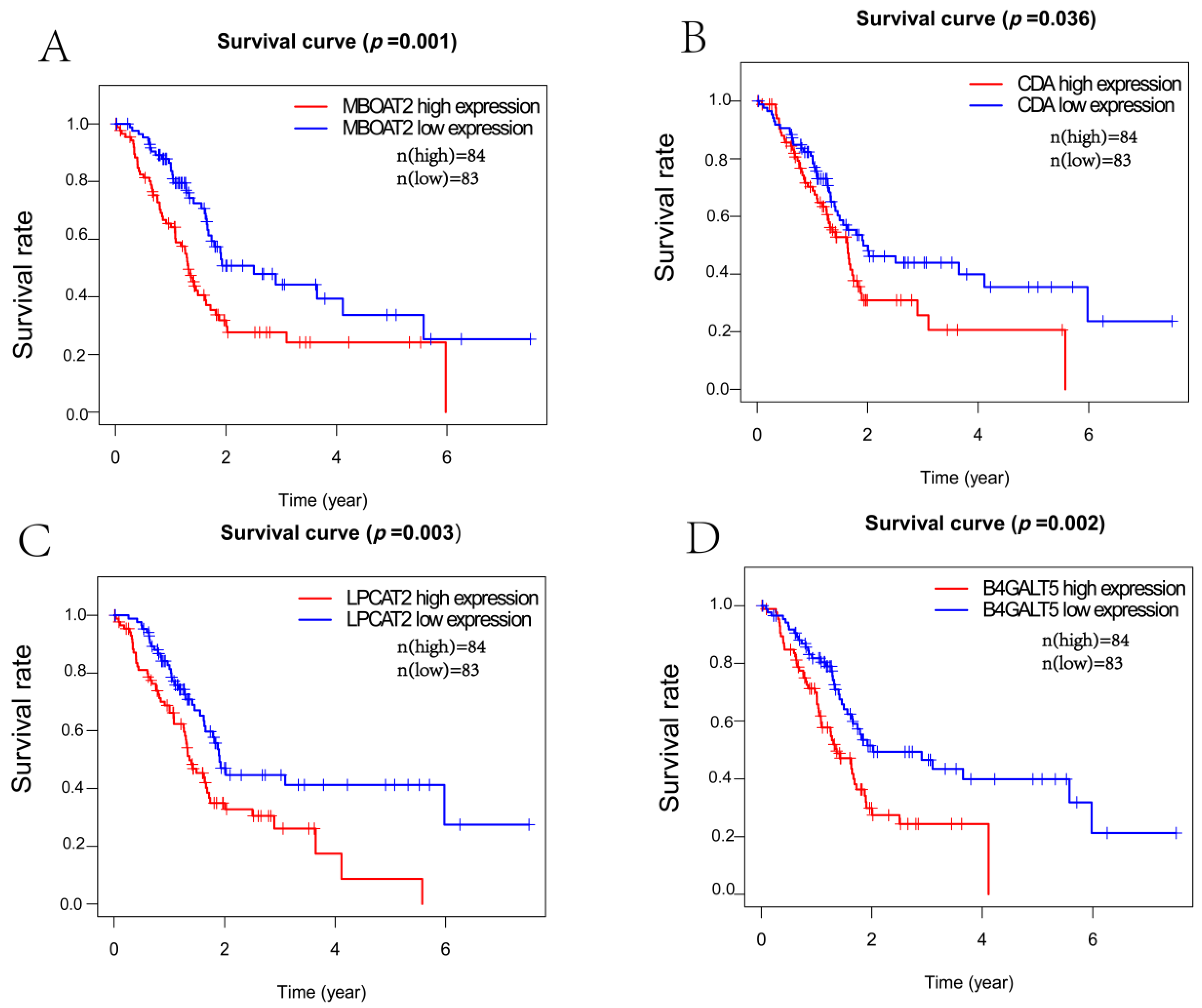
2.3. Survival Analysis
2.4. Cell Experiment
2.5. Clinical Prognostic Analysis
2.6. Immune Infiltration Assay
2.7. PPI Protein Network Interaction Analysis
2.8. Statistical Analysis
3. Results
3.1. DERGs Identification
3.2. GO and KEGG Analysis
3.3. Survival Analysis
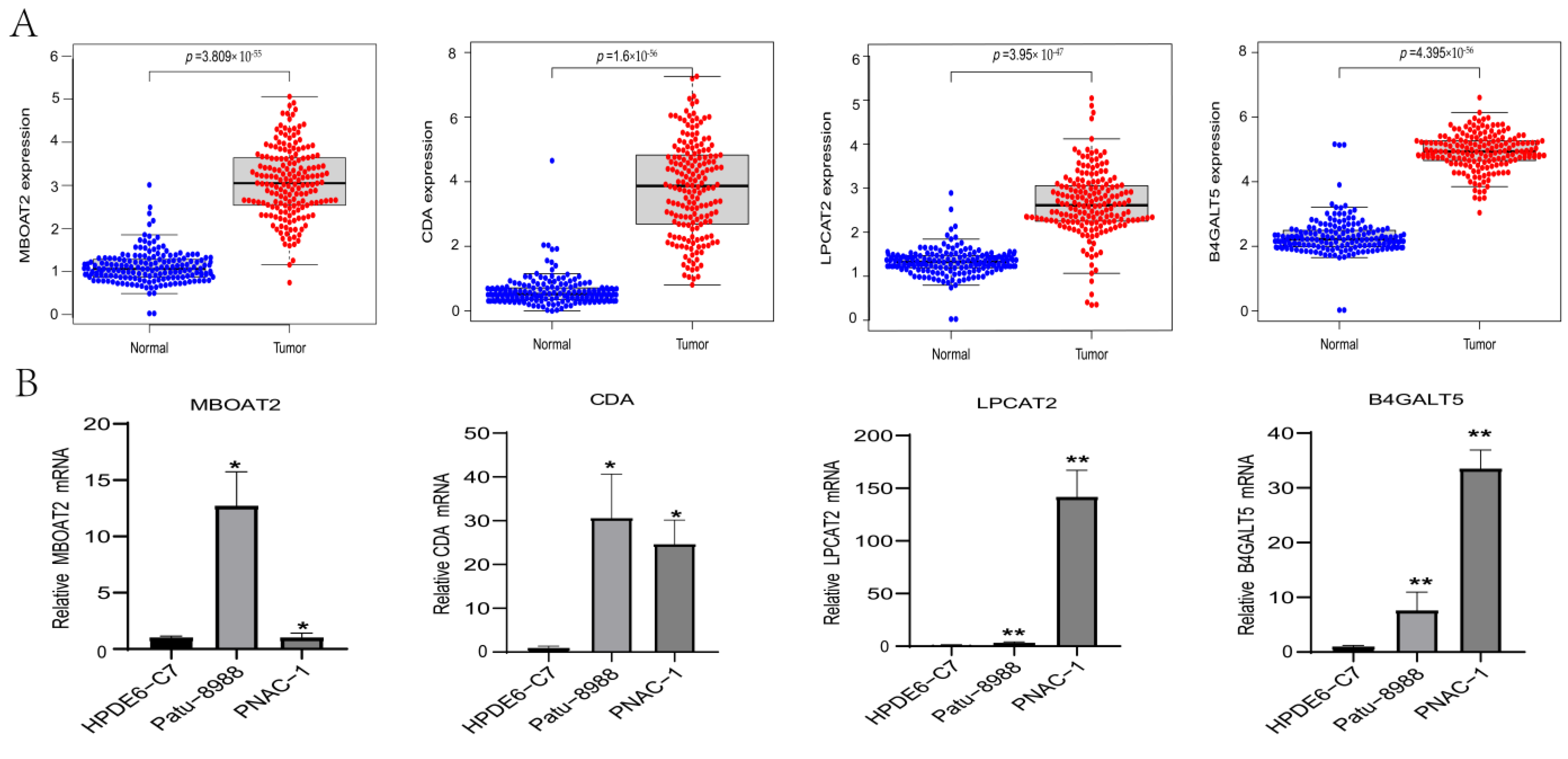
3.4. Expression Level Analysis
3.5. Clinical Prognostic Analysis
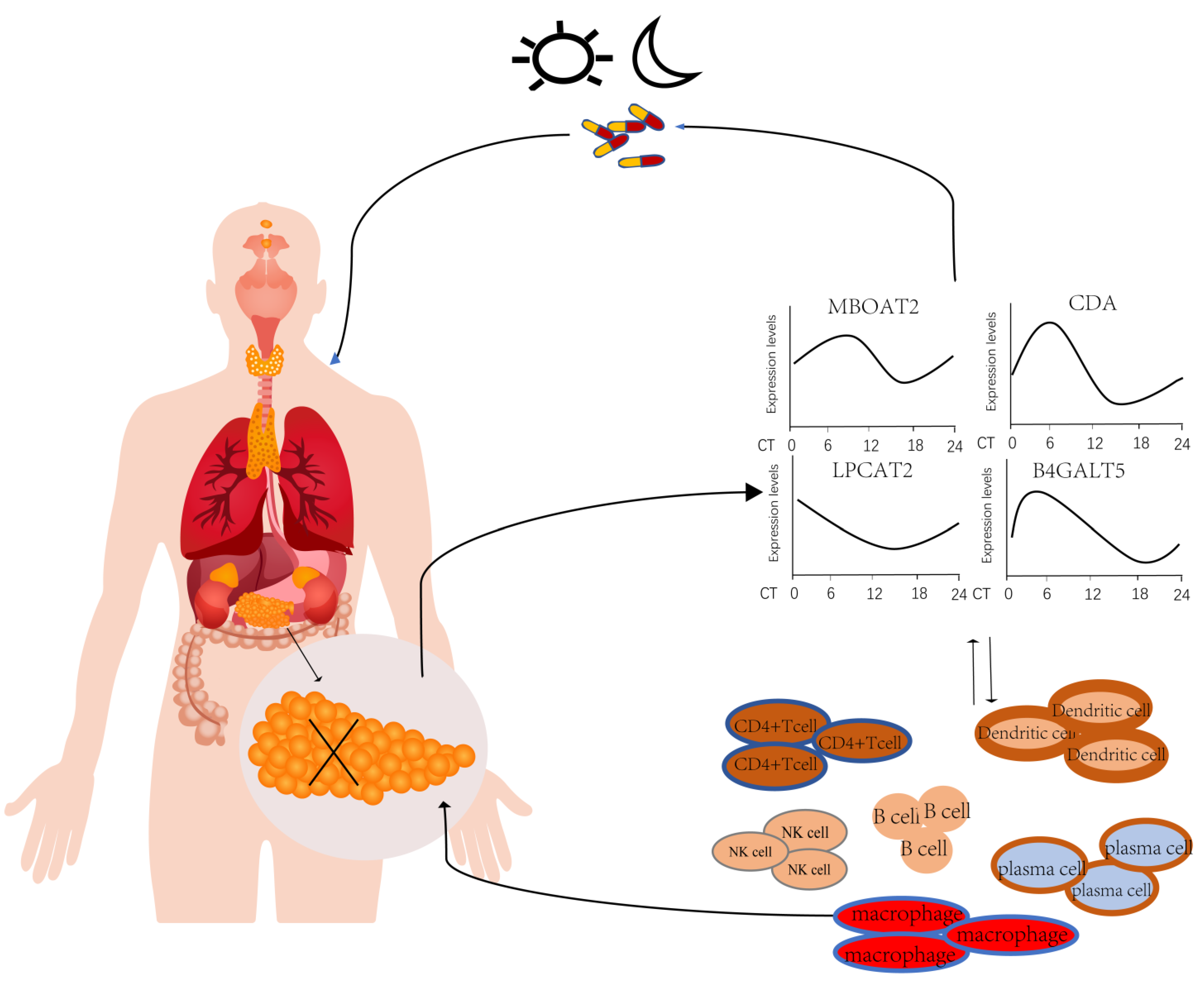
3.6. Immune Infiltration Analysis
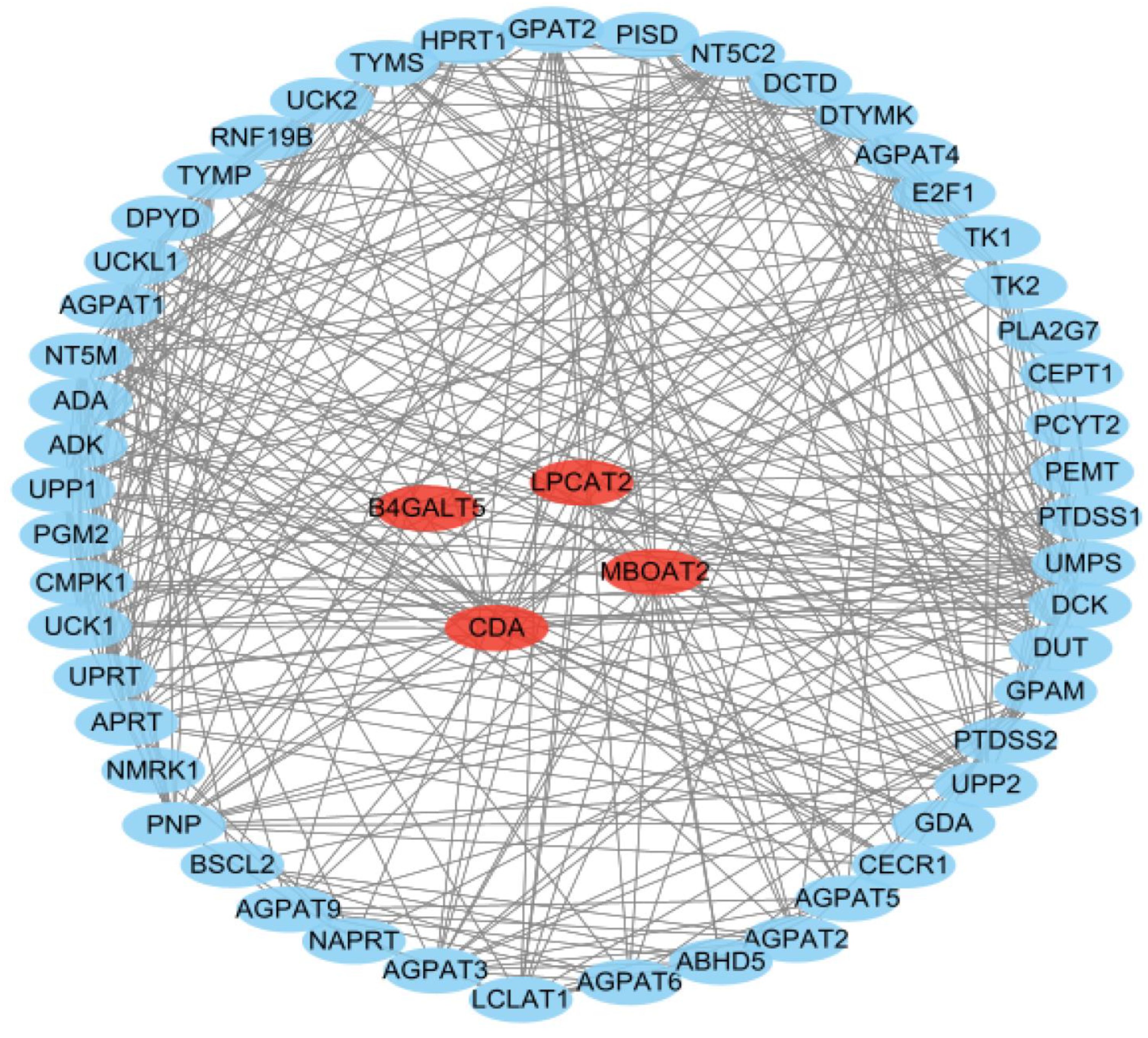
3.7. PPI Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ansari, D.; Tingstedt, B.; Andersson, B.; Holmquist, F.; Sturesson, C.; Williamsson, C.; Sasor, A.; Borg, D.; Bauden, M.; Andersson, R. Pancreatic cancer: Yesterday, today and tomorrow. Future Oncol. 2016, 12, 1929–1946. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Bao, X.; Wang, H.; Zhu, S.; Liu, Z.; Chen, Q.; Ai, K.; Shi, B. TRPM2 promotes pancreatic cancer by PKC/MAPK pathway. Cell Death Dis. 2021, 12, 585. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zheng, Y.; Yang, F.; Zhu, L.; Zhu, X.Q.; Wang, Z.F.; Wu, X.L.; Zhou, C.H.; Yan, J.Y.; Hu, B.Y.; et al. The molecular biology of pancreatic adenocarcinoma: Translational challenges and clinical perspectives. Signal Transduct. Target. Ther. 2021, 6, 249. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.; Herman, J.; Schulick, R.; Hruban, R.H.; Goggins, M. Pancreatic cancer. Lancet 2011, 378, 607–620. [Google Scholar] [CrossRef]
- Roenneberg, T.; Merrow, M. The Circadian Clock and Human Health. Curr. Biol. 2016, 26, R432–R443. [Google Scholar] [CrossRef]
- Wang, X.; Yang, G.; Wang, Q.; Zhao, Y.; Ding, K.; Ji, C.; Shi, Z.; Li, H.; Li, Y.; Li, S. C1R, CCL2, and TNFRSF1A Genes in Coronavirus Disease-COVID-19 Pathway Serve as Novel Molecular Biomarkers of GBM Prognosis and Immune Infiltration. Dis. Markers 2022, 2022, 8602068. [Google Scholar] [CrossRef]
- Liu, H.; Gao, Y.; Hu, S.; Fan, Z.; Wang, X.; Li, S. Bioinformatics Analysis of Differentially Expressed Rhythm Genes in Liver Hepatocellular Carcinoma. Front. Genet. 2021, 12, 680528. [Google Scholar] [CrossRef]
- Li, S.; Wang, X.; Wang, Q.; Ding, K.; Chen, X.; Zhao, Y.; Gao, Y.; Wang, Y. Effects and Prognostic Values of Circadian Genes CSNK1E/GNA11/KLF9/THRAP3 in Kidney Renal Clear Cell Carcinoma via a Comprehensive Analysis. Bioengineering 2022, 9, 306. [Google Scholar] [CrossRef]
- Carbone, A.; De Santis, E.; Cela, O.; Giambra, V.; Miele, L.; Marrone, G.; Grieco, A.; Buschbeck, M.; Capitanio, N.; Mazza, T.; et al. The Histone Variant MacroH2A1 Impacts Circadian Gene Expression and Cell Phenotype in an In Vitro Model of Hepatocellular Carcinoma. Biomedicines 2021, 9, 1057. [Google Scholar] [CrossRef]
- Emura, T.; Matsui, S.; Chen, H.Y. compound.Cox: Univariate feature selection and compound covariate for predicting survival. Comput. Methods Programs Biomed. 2019, 168, 21–37. [Google Scholar] [CrossRef]
- Sancar, A.; Van Gelder, R.N. Clocks, cancer, and chronochemotherapy. Science 2021, 371, eabb0738. [Google Scholar] [CrossRef] [PubMed]
- Winter, S.L.; Bosnoyan-Collins, L.; Pinnaduwage, D.; Andrulis, I.L. Expression of the circadian clock genes Per1 and Per2 in sporadic and familial breast tumors. Neoplasia 2007, 9, 797–800. [Google Scholar] [CrossRef] [PubMed]
- Yeh, K.T.; Yang, M.Y.; Liu, T.C.; Chen, J.C.; Chan, W.L.; Lin, S.F.; Chang, J.G. Abnormal expression of period 1 (PER1) in endometrial carcinoma. J. Pathol. 2005, 206, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Qu, M.; Zhang, G.; Qu, H.; Vu, A.; Wu, R.; Tsukamoto, H.; Jia, Z.; Huang, W.; Lenz, H.J.; Rich, J.N.; et al. Circadian regulator BMAL1::CLOCK promotes cell proliferation in hepatocellular carcinoma by controlling apoptosis and cell cycle. Proc. Natl. Acad. Sci. USA 2023, 120, e2214829120. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.K.; Long, N.P.; Jung, J.; Kim, T.J.; Na, E.; Kang, Y.P.; Kwon, S.W.; Jang, J. Integrative lipidomic and transcriptomic analysis of X-linked adrenoleukodystrophy reveals distinct lipidome signatures between adrenomyeloneuropathy and childhood cerebral adrenoleukodystrophy. Biochem. Biophys. Res. Commun. 2019, 508, 563–569. [Google Scholar] [CrossRef]
- Sonnenschein, K.; Wilczek, A.L.; de Gonzalo-Calvo, D.; Pfanne, A.; Derda, A.A.; Zwadlo, C.; Bavendiek, U.; Bauersachs, J.; Fiedler, J.; Thum, T. Serum circular RNAs act as blood-based biomarkers for hypertrophic obstructive cardiomyopathy. Sci. Rep. 2019, 9, 20350. [Google Scholar] [CrossRef]
- Schmied, M.C.; Zehetmayer, S.; Reindl, M.; Ehling, R.; Bajer-Kornek, B.; Leutmezer, F.; Zebenholzer, K.; Hotzy, C.; Lichtner, P.; Meitinger, T.; et al. Replication study of multiple sclerosis (MS) susceptibility alleles and correlation of DNA-variants with disease features in a cohort of Austrian MS patients. Neurogenetics 2012, 13, 181–187. [Google Scholar] [CrossRef]
- Li, Z.; Zhuang, H.; Chen, X.; Zhang, Y.; Ma, Z.; Wang, S.; Yan, Q.; Zhou, Z.; Huang, S.; Zhang, C.; et al. Identification of MBOAT2 as an Unfavorable Biomarker Correlated with KRAS Activation and Reduced CD8(+) T-Cell Infiltration in Pancreatic Cancer. J. Oncol. 2022, 2022, 4269733. [Google Scholar] [CrossRef]
- Xie, L.Y.; Huang, H.Y.; Fang, T.; Liang, J.Y.; Hao, Y.L.; Zhang, X.J.; Xie, Y.X.; Wang, C.; Tan, Y.H.; Zeng, L. A Prognostic Survival Model of Pancreatic Adenocarcinoma Based on Metabolism-Related Gene Expression. Front. Genet. 2022, 13, 804190. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, K.; Cui, J.; Xiong, J.; Wu, H.; Peng, T.; Guo, Y. Circ-MBOAT2 knockdown represses tumor progression and glutamine catabolism by miR-433-3p/GOT1 axis in pancreatic cancer. J. Exp. Clin. Cancer Res. 2021, 40, 124. [Google Scholar] [CrossRef]
- Frances, A.; Cordelier, P. The Emerging Role of Cytidine Deaminase in Human Diseases: A New Opportunity for Therapy? Mol. Ther. 2020, 28, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.F.; Feng, Y.F.; Chen, Q.L.; Zhang, Q.K. CDA gene silencing regulated the proliferation and apoptosis of chronic myeloid leukemia K562 cells. Cancer Cell Int. 2018, 18, 96. [Google Scholar] [CrossRef] [PubMed]
- Abraham, A.; Varatharajan, S.; Abbas, S.; Zhang, W.; Shaji, R.V.; Ahmed, R.; Abraham, A.; George, B.; Srivastava, A.; Chandy, M.; et al. Cytidine deaminase genetic variants influence RNA expression and cytarabine cytotoxicity in acute myeloid leukemia. Pharmacogenomics 2012, 13, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.W.; van der Noort, V.; van der Straaten, T.; Honkoop, A.H.; Peters, G.J.; Guchelaar, H.J.; Boven, E. Single-nucleotide polymorphisms in the genes of CES2, CDA and enzymatic activity of CDA for prediction of the efficacy of capecitabine-containing chemotherapy in patients with metastatic breast cancer. Pharmacol. Res. 2018, 128, 122–129. [Google Scholar] [CrossRef]
- Yuan, M.H.; Wei, L.X.; Zhou, R.S.; Xu, H.F.; Wang, J.Y.; Bai, Q.R. Therapeutic effects of adenovirus-mediated CD and NIS expression combined with Na(131)I/5-FC on human thyroid cancer. Oncol. Lett. 2017, 14, 7431–7436. [Google Scholar] [CrossRef]
- Zhou, M.; Wan, H.Y.; Gao, B.L.; Ding, Y.J.; Jun, R.X. Genetic polymorphisms of XPD and CDA and lung cancer risk. Oncol. Lett. 2012, 4, 247–251. [Google Scholar] [CrossRef]
- Hamano, F.; Matoba, K.; Hashidate-Yoshida, T.; Suzuki, T.; Miura, K.; Hishikawa, D.; Harayama, T.; Yuki, K.; Kita, Y.; Noda, N.N.; et al. Mutagenesis and homology modeling reveal a predicted pocket of lysophosphatidylcholine acyltransferase 2 to catch Acyl-CoA. FASEB J. 2021, 35, e21501. [Google Scholar] [CrossRef]
- Abate, W.; Alrammah, H.; Kiernan, M.; Tonks, A.J.; Jackson, S.K. Lysophosphatidylcholine acyltransferase 2 (LPCAT2) co-localises with TLR4 and regulates macrophage inflammatory gene expression in response to LPS. Sci. Rep. 2020, 10, 10355. [Google Scholar] [CrossRef]
- Agarwal, A.K.; Garg, A. Enzymatic activity of the human 1-acylglycerol-3-phosphate-O-acyltransferase isoform 11: Upregulated in breast and cervical cancers. J. Lipid Res. 2010, 51, 2143–2152. [Google Scholar] [CrossRef]
- Cotte, A.K.; Aires, V.; Ghiringhelli, F.; Delmas, D. LPCAT2 controls chemoresistance in colorectal cancer. Mol. Cell. Oncol. 2018, 5, e1448245. [Google Scholar] [CrossRef]
- Ma, B.; Jiang, H.; Wen, D.; Hu, J.; Han, L.; Liu, W.; Xu, W.; Shi, X.; Wei, W.; Liao, T.; et al. Transcriptome Analyses Identify a Metabolic Gene Signature Indicative of Dedifferentiation of Papillary Thyroid Cancer. J. Clin. Endocrinol. Metab. 2019, 104, 3713–3725. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhao, B.; Chen, X.; Geng, X.; Zhang, Z. Circ_0009910 sponges miR-491-5p to promote acute myeloid leukemia progression through modulating B4GALT5 expression and PI3K/AKT signaling pathway. Int. J. Lab. Hematol. 2022, 44, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Li, Z.; Wu, Q.; Liu, H.; Sun, Z.; Wu, Y.; Luo, J. B4GALT5 high expression associated with poor prognosis of hepatocellular carcinoma. BMC Cancer 2022, 22, 392. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wu, Y.; Jia, G.; Zhu, Q.; Li, D.; Xie, K. A signature based on glycosyltransferase genes provides a promising tool for the prediction of prognosis and immunotherapy responsiveness in ovarian cancer. J. Ovarian Res. 2023, 16, 5. [Google Scholar] [CrossRef] [PubMed]
- Narayan, G.; Murty, V.V. Integrative genomic approaches in cervical cancer: Implications for molecular pathogenesis. Future Oncol. 2010, 6, 1643–1652. [Google Scholar] [CrossRef]


| Parameter | Univariate Analysis | ||
|---|---|---|---|
| HR | 95% CI | p | |
| age | 1.03 | 1.01−1.05 | <0.05 |
| gender | 0.76 | 0.49−1.17 | >0.05 |
| grade | 1.17 | 1.08−1.26 | <0.005 |
| stage | 0.89 | 0.46−1.70 | >0.05 |
| T | 1.60 | 0.84−3.04 | >0.05 |
| N | 1.11 | 0.89−1.38 | >0.05 |
| MBOAT2 | 1.56 | 1.18−2.07 | <0.005 |
| CDA | 1.36 | 1.14−1.61 | <0.005 |
| LPCAT2 | 1.74 | 1.27−2.40 | <0.005 |
| B4GALT5 | 1.67 | 1.10−2.55 | <0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Zhou, S.; Hu, X.; Wang, X.; Wu, X.; Huai, Z.; Gao, Y.; Li, S. Circadian Genes MBOAT2/CDA/LPCAT2/B4GALT5 in the Metabolic Pathway Serve as New Biomarkers of PACA Prognosis and Immune Infiltration. Life 2023, 13, 1116. https://doi.org/10.3390/life13051116
Wang Q, Zhou S, Hu X, Wang X, Wu X, Huai Z, Gao Y, Li S. Circadian Genes MBOAT2/CDA/LPCAT2/B4GALT5 in the Metabolic Pathway Serve as New Biomarkers of PACA Prognosis and Immune Infiltration. Life. 2023; 13(5):1116. https://doi.org/10.3390/life13051116
Chicago/Turabian StyleWang, Qingqing, Shuning Zhou, Xinyi Hu, Xianggang Wang, Xue Wu, Ziyou Huai, Yu Gao, and Shujing Li. 2023. "Circadian Genes MBOAT2/CDA/LPCAT2/B4GALT5 in the Metabolic Pathway Serve as New Biomarkers of PACA Prognosis and Immune Infiltration" Life 13, no. 5: 1116. https://doi.org/10.3390/life13051116
APA StyleWang, Q., Zhou, S., Hu, X., Wang, X., Wu, X., Huai, Z., Gao, Y., & Li, S. (2023). Circadian Genes MBOAT2/CDA/LPCAT2/B4GALT5 in the Metabolic Pathway Serve as New Biomarkers of PACA Prognosis and Immune Infiltration. Life, 13(5), 1116. https://doi.org/10.3390/life13051116







