Gut Microbiota in Autophagy Regulation: New Therapeutic Perspective in Neurodegeneration
Abstract
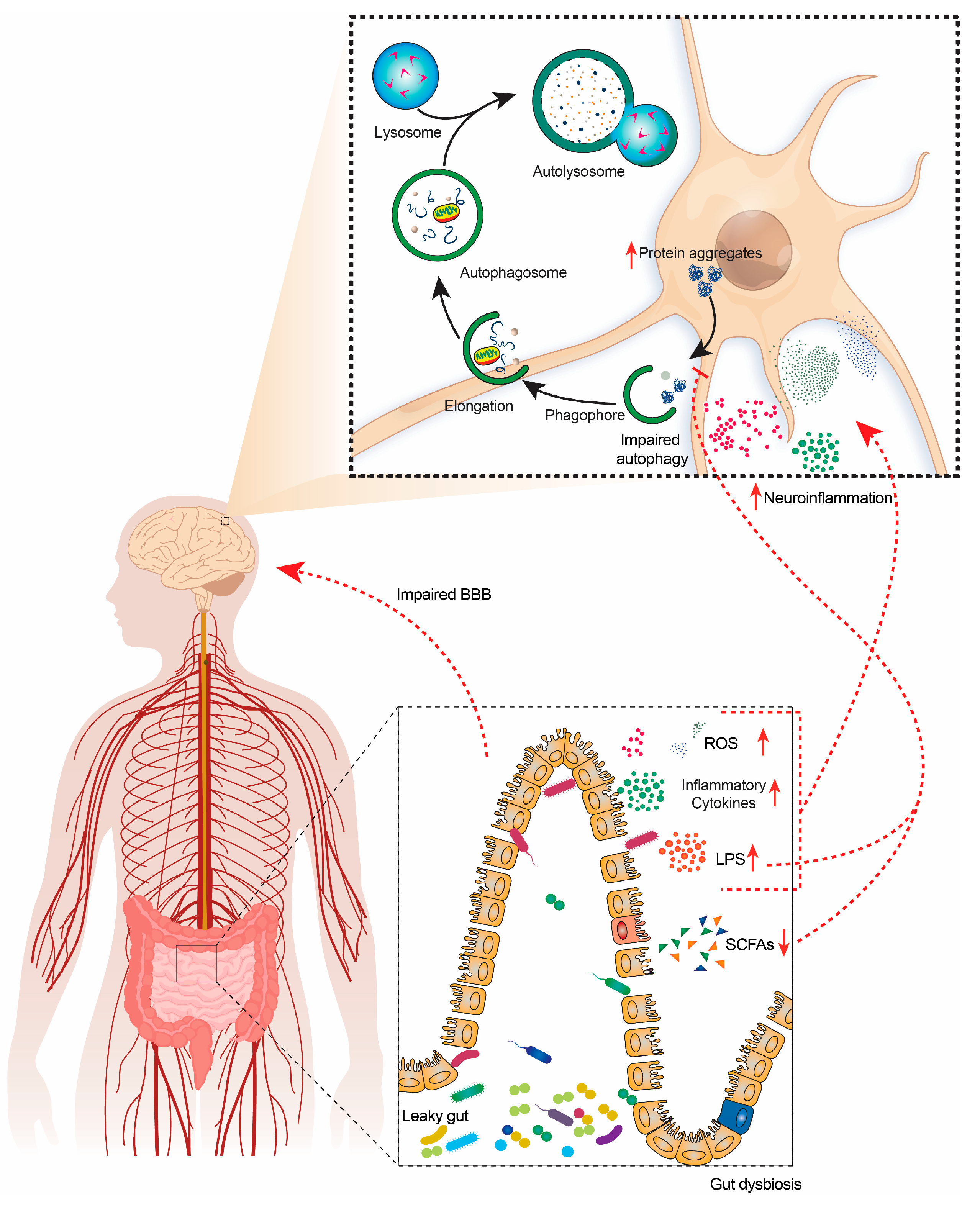
1. Introduction
2. Gut Microbiota (GM) and Brain Relationship
3. Autophagy and Neurodegeneration
4. Relation of Gut Microbiota (GM) and Autophagy in Neurodegeneration
5. Role of Gut Microbiota (GM) and Metabolites in NDD Pathogenesis
5.1. Parkinson’s Disease (PD)
5.2. Alzheimer’s Disease (AD)
5.3. Huntington’s Disease (HD)
5.4. Amyotrophic Lateral Sclerosis (ALS)
5.5. Multiple Sclerosis (MS)
6. Therapeutic Interventions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Alvarez, A.-S.; de Vos, W.M. The Gut Microbiota in the First Decade of Life. Trends Microbiol. 2019, 27, 997–1010. [Google Scholar] [CrossRef] [PubMed]
- Moloney, E.B.; de Winter, F.; Verhaagen, J. ALS as a distal axonopathy: Molecular mechanisms affecting neuromuscular junction stability in the presymptomatic stages of the disease. Front. Neurosci. 2014, 8, 252. [Google Scholar] [CrossRef] [PubMed]
- Guinane, C.M.; Cotter, P.D. Role of the gut microbiota in health and chronic gastrointestinal disease: Understanding a hidden metabolic organ. Ther. Adv. Gastroenterol. 2013, 6, 295–308. [Google Scholar] [CrossRef]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, Y.; Wang, Z.; Xie, G.; Liu, M.; Yuan, B.; Chai, H.; Wang, W.; Cheng, P. Implications of Gut Microbiota in Neurodegenerative Diseases. Front. Immunol. 2022, 13, 785644. [Google Scholar] [CrossRef]
- Ávila, P.R.M.; Fiorot, M.; Michels, M.; Dominguini, D.; Abatti, M.; Vieira, A.; de Moura, A.B.; Behenck, J.P.; Borba, L.A.; Botelho, M.E.M. Effects of microbiota transplantation and the role of the vagus nerve in gut–brain axis in animals subjected to chronic mild stress. J. Affect. Disord. 2020, 277, 410–416. [Google Scholar] [CrossRef]
- Chidambaram, S.B.; Essa, M.M.; Rathipriya, A.G.; Bishir, M.; Ray, B.; Mahalakshmi, A.M.; Tousif, A.H.; Sakharkar, M.K.; Kashyap, R.S.; Friedland, R.P.; et al. Gut dysbiosis, defective autophagy and altered immune responses in neurodegenerative diseases: Tales of a vicious cycle. Pharmacol. Ther. 2022, 231, 107988. [Google Scholar] [CrossRef]
- Zeng, Q.; Junli, G.; Liu, X.; Chen, C.; Sun, X.; Li, H.; Zhou, Y.; Cui, C.; Wang, Y.; Yang, Y.; et al. Gut dysbiosis and lack of short chain fatty acids in a Chinese cohort of patients with multiple sclerosis. Neurochem. Int. 2019, 129, 104468. [Google Scholar] [CrossRef]
- Wu, S.; Yi, J.; Zhang, Y.-g.; Zhou, J.; Sun, J. Leaky intestine and impaired microbiome in an amyotrophic lateral sclerosis mouse model. Physiol. Rep. 2015, 3, e12356. [Google Scholar] [CrossRef]
- Wang, Q.; Luo, Y.; Ray Chaudhuri, K.; Reynolds, R.; Tan, E.-K.; Pettersson, S. The role of gut dysbiosis in Parkinson’s disease: Mechanistic insights and therapeutic options. Brain 2021, 144, 2571–2593. [Google Scholar] [CrossRef]
- Liu, S.; Gao, J.; Zhu, M.; Liu, K.; Zhang, H.-L. Gut Microbiota and Dysbiosis in Alzheimer’s Disease: Implications for Pathogenesis and Treatment. Mol. Neurobiol. 2020, 57, 5026–5043. [Google Scholar] [CrossRef]
- Wasser, C.I.; Mercieca, E.-C.; Kong, G.; Hannan, A.J.; McKeown, S.J.; Glikmann-Johnston, Y.; Stout, J.C. Gut dysbiosis in Huntington’s disease: Associations among gut microbiota, cognitive performance and clinical outcomes. Brain Commun. 2020, 2, fcaa110. [Google Scholar] [CrossRef]
- Ohsumi, Y. Historical landmarks of autophagy research. Cell Res. 2014, 24, 9–23. [Google Scholar] [CrossRef]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef]
- Ma, X.; Lu, C.; Chen, Y.; Li, S.; Ma, N.; Tao, X.; Li, Y.; Wang, J.; Zhou, M.; Yan, Y.B.; et al. CCT2 is an aggrephagy receptor for clearance of solid protein aggregates. Cell 2022, 185, 1325–1345.e1322. [Google Scholar] [CrossRef]
- Bourdenx, M.; Koulakiotis, N.S.; Sanoudou, D.; Bezard, E.; Dehay, B.; Tsarbopoulos, A. Protein aggregation and neurodegeneration in prototypical neurodegenerative diseases: Examples of amyloidopathies, tauopathies and synucleinopathies. Prog. Neurobiol. 2017, 155, 171–193. [Google Scholar] [CrossRef]
- Aziz, M.N.M.; Kumar, J.; Muhammad Nawawi, K.N.; Raja Ali, R.A.; Mokhtar, N.M. Irritable Bowel Syndrome, Depression, and Neurodegeneration: A Bidirectional Communication from Gut to Brain. Nutrients 2021, 13, 3061. [Google Scholar] [CrossRef]
- Kesika, P.; Suganthy, N.; Sivamaruthi, B.S.; Chaiyasut, C. Role of gut-brain axis, gut microbial composition, and probiotic intervention in Alzheimer’s disease. Life Sci. 2021, 264, 118627. [Google Scholar] [CrossRef]
- Roy Sarkar, S.; Banerjee, S. Gut microbiota in neurodegenerative disorders. J. Neuroimmunol. 2019, 328, 98–104. [Google Scholar] [CrossRef]
- Russo, R.; Cristiano, C.; Avagliano, C.; De Caro, C.; La Rana, G.; Raso, G.M.; Canani, R.B.; Meli, R.; Calignano, A. Gut-brain axis: Role of lipids in the regulation of inflammation, pain and CNS diseases. Curr. Med. Chem. 2018, 25, 3930–3952. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; Leyrolle, Q.; Koistinen, V.; Kärkkäinen, O.; Layé, S.; Delzenne, N.; Hanhineva, K. Microbiota-derived metabolites as drivers of gut-brain communication. Gut Microbes 2022, 14, 2102878. [Google Scholar] [CrossRef] [PubMed]
- Rogers, G.B.; Keating, D.J.; Young, R.L.; Wong, M.L.; Licinio, J.; Wesselingh, S. From gut dysbiosis to altered brain function and mental illness: Mechanisms and pathways. Mol. Psychiatry 2016, 21, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, C.; Garagnani, P.; Parini, P.; Giuliani, C.; Santoro, A. Inflammaging: A new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 2018, 14, 576–590. [Google Scholar] [CrossRef]
- Wu, M.; Luo, Q.; Nie, R.; Yang, X.; Tang, Z.; Chen, H. Potential implications of polyphenols on aging considering oxidative stress, inflammation, autophagy, and gut microbiota. Crit. Rev. Food Sci. Nutr. 2021, 61, 2175–2193. [Google Scholar] [CrossRef]
- Lapaquette, P.; Bizeau, J.B.; Acar, N.; Bringer, M.A. Reciprocal interactions between gut microbiota and autophagy. World J. Gastroenterol. 2021, 27, 8283–8301. [Google Scholar] [CrossRef]
- Castelli, V.; d’Angelo, M.; Quintiliani, M.; Benedetti, E.; Cifone, M.G.; Cimini, A. The emerging role of probiotics in neurodegenerative diseases: New hope for Parkinson’s disease? Neural Regen. Res. 2021, 16, 628–634. [Google Scholar] [CrossRef]
- Shoubridge, A.P.; Fourrier, C.; Choo, J.M.; Proud, C.G.; Sargeant, T.J.; Rogers, G.B. Gut Microbiome Regulation of Autophagic Flux and Neurodegenerative Disease Risks. Front. Microbiol. 2021, 12, 817433. [Google Scholar] [CrossRef]
- Yin, Y.; Sun, G.; Li, E.; Kiselyov, K.; Sun, D. ER stress and impaired autophagy flux in neuronal degeneration and brain injury. Ageing Res. Rev. 2017, 34, 3–14. [Google Scholar] [CrossRef]
- Hoyles, L.; Snelling, T.; Umlai, U.-K.; Nicholson, J.K.; Carding, S.R.; Glen, R.C.; McArthur, S. Microbiome–host systems interactions: Protective effects of propionate upon the blood–brain barrier. Microbiome 2018, 6, 1–13. [Google Scholar] [CrossRef]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef]
- Cheng, S.; Ma, X.; Geng, S.; Jiang, X.; Li, Y.; Hu, L.; Li, J.; Wang, Y.; Han, X. Fecal microbiota transplantation beneficially regulates intestinal mucosal autophagy and alleviates gut barrier injury. Msystems 2018, 3, e00137-18. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, B.; Xu, H.; Tang, L.; Li, Y.; Gong, L.; Wang, Y.; Li, W. Probiotic Bacillus attenuates oxidative stress-induced intestinal injury via p38-mediated autophagy. Front. Microbiol. 2019, 10, 2185. [Google Scholar] [CrossRef]
- Mitra, S.; Dash, R.; Nishan, A.A.; Habiba, S.U.; Moon, I.S. Brain modulation by the gut microbiota: From disease to therapy. J. Adv. Res. 2022. [Google Scholar] [CrossRef]
- Montiel-Castro, A.; González-Cervantes, R.; Bravo-Ruiseco, G.; Pacheco-Lopez, G. The microbiota-gut-brain axis: Neurobehavioral correlates, health and sociality. Front. Integr. Neurosci. 2013, 7, 70. [Google Scholar] [CrossRef]
- Cryan, J.F.; Dinan, T.G. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef]
- Wang, H.-X.; Wang, Y.-P.; Chen, X. Gut Microbiota-brain Axis. Chin. Med. J. 2016, 129, 2373–2380. [Google Scholar] [CrossRef]
- Schemann, M.; Neunlist, M. The human enteric nervous system. Neurogastroenterol. Motil. 2004, 16, 55–59. [Google Scholar] [CrossRef]
- Sherwin, E.; Sandhu, K.V.; Dinan, T.G.; Cryan, J.F. May the Force Be With You: The Light and Dark Sides of the Microbiota–Gut–Brain Axis in Neuropsychiatry. CNS Drugs 2016, 30, 1019–1041. [Google Scholar] [CrossRef]
- Dinan, T.G.; Stilling, R.M.; Stanton, C.; Cryan, J.F. Collective unconscious: How gut microbes shape human behavior. J. Psychiatr. Res. 2015, 63, 1–9. [Google Scholar] [CrossRef]
- Browning, K.N.; Verheijden, S.; Boeckxstaens, G.E. The Vagus Nerve in Appetite Regulation, Mood, and Intestinal Inflammation. Gastroenterology 2017, 152, 730–744. [Google Scholar] [CrossRef] [PubMed]
- Sherwin, E.; Dinan, T.G.; Cryan, J.F. Recent developments in understanding the role of the gut microbiota in brain health and disease. Ann. N. Y. Acad. Sci. 2018, 1420, 5–25. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Haq, R.; Schlachetzki, J.C.M.; Glass, C.K.; Mazmanian, S.K. Microbiome–microglia connections via the gut–brain axis. J. Exp. Med. 2018, 216, 41–59. [Google Scholar] [CrossRef] [PubMed]
- Pascale, A.; Marchesi, N.; Marelli, C.; Coppola, A.; Luzi, L.; Govoni, S.; Giustina, A.; Gazzaruso, C. Microbiota and metabolic diseases. Endocrine 2018, 61, 357–371. [Google Scholar] [CrossRef]
- Nagashima, H.; Morio, Y.; Meshitsuka, S.; Yamane, K.; Nanjo, Y.; Teshima, R. High-resolution nuclear magnetic resonance spectroscopic study of metabolites in the cerebrospinal fluid of patients with cervical myelopathy and lumbar radiculopathy. Eur. Spine J. 2010, 19, 1363–1368. [Google Scholar] [CrossRef]
- Foster, J.A.; Lyte, M.; Meyer, E.; Cryan, J.F. Gut Microbiota and Brain Function: An Evolving Field in Neuroscience. Int. J. Neuropsychopharmacol. 2016, 19, pyv114. [Google Scholar] [CrossRef]
- Mu, C.; Yang, Y.; Zhu, W. Gut Microbiota: The Brain Peacekeeper. Front. Microbiol. 2016, 7, 345. [Google Scholar] [CrossRef]
- Das, B.; Nair, G.B. Homeostasis and dysbiosis of the gut microbiome in health and disease. J. Biosci. 2019, 44, 117. [Google Scholar] [CrossRef]
- Spielman, L.J.; Gibson, D.L.; Klegeris, A. Unhealthy gut, unhealthy brain: The role of the intestinal microbiota in neurodegenerative diseases. Neurochem. Int. 2018, 120, 149–163. [Google Scholar] [CrossRef]
- Carino, A.; Marchianò, S.; Biagioli, M.; Scarpelli, P.; Bordoni, M.; Di Giorgio, C.; Roselli, R.; Fiorucci, C.; Monti, M.C.; Distrutti, E.; et al. The bile acid activated receptors GPBAR1 and FXR exert antagonistic effects on autophagy. FASEB J. 2021, 35, e21271. [Google Scholar] [CrossRef]
- Iannucci, L.F.; Sun, J.; Singh, B.K.; Zhou, J.; Kaddai, V.A.; Lanni, A.; Yen, P.M.; Sinha, R.A. Short chain fatty acids induce UCP2-mediated autophagy in hepatic cells. Biochem. Biophys Res. Commun. 2016, 480, 461–467. [Google Scholar] [CrossRef]
- Nikoletopoulou, V.; Papandreou, M.E.; Tavernarakis, N. Autophagy in the physiology and pathology of the central nervous system. Cell Death Differ. 2015, 22, 398–407. [Google Scholar] [CrossRef]
- Cavallucci, V.; Fidaleo, M.; Pani, G. Nutrients and neurogenesis: The emerging role of autophagy and gut microbiota. Curr. Opin. Pharmacol. 2020, 50, 46–52. [Google Scholar] [CrossRef]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. Embo J 2017, 36, 1811–1836. [Google Scholar] [CrossRef]
- Boya, P.; Reggiori, F.; Codogno, P. Emerging regulation and functions of autophagy. Nat Cell Biol 2013, 15, 713–720. [Google Scholar] [CrossRef]
- Butsch, T.J.; Ghosh, B.; Bohnert, K.A. Organelle-Specific Autophagy in Cellular Aging and Rejuvenation. Adv. Geriatr. Med. Res. 2021, 3. [Google Scholar] [CrossRef]
- Lamark, T.; Johansen, T. Aggrephagy: Selective Disposal of Protein Aggregates by Macroautophagy. Int. J. Cell Biol. 2012, 2012, 736905. [Google Scholar] [CrossRef]
- Simonsen, A.; Wollert, T. Don’t forget to be picky—Selective autophagy of protein aggregates in neurodegenerative diseases. Curr. Opin. Cell Biol. 2022, 75, 102064. [Google Scholar] [CrossRef]
- Wileman, T. Autophagy as a defence against intracellular pathogens. Essays Biochem. 2013, 55, 153–163. [Google Scholar] [CrossRef]
- Mizushima, N.; White, E.; Rubinsztein, D.C. Breakthroughs and bottlenecks in autophagy research. Trends Mol. Med. 2021, 27, 835–838. [Google Scholar] [CrossRef]
- Johansen, T.; Lamark, T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011, 7, 279–296. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, G.; Holtzman, D.M. Amyloid-β and Tau at the Crossroads of Alzheimer’s Disease. In Tau Biology; Takashima, A., Wolozin, B., Buee, L., Eds.; Springer: Singapore, 2019; pp. 187–203. [Google Scholar]
- Recasens, A.; Dehay, B. Alpha-synuclein spreading in Parkinson’s disease. Front. Neuroanat. 2014, 8, 159. [Google Scholar] [CrossRef] [PubMed]
- Dani, M.; Brooks, D.J.; Edison, P. Tau imaging in neurodegenerative diseases. Eur. J. Nucl. Med. Mol. Imaging 2016, 43, 1139–1150. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.B.; Lee, V.M.Y.; Trojanowski, J.Q. Gains or losses: Molecular mechanisms of TDP43-mediated neurodegeneration. Nat. Rev. Neurosci. 2012, 13, 38–50. [Google Scholar] [CrossRef]
- Deng, H.; Gao, K.; Jankovic, J. The role of FUS gene variants in neurodegenerative diseases. Nat. Rev. Neurol. 2014, 10, 337–348. [Google Scholar] [CrossRef]
- Saudou, F.; Humbert, S. The Biology of Huntingtin. Neuron 2016, 89, 910–926. [Google Scholar] [CrossRef]
- Conway, O.; Akpinar, H.A.; Rogov, V.V.; Kirkin, V. Selective Autophagy Receptors in Neuronal Health and Disease. J. Mol. Biol. 2020, 432, 2483–2509. [Google Scholar] [CrossRef]
- Dash, R.; Ali, M.C.; Jahan, I.; Munni, Y.A.; Mitra, S.; Hannan, M.A.; Timalsina, B.; Oktaviani, D.F.; Choi, H.J.; Moon, I.S. Emerging potential of cannabidiol in reversing proteinopathies. Ageing Res. Rev. 2021, 65, 101209. [Google Scholar] [CrossRef]
- Dash, R.; Jahan, I.; Ali, M.C.; Mitra, S.; Munni, Y.A.; Timalsina, B.; Hannan, M.A.; Moon, I.S. Potential roles of natural products in the targeting of proteinopathic neurodegenerative diseases. Neurochem. Int. 2021, 145, 105011. [Google Scholar] [CrossRef]
- Dash, R.; Mitra, S.; Ali, M.C.; Oktaviani, D.F.; Hannan, M.A.; Choi, S.M.; Moon, I.S. Phytosterols: Targeting Neuroinflammation in Neurodegeneration. Curr. Pharm. Des. 2021, 27, 383–401. [Google Scholar] [CrossRef]
- Hannan, M.A.; Rahman, M.A.; Rahman, M.S.; Sohag, A.A.M.; Dash, R.; Hossain, K.S.; Farjana, M.; Uddin, M.J. Intermittent fasting, a possible priming tool for host defense against SARS-CoV-2 infection: Crosstalk among calorie restriction, autophagy and immune response. Immunol. Lett. 2020, 226, 38–45. [Google Scholar] [CrossRef]
- Mitra, S.; Dash, R.; Munni, Y.A.; Selsi, N.J.; Akter, N.; Uddin, M.N.; Mazumder, K.; Moon, I.S. Natural Products Targeting Hsp90 for a Concurrent Strategy in Glioblastoma and Neurodegeneration. Metabolites 2022, 12, 1153. [Google Scholar] [CrossRef]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458–467. [Google Scholar] [CrossRef]
- Lynch-Day, M.A.; Klionsky, D.J. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010, 584, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, L.; Che, K.H.; Funderburk, S.F.; Pan, L.; Pan, N.; Zhang, M.; Yue, Z.; Zhao, Y. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat. Commun. 2012, 3, 662. [Google Scholar] [CrossRef]
- Nishida, Y.; Arakawa, S.; Fujitani, K.; Yamaguchi, H.; Mizuta, T.; Kanaseki, T.; Komatsu, M.; Otsu, K.; Tsujimoto, Y.; Shimizu, S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 2009, 461, 654–658. [Google Scholar] [CrossRef]
- Jahreiss, L.; Menzies, F.M.; Rubinsztein, D.C. The Itinerary of Autophagosomes: From Peripheral Formation to Kiss-and-Run Fusion with Lysosomes. Traffic 2008, 9, 574–587. [Google Scholar] [CrossRef] [PubMed]
- Hannan, M.A.; Dash, R.; Sohag, A.A.M.; Haque, M.N.; Moon, I.S. Neuroprotection Against Oxidative Stress: Phytochemicals Targeting TrkB Signaling and the Nrf2-ARE Antioxidant System. Front. Mol. Neurosci. 2020, 13, 116. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.S.U.; Samanta, S.; Dash, R.; Karpiński, T.M.; Habibi, E.; Sadiq, A.; Ahmadi, A.; Bunagu, S. The neuroprotective effects of fisetin, a natural flavonoid in neurodegenerative diseases: Focus on the role of oxidative stress. Front Pharm. 2022, 13, 1015835. [Google Scholar] [CrossRef]
- Mitra, S.; Dash, R.; Sohel, M.; Chowdhury, A.; Munni, Y.A.; Ali, M.C.; Hannan, M.A.; Islam, M.T.; Moon, I.S. Targeting estrogen signaling in the radiation-induced neurodegeneration: Possible role of phytoestrogens. Curr. Neuropharmacol. 2022. [Google Scholar] [CrossRef]
- Rahman, M.A.; Dash, R.; Sohag, A.A.M.; Alam, M.; Rhim, H.; Ha, H.; Moon, I.S.; Uddin, M.J.; Hannan, M.A. Prospects of Marine Sterols against Pathobiology of Alzheimer’s Disease: Pharmacological Insights and Technological Advances. Mar. Drugs 2021, 19, 167. [Google Scholar] [CrossRef] [PubMed]
- Ripon, M.K.H.; Lee, H.; Dash, R.; Choi, H.J.; Oktaviani, D.F.; Moon, I.S.; Haque, M.N. N-acetyl-D-glucosamine kinase binds dynein light chain roadblock 1 and promotes protein aggregate clearance. Cell Death Dis. 2020, 11, 619. [Google Scholar] [CrossRef]
- Winslow, A.R.; Chen, C.W.; Corrochano, S.; Acevedo-Arozena, A.; Gordon, D.E.; Peden, A.A.; Lichtenberg, M.; Menzies, F.M.; Ravikumar, B.; Imarisio, S.; et al. α-Synuclein impairs macroautophagy: Implications for Parkinson’s disease. J Cell Biol. 2010, 190, 1023–1037. [Google Scholar] [CrossRef]
- Merenlender-Wagner, A.; Malishkevich, A.; Shemer, Z.; Udawela, M.; Gibbons, A.; Scarr, E.; Dean, B.; Levine, J.; Agam, G.; Gozes, I. Autophagy has a key role in the pathophysiology of schizophrenia. Mol. Psychiatry 2015, 20, 126–132. [Google Scholar] [CrossRef]
- Kim, H.J.; Cho, M.H.; Shim, W.H.; Kim, J.K.; Jeon, E.Y.; Kim, D.H.; Yoon, S.Y. Deficient autophagy in microglia impairs synaptic pruning and causes social behavioral defects. Mol. Psychiatry 2017, 22, 1576–1584. [Google Scholar] [CrossRef]
- Sakai, M.; Yu, Z.; Hirayama, R.; Nakasato, M.; Kikuchi, Y.; Ono, C.; Komatsu, H.; Nakanishi, M.; Yoshii, H.; Stellwagen, D.; et al. Deficient Autophagy in Microglia Aggravates Repeated Social Defeat Stress-Induced Social Avoidance. Neural Plast. 2022, 2022, 7503553. [Google Scholar] [CrossRef]
- Chen, A.; Xiong, L.-J.; Tong, Y.; Mao, M. Neuroprotective effect of brain-derived neurotrophic factor mediated by autophagy through the PI3K/Akt/mTOR pathway. Mol. Med. Rep. 2013, 8, 1011–1016. [Google Scholar] [CrossRef]
- Jing, C.h.; Wang, L.; Liu, P.p.; Wu, C.; Ruan, D.; Chen, G. Autophagy activation is associated with neuroprotection against apoptosis via a mitochondrial pathway in a rat model of subarachnoid hemorrhage. Neuroscience 2012, 213, 144–153. [Google Scholar] [CrossRef]
- Winslow, A.R.; Rubinsztein, D.C. Autophagy in neurodegeneration and development. Biochim. Et Biophys. Acta (BBA)—Mol. Basis Dis. 2008, 1782, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Larabi, A.; Barnich, N.; Nguyen, H.T.T. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy 2020, 16, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Elfil, M.; Kamel, S.; Kandil, M.; Koo, B.B.; Schaefer, S.M. Implications of the Gut Microbiome in Parkinson’s Disease. Mov. Disord. 2020, 35, 921–933. [Google Scholar] [CrossRef]
- Toh, T.S.; Chong, C.W.; Lim, S.-Y.; Bowman, J.; Cirstea, M.; Lin, C.-H.; Chen, C.-C.; Appel-Cresswell, S.; Finlay, B.B.; Tan, A.H. Gut microbiome in Parkinson’s disease: New insights from meta-analysis. Park. Relat. Disord. 2022, 94, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Boertien, J.M.; Pereira, P.A.B.; Aho, V.T.E.; Scheperjans, F. Increasing Comparability and Utility of Gut Microbiome Studies in Parkinson’s Disease: A Systematic Review. J. Park. Dis. 2019, 9, S297–S312. [Google Scholar] [CrossRef]
- Pavan, S.; Prabhu, A.N.; Prasad Gorthi, S.; Das, B.; Mutreja, A.; Shetty, V.; Ramamurthy, T.; Ballal, M. Exploring the multifactorial aspects of Gut Microbiome in Parkinson’s Disease. Folia Microbiol. 2022, 67, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Wallen, Z.D.; Demirkan, A.; Twa, G.; Cohen, G.; Dean, M.N.; Standaert, D.G.; Sampson, T.R.; Payami, H. Metagenomics of Parkinson’s disease implicates the gut microbiome in multiple disease mechanisms. Nat. Commun. 2022, 13, 6958. [Google Scholar] [CrossRef]
- Hisahara, S.; Shimohama, S. Toxin-Induced and Genetic Animal Models of Parkinson’s Disease. Park. Dis. 2011, 2011, 951709. [Google Scholar] [CrossRef] [PubMed]
- Lai, F.; Jiang, R.; Xie, W.; Liu, X.; Tang, Y.; Xiao, H.; Gao, J.; Jia, Y.; Bai, Q. Intestinal Pathology and Gut Microbiota Alterations in a Methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) Mouse Model of Parkinson’s Disease. Neurochem. Res. 2018, 43, 1986–1999. [Google Scholar] [CrossRef]
- Tasselli, M.; Chaumette, T.; Paillusson, S.; Monnet, Y.; Lafoux, A.; Huchet-Cadiou, C.; Aubert, P.; Hunot, S.; Derkinderen, P.; Neunlist, M. Effects of oral administration of rotenone on gastrointestinal functions in mice. Neurogastroenterol. Motil. 2013, 25, e183–e193. [Google Scholar] [CrossRef]
- Inden, M.; Kitamura, Y.; Takeuchi, H.; Yanagida, T.; Takata, K.; Kobayashi, Y.; Taniguchi, T.; Yoshimoto, K.; Kaneko, M.; Okuma, Y.; et al. Neurodegeneration of mouse nigrostriatal dopaminergic system induced by repeated oral administration of rotenone is prevented by 4-phenylbutyrate, a chemical chaperone. J. Neurochem. 2007, 101, 1491–1504. [Google Scholar] [CrossRef]
- Dodiya, H.B.; Forsyth, C.B.; Voigt, R.M.; Engen, P.A.; Patel, J.; Shaikh, M.; Green, S.J.; Naqib, A.; Roy, A.; Kordower, J.H.; et al. Chronic stress-induced gut dysfunction exacerbates Parkinson’s disease phenotype and pathology in a rotenone-induced mouse model of Parkinson’s disease. Neurobiol. Dis. 2020, 135, 104352. [Google Scholar] [CrossRef]
- Yang, X.; Qian, Y.; Xu, S.; Song, Y.; Xiao, Q. Longitudinal Analysis of Fecal Microbiome and Pathologic Processes in a Rotenone Induced Mice Model of Parkinson’s Disease. Front. Aging Neurosci. 2018, 9, 441. [Google Scholar] [CrossRef]
- Bhattarai, Y.; Si, J.; Pu, M.; Ross, O.A.; McLean, P.J.; Till, L.; Moor, W.; Grover, M.; Kandimalla, K.K.; Margolis, K.G.; et al. Role of gut microbiota in regulating gastrointestinal dysfunction and motor symptoms in a mouse model of Parkinson’s disease. Gut Microbes 2021, 13, 1866974. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Du, Z.-R.; Wang, X.; Luk, K.-H.; Chan, C.-H.; Cao, X.; Zhao, Q.; Zhao, F.; Wong, W.-T.; Wong, K.-H.; et al. Colonic Dopaminergic Neurons Changed Reversely With Those in the Midbrain via Gut Microbiota-Mediated Autophagy in a Chronic Parkinson’s Disease Mice Model. Front. Aging Neurosci. 2021, 13, 649627. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Qian, Y.; He, X.; Mo, C.; Yang, X.; Xiao, Q. Sodium butyrate attenuates rotenone-induced toxicity by activation of autophagy through epigenetically regulating PGC-1α expression in PC12 cells. Brain Res. 2022, 1776, 147749. [Google Scholar] [CrossRef] [PubMed]
- Qiao, C.-M.; Sun, M.-F.; Jia, X.-B.; Shi, Y.; Zhang, B.-P.; Zhou, Z.-L.; Zhao, L.-P.; Cui, C.; Shen, Y.-Q. Sodium butyrate causes α-synuclein degradation by an Atg5-dependent and PI3K/Akt/mTOR-related autophagy pathway. Exp. Cell Res. 2020, 387, 111772. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.; Song, X.; Wang, Z.; Jia, J.; Qing, S.; Huang, L.; Wang, Y.; Wang, S.; Ren, Z.; et al. The intestinal microbial metabolite nicotinamide n-oxide prevents herpes simplex encephalitis via activating mitophagy in microglia. Gut Microbes 2022, 14, 2096989. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Zhu, Y.; Guo, M.; Yin, X.; Liang, M.; Lou, X.; Chen, J.; Zhou, L.; Fan, D.; Shi, L.; et al. The potential mechanism of BPF-induced neurotoxicity in adult zebrafish: Correlation between untargeted metabolomics and gut microbiota. Sci. Total Environ. 2022, 839, 156221. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, B.; Wang, Q.; Liu, Y.; Liu, X.; Wu, B.; Lu, G. Intestinal toxicity and microbial community disorder induced by bisphenol F and bisphenol S in zebrafish. Chemosphere 2021, 280, 130711. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, C.; He, D.; Jiang, N.; Bai, Y.; Xin, Y. Rapamycin and MCC950 modified gut microbiota in experimental autoimmune encephalomyelitis mouse by brain gut axis. Life Sci. 2020, 253, 117747. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, X.; Ta, X.; Luo, M.; Chang, X.; Wang, H. Fecal microbiome transplantation attenuates manganese-induced neurotoxicity through regulation of the apelin signaling pathway by inhibition of autophagy in mouse brain. Ecotoxicol. Environ. Saf. 2022, 242, 113925. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, X.; Zhang, J.; Xia, Y.; Qiu, J.; Wang, T.; Qiu, Y.; Qin, X.; Wang, B.; Zou, Z.; et al. Heterozygous disruption of beclin 1 mitigates arsenite-induced neurobehavioral deficits via reshaping gut microbiota-brain axis. J. Hazard. Mater. 2020, 398, 122748. [Google Scholar] [CrossRef]
- Martin, P.K.; Marchiando, A.; Xu, R.; Rudensky, E.; Yeung, F.; Schuster, S.L.; Kernbauer, E.; Cadwell, K. Autophagy proteins suppress protective type I interferon signalling in response to the murine gut microbiota. Nat. Microbiol. 2018, 3, 1131–1141. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, H.; Zeng, M.; Tang, X.; Zhu, X.; Guo, Y.; Qi, L.; Xie, Y.; Zhang, M.; Chen, D. Maternal high sugar and fat diet benefits offspring brain function via targeting on the gut-brain axis. Aging (Albany NY) 2021, 13, 10240–10274. [Google Scholar] [CrossRef]
- Chen, P.; Chen, F.; Lei, J.; Li, Q.; Zhou, B. Activation of the miR-34a-Mediated SIRT1/mTOR Signaling Pathway by Urolithin A Attenuates D-Galactose-Induced Brain Aging in Mice. Neurotherapeutics 2019, 16, 1269–1282. [Google Scholar] [CrossRef]
- Bonfili, L.; Cecarini, V.; Berardi, S.; Scarpona, S.; Suchodolski, J.S.; Nasuti, C.; Fiorini, D.; Boarelli, M.C.; Rossi, G.; Eleuteri, A.M. Microbiota modulation counteracts Alzheimer’s disease progression influencing neuronal proteolysis and gut hormones plasma levels. Sci. Rep. 2017, 7, 2426. [Google Scholar] [CrossRef]
- Bonfili, L.; Cecarini, V.; Cuccioloni, M.; Angeletti, M.; Berardi, S.; Scarpona, S.; Rossi, G.; Eleuteri, A.M. SLAB51 Probiotic Formulation Activates SIRT1 Pathway Promoting Antioxidant and Neuroprotective Effects in an AD Mouse Model. Mol. Neurobiol. 2018, 55, 7987–8000. [Google Scholar] [CrossRef]
- Chen, D.; Yang, X.; Yang, J.; Lai, G.; Yong, T.; Tang, X.; Shuai, O.; Zhou, G.; Xie, Y.; Wu, Q. Prebiotic Effect of Fructooligosaccharides from Morinda officinalis on Alzheimer’s Disease in Rodent Models by Targeting the Microbiota-Gut-Brain Axis. Front. Aging Neurosci. 2017, 9, 403. [Google Scholar] [CrossRef]
- Wolters, E.; Braak, H. Parkinson’s disease: Premotor clinico-pathological correlations. J. Neural. Transm. Suppl. 2006, 70, 309–319. [Google Scholar] [CrossRef]
- Braak, H.; Braak, E. Pathoanatomy of Parkinson’s disease. J. Neurol. 2000, 247 (Suppl. S2), II3–II10. [Google Scholar] [CrossRef]
- Hawkes, C.H.; Del Tredici, K.; Braak, H. Parkinson’s disease: A dual-hit hypothesis. Neuropathol. Appl. Neurobiol. 2007, 33, 599–614. [Google Scholar] [CrossRef]
- Zhang, H.; Tong, R.; Bai, L.; Shi, J.; Ouyang, L. Emerging targets and new small molecule therapies in Parkinson’s disease treatment. Bioorg. Med. Chem. 2016, 24, 1419–1430. [Google Scholar] [CrossRef] [PubMed]
- Warren, N.; O’Gorman, C.; Lehn, A.; Siskind, D. Dopamine dysregulation syndrome in Parkinson’s disease: A systematic review of published cases. J. Neurol. Neurosurg. Psychiatry 2017, 88, 1060–1064. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T.R.; Debelius, J.W.; Thron, T.; Janssen, S.; Shastri, G.G.; Ilhan, Z.E.; Challis, C.; Schretter, C.E.; Rocha, S.; Gradinaru, V.; et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 2016, 167, 1469–1480.e1412. [Google Scholar] [CrossRef] [PubMed]
- Su, A.; Gandhy, R.; Barlow, C.; Triadafilopoulos, G. A practical review of gastrointestinal manifestations in Parkinson’s disease. Park. Relat. Disord. 2017, 39, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Edwards, L.L.; Pfeiffer, R.F.; Quigley, E.M.; Hofman, R.; Balluff, M. Gastrointestinal symptoms in Parkinson’s disease. Mov. Disord. 1991, 6, 151–156. [Google Scholar] [CrossRef]
- Savica, R.; Carlin, J.M.; Grossardt, B.R.; Bower, J.H.; Ahlskog, J.E.; Maraganore, D.M.; Bharucha, A.E.; Rocca, W.A. Medical records documentation of constipation preceding Parkinson disease: A case-control study. Neurology 2009, 73, 1752–1758. [Google Scholar] [CrossRef]
- Scheperjans, F.; Aho, V.; Pereira, P.A.; Koskinen, K.; Paulin, L.; Pekkonen, E.; Haapaniemi, E.; Kaakkola, S.; Eerola-Rautio, J.; Pohja, M.; et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord. 2015, 30, 350–358. [Google Scholar] [CrossRef]
- González-Arancibia, C.; Urrutia-Piñones, J.; Illanes-González, J.; Martinez-Pinto, J.; Sotomayor-Zárate, R.; Julio-Pieper, M.; Bravo, J.A. Do your gut microbes affect your brain dopamine? Psychopharmacology 2019, 236, 1611–1622. [Google Scholar] [CrossRef]
- Altschuler, E. Gastric Helicobacter pylori infection as a cause of idiopathic Parkinson disease and non-arteric anterior optic ischemic neuropathy. Med. Hypotheses 1996, 47, 413–414. [Google Scholar] [CrossRef]
- Queipo-Ortuño, M.I.; Seoane, L.M.; Murri, M.; Pardo, M.; Gomez-Zumaquero, J.M.; Cardona, F.; Casanueva, F.; Tinahones, F.J. Gut microbiota composition in male rat models under different nutritional status and physical activity and its association with serum leptin and ghrelin levels. PLoS ONE 2013, 8, e65465. [Google Scholar] [CrossRef]
- Chang, R.C.-C. Advanced Understanding of Neurodegenerative Diseases; BoD–Books on Demand: Norderstedt, Germany, 2011. [Google Scholar]
- Minato, T.; Maeda, T.; Fujisawa, Y.; Tsuji, H.; Nomoto, K.; Ohno, K.; Hirayama, M. Progression of Parkinson’s disease is associated with gut dysbiosis: Two-year follow-up study. PLoS ONE 2017, 12, e0187307. [Google Scholar] [CrossRef]
- Tagliabue, A.; Elli, M. The role of gut microbiota in human obesity: Recent findings and future perspectives. Nutr. Metab. Cardiovasc. Dis. 2013, 23, 160–168. [Google Scholar] [CrossRef]
- Unger, M.M.; Spiegel, J.; Dillmann, K.U.; Grundmann, D.; Philippeit, H.; Bürmann, J.; Faßbender, K.; Schwiertz, A.; Schäfer, K.H. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Park. Relat. Disord. 2016, 32, 66–72. [Google Scholar] [CrossRef]
- Bedarf, J.R.; Hildebrand, F.; Coelho, L.P.; Sunagawa, S.; Bahram, M.; Goeser, F.; Bork, P.; Wüllner, U. Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naïve Parkinson’s disease patients. Genome Med. 2017, 9, 39. [Google Scholar] [CrossRef]
- Cásedas, G.; Les, F.; Choya-Foces, C.; Hugo, M.; López, V. The Metabolite Urolithin-A Ameliorates Oxidative Stress in Neuro-2a Cells, Becoming a Potential Neuroprotective Agent. Antioxidants 2020, 9, 177. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, J.; Qiu, J.; Wang, L.; Zhuo, J.; Wang, B.; Sun, D.; Yu, S.; Lou, H. Urolithin A protects dopaminergic neurons in experimental models of Parkinson’s disease by promoting mitochondrial biogenesis through the SIRT1/PGC-1α signaling pathway. Food Funct. 2022, 13, 375–385. [Google Scholar] [CrossRef]
- Kujawska, M.; Jourdes, M.; Kurpik, M.; Szulc, M.; Szaefer, H.; Chmielarz, P.; Kreiner, G.; Krajka-Kuźniak, V.; Mikołajczak, P.; Teissedre, P.L.; et al. Neuroprotective Effects of Pomegranate Juice against Parkinson’s Disease and Presence of Ellagitannins-Derived Metabolite-Urolithin A-In the Brain. Int. J. Mol. Sci. 2019, 21, 202. [Google Scholar] [CrossRef]
- Hazen, S.L.; Brown, J.M. Eggs as a dietary source for gut microbial production of trimethylamine-N-oxide. Am. J. Clin. Nutr. 2014, 100, 741–743. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef]
- Jamal, S.; Kumari, A.; Singh, A.; Goyal, S.; Grover, A. Conformational Ensembles of α-Synuclein Derived Peptide with Different Osmolytes from Temperature Replica Exchange Sampling. Front. Neurosci. 2017, 11, 684. [Google Scholar] [CrossRef]
- Chung, S.J.; Rim, J.H.; Ji, D.; Lee, S.; Yoo, H.S.; Jung, J.H.; Baik, K.; Choi, Y.; Ye, B.S.; Sohn, Y.H.; et al. Gut microbiota-derived metabolite trimethylamine N-oxide as a biomarker in early Parkinson’s disease. Nutrition 2021, 83, 111090. [Google Scholar] [CrossRef] [PubMed]
- Stefanova, N.; Fellner, L.; Reindl, M.; Masliah, E.; Poewe, W.; Wenning, G.K. Toll-like receptor 4 promotes α-synuclein clearance and survival of nigral dopaminergic neurons. Am. J. Pathol. 2011, 179, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Miranda, A.F.; Boegman, R.J.; Beninger, R.J.; Jhamandas, K. Protection against quinolinic acid-mediated excitotoxicity in nigrostriatal dopaminergic neurons by endogenous kynurenic acid. Neuroscience 1997, 78, 967–975. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, J.G.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef]
- da Silva, F.L.; Coelho Cerqueira, E.; de Freitas, M.S.; Gonçalves, D.L.; Costa, L.T.; Follmer, C. Vitamins K interact with N-terminus α-synuclein and modulate the protein fibrillization in vitro. Exploring the interaction between quinones and α-synuclein. Neurochem. Int. 2013, 62, 103–112. [Google Scholar] [CrossRef]
- Weller, J.; Budson, A. Current understanding of Alzheimer’s disease diagnosis and treatment. F1000Res 2018, 7, 1161. [Google Scholar] [CrossRef]
- Nisbet, R.M.; Götz, J. Amyloid-β and Tau in Alzheimer’s Disease: Novel Pathomechanisms and Non-Pharmacological Treatment Strategies. J. Alzheimers Dis. 2018, 64, S517–S527. [Google Scholar] [CrossRef]
- Chen, Y.; Fang, L.; Chen, S.; Zhou, H.; Fan, Y.; Lin, L.; Li, J.; Xu, J.; Chen, Y.; Ma, Y.; et al. Gut Microbiome Alterations Precede Cerebral Amyloidosis and Microglial Pathology in a Mouse Model of Alzheimer’s Disease. Biomed Res. Int. 2020, 2020, 8456596. [Google Scholar] [CrossRef]
- Gu, X.; Zhou, J.; Zhou, Y.; Wang, H.; Si, N.; Ren, W.; Zhao, W.; Fan, X.; Gao, W.; Wei, X.; et al. Huanglian Jiedu decoction remodels the periphery microenvironment to inhibit Alzheimer’s disease progression based on the “brain-gut” axis through multiple integrated omics. Alzheimers Res. Ther. 2021, 13, 44. [Google Scholar] [CrossRef]
- Emery, D.C.; Shoemark, D.K.; Batstone, T.E.; Waterfall, C.M.; Coghill, J.A.; Cerajewska, T.L.; Davies, M.; West, N.X.; Allen, S.J. 16S rRNA Next Generation Sequencing Analysis Shows Bacteria in Alzheimer’s Post-Mortem Brain. Front. Aging Neurosci. 2017, 9, 195. [Google Scholar] [CrossRef]
- Readhead, B.; Haure-Mirande, J.V.; Funk, C.C.; Richards, M.A.; Shannon, P.; Haroutunian, V.; Sano, M.; Liang, W.S.; Beckmann, N.D.; Price, N.D.; et al. Multiscale Analysis of Independent Alzheimer’s Cohorts Finds Disruption of Molecular, Genetic, and Clinical Networks by Human Herpesvirus. Neuron 2018, 99, 64–82.e67. [Google Scholar] [CrossRef]
- Zheng, J.; Zheng, S.J.; Cai, W.J.; Yu, L.; Yuan, B.F.; Feng, Y.Q. Stable isotope labeling combined with liquid chromatography-tandem mass spectrometry for comprehensive analysis of short-chain fatty acids. Anal. Chim. Acta 2019, 1070, 51–59. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.; Xiayu, X.; Shi, C.; Chen, W.; Song, N.; Fu, X.; Zhou, R.; Xu, Y.F.; Huang, L.; et al. Altered Gut Microbiota in a Mouse Model of Alzheimer’s Disease. J. Alzheimers Dis. 2017, 60, 1241–1257. [Google Scholar] [CrossRef]
- Park, J.; Kim, C.H. Regulation of common neurological disorders by gut microbial metabolites. Exp. Mol. Med. 2021, 53, 1821–1833. [Google Scholar] [CrossRef]
- Nagpal, R.; Neth, B.J.; Wang, S.; Craft, S.; Yadav, H. Modified Mediterranean-ketogenic diet modulates gut microbiome and short-chain fatty acids in association with Alzheimer’s disease markers in subjects with mild cognitive impairment. EBioMedicine 2019, 47, 529–542. [Google Scholar] [CrossRef]
- Ferrante, R.J.; Kubilus, J.K.; Lee, J.; Ryu, H.; Beesen, A.; Zucker, B.; Smith, K.; Kowall, N.W.; Ratan, R.R.; Luthi-Carter, R.; et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington’s disease mice. J. Neurosci. 2003, 23, 9418–9427. [Google Scholar] [CrossRef]
- Ho, L.; Ono, K.; Tsuji, M.; Mazzola, P.; Singh, R.; Pasinetti, G.M. Protective roles of intestinal microbiota derived short chain fatty acids in Alzheimer’s disease-type beta-amyloid neuropathological mechanisms. Expert. Rev. Neurother. 2018, 18, 83–90. [Google Scholar] [CrossRef]
- Gong, Z.; Huang, J.; Xu, B.; Ou, Z.; Zhang, L.; Lin, X.; Ye, X.; Kong, X.; Long, D.; Sun, X.; et al. Urolithin A attenuates memory impairment and neuroinflammation in APP/PS1 mice. J. Neuroinflamm. 2019, 16, 62. [Google Scholar] [CrossRef]
- Ballesteros-Álvarez, J.; Nguyen, W.; Sivapatham, R.; Rane, A.; Andersen, J.K. Urolithin A reduces amyloid-beta load and improves cognitive deficits uncorrelated with plaque burden in a mouse model of Alzheimer’s disease. Geroscience 2022, 45, 1095–1113. [Google Scholar] [CrossRef]
- Ali, T.; Kim, T.; Rehman, S.U.; Khan, M.S.; Amin, F.U.; Khan, M.; Ikram, M.; Kim, M.O. Natural Dietary Supplementation of Anthocyanins via PI3K/Akt/Nrf2/HO-1 Pathways Mitigate Oxidative Stress, Neurodegeneration, and Memory Impairment in a Mouse Model of Alzheimer’s Disease. Mol. Neurobiol. 2018, 55, 6076–6093. [Google Scholar] [CrossRef]
- Winter, A.N.; Bickford, P.C. Anthocyanins and Their Metabolites as Therapeutic Agents for Neurodegenerative Disease. Antioxidants 2019, 8, 333. [Google Scholar] [CrossRef] [PubMed]
- Maitre, M.; Klein, C.; Patte-Mensah, C.; Mensah-Nyagan, A.G. Tryptophan metabolites modify brain Aβ peptide degradation: A role in Alzheimer’s disease? Prog. Neurobiol. 2020, 190, 101800. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Tai, J.; Xu, H.; Lu, X.; Meng, D. Xanthoceraside Could Ameliorate Alzheimer’s Disease Symptoms of Rats by Affecting the Gut Microbiota Composition and Modulating the Endogenous Metabolite Levels. Front. Pharm. 2019, 10, 1035. [Google Scholar] [CrossRef] [PubMed]
- Drewes, G.; Ebneth, A.; Preuss, U.; Mandelkow, E.M.; Mandelkow, E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 1997, 89, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Nagase, Y.; Hirose, J.; Tokuyama, N.; Yasui, T.; Kadono, Y.; Ueki, K.; Kadowaki, T.; Nakamura, K.; Tanaka, S. Regulation of bone resorption and sealing zone formation in osteoclasts occurs through protein kinase B-mediated microtubule stabilization. J. Bone Min. Res. 2013, 28, 1191–1202. [Google Scholar] [CrossRef]
- Tseng, H.C.; Graves, D.J. Natural methylamine osmolytes, trimethylamine N-oxide and betaine, increase tau-induced polymerization of microtubules. Biochem. Biophys. Res. Commun. 1998, 250, 726–730. [Google Scholar] [CrossRef]
- Allison, A.C. The possible role of vitamin K deficiency in the pathogenesis of Alzheimer’s disease and in augmenting brain damage associated with cardiovascular disease. Med. Hypotheses 2001, 57, 151–155. [Google Scholar] [CrossRef]
- Douaud, G.; Refsum, H.; de Jager, C.A.; Jacoby, R.; Nichols, T.E.; Smith, S.M.; Smith, A.D. Preventing Alzheimer’s disease-related gray matter atrophy by B-vitamin treatment. Proc. Natl. Acad. Sci. USA 2013, 110, 9523–9528. [Google Scholar] [CrossRef]
- Walker, F.O. Huntington’s disease. Lancet 2007, 369, 218–228. [Google Scholar] [CrossRef]
- Nayak, A.; Ansar, R.; Verma, S.K.; Bonifati, D.M.; Kishore, U. Huntington’s Disease: An Immune Perspective. Neurol. Res. Int. 2011, 2011, 563784. [Google Scholar] [CrossRef]
- Kong, G.; Ellul, S.; Narayana, V.K.; Kanojia, K.; Ha, H.T.T.; Li, S.; Renoir, T.; Cao, K.L.; Hannan, A.J. An integrated metagenomics and metabolomics approach implicates the microbiota-gut-brain axis in the pathogenesis of Huntington’s disease. Neurobiol. Dis. 2021, 148, 105199. [Google Scholar] [CrossRef]
- Kong, G.; Cao, K.L.; Judd, L.M.; Li, S.; Renoir, T.; Hannan, A.J. Microbiome profiling reveals gut dysbiosis in a transgenic mouse model of Huntington’s disease. Neurobiol. Dis. 2020, 135, 104268. [Google Scholar] [CrossRef]
- Du, G.; Dong, W.; Yang, Q.; Yu, X.; Ma, J.; Gu, W.; Huang, Y. Altered Gut Microbiota Related to Inflammatory Responses in Patients With Huntington’s Disease. Front. Immunol. 2020, 11, 603594. [Google Scholar] [CrossRef]
- Stan, T.L.; Soylu-Kucharz, R.; Burleigh, S.; Prykhodko, O.; Cao, L.; Franke, N.; Sjögren, M.; Haikal, C.; Hållenius, F.; Björkqvist, M. Increased intestinal permeability and gut dysbiosis in the R6/2 mouse model of Huntington’s disease. Sci. Rep. 2020, 10, 18270. [Google Scholar] [CrossRef]
- Stoy, N.; Mackay, G.M.; Forrest, C.M.; Christofides, J.; Egerton, M.; Stone, T.W.; Darlington, L.G. Tryptophan metabolism and oxidative stress in patients with Huntington’s disease. J. Neurochem. 2005, 93, 611–623. [Google Scholar] [CrossRef]
- Sun, X.; Zhu, J.; Sun, X.Y.; Ji, M.; Yu, X.L.; Liu, R.T. Ellagic acid rescues motor and cognitive deficits in the R6/2 mouse model of Huntington’s disease by lowering mutant huntingtin protein. Food Funct. 2020, 11, 1334–1348. [Google Scholar] [CrossRef]
- Rosen, D.R.; Siddique, T.; Patterson, D.; Figlewicz, D.A.; Sapp, P.; Hentati, A.; Donaldson, D.; Goto, J.; O’Regan, J.P.; Deng, H.X.; et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993, 362, 59–62. [Google Scholar] [CrossRef]
- Nicholson, K.; Bjornevik, K.; Abu-Ali, G.; Chan, J.; Cortese, M.; Dedi, B.; Jeon, M.; Xavier, R.; Huttenhower, C.; Ascherio, A.; et al. The human gut microbiota in people with amyotrophic lateral sclerosis. Amyotroph Lateral Scler Front. Degener 2021, 22, 186–194. [Google Scholar] [CrossRef]
- Ngo, S.T.; Restuadi, R.; McCrae, A.F.; Van Eijk, R.P.; Garton, F.; Henderson, R.D.; Wray, N.R.; McCombe, P.A.; Steyn, F.J. Progression and survival of patients with motor neuron disease relative to their fecal microbiota. Amyotroph Lateral Scler Front. Degener 2020, 21, 549–562. [Google Scholar] [CrossRef]
- Schwiertz, A.; Taras, D.; Schäfer, K.; Beijer, S.; Bos, N.A.; Donus, C.; Hardt, P.D. Microbiota and SCFA in lean and overweight healthy subjects. Obesity 2010, 18, 190–195. [Google Scholar] [CrossRef]
- O’Reilly, É.J.; Wang, M.; Adami, H.O.; Alonso, A.; Bernstein, L.; van den Brandt, P.; Buring, J.; Daugherty, S.; Deapen, D.; Freedman, D.M.; et al. Prediagnostic body size and risk of amyotrophic lateral sclerosis death in 10 studies. Amyotroph Lateral Scler Front. Degener 2018, 19, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Paganoni, S.; Deng, J.; Jaffa, M.; Cudkowicz, M.E.; Wills, A.M. Body mass index, not dyslipidemia, is an independent predictor of survival in amyotrophic lateral sclerosis. Muscle Nerve 2011, 44, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Blacher, E.; Bashiardes, S.; Shapiro, H.; Rothschild, D.; Mor, U.; Dori-Bachash, M.; Kleimeyer, C.; Moresi, C.; Harnik, Y.; Zur, M.; et al. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature 2019, 572, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Iłzecka, J.; Kocki, T.; Stelmasiak, Z.; Turski, W.A. Endogenous protectant kynurenic acid in amyotrophic lateral sclerosis. Acta Neurol. Scand 2003, 107, 412–418. [Google Scholar] [CrossRef]
- Sospedra, M.; Martin, R. Immunology of multiple sclerosis. Annu. Rev. Immunol. 2005, 23, 683–747. [Google Scholar] [CrossRef]
- Chen, J.; Chia, N.; Kalari, K.R.; Yao, J.Z.; Novotna, M.; Paz Soldan, M.M.; Luckey, D.H.; Marietta, E.V.; Jeraldo, P.R.; Chen, X.; et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci. Rep. 2016, 6, 28484. [Google Scholar] [CrossRef]
- Yokote, H.; Miyake, S.; Croxford, J.L.; Oki, S.; Mizusawa, H.; Yamamura, T. NKT cell-dependent amelioration of a mouse model of multiple sclerosis by altering gut flora. Am. J. Pathol. 2008, 173, 1714–1723. [Google Scholar] [CrossRef]
- Hirschberg, S.; Gisevius, B.; Duscha, A.; Haghikia, A. Implications of Diet and The Gut Microbiome in Neuroinflammatory and Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 3109. [Google Scholar] [CrossRef]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef]
- Jangi, S.; Gandhi, R.; Cox, L.M.; Li, N.; von Glehn, F.; Yan, R.; Patel, B.; Mazzola, M.A.; Liu, S.; Glanz, B.L.; et al. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 2016, 7, 12015. [Google Scholar] [CrossRef]
- Gaetani, L.; Boscaro, F.; Pieraccini, G.; Calabresi, P.; Romani, L.; Di Filippo, M.; Zelante, T. Host and Microbial Tryptophan Metabolic Profiling in Multiple Sclerosis. Front. Immunol. 2020, 11, 157. [Google Scholar] [CrossRef]
- Nourbakhsh, B.; Bhargava, P.; Tremlett, H.; Hart, J.; Graves, J.; Waubant, E. Altered tryptophan metabolism is associated with pediatric multiple sclerosis risk and course. Ann. Clin. Transl. Neurol. 2018, 5, 1211–1221. [Google Scholar] [CrossRef]
- Lieben, C.K.; Blokland, A.; Deutz, N.E.; Jansen, W.; Han, G.; Hupperts, R.M. Intake of tryptophan-enriched whey protein acutely enhances recall of positive loaded words in patients with multiple sclerosis. Clin. Nutr. 2018, 37, 321–328. [Google Scholar] [CrossRef]
- Rothhammer, V.; Mascanfroni, I.D.; Bunse, L.; Takenaka, M.C.; Kenison, J.E.; Mayo, L.; Chao, C.C.; Patel, B.; Yan, R.; Blain, M.; et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016, 22, 586–597. [Google Scholar] [CrossRef]
- Rothhammer, V.; Borucki, D.M.; Tjon, E.C.; Takenaka, M.C.; Chao, C.C.; Ardura-Fabregat, A.; de Lima, K.A.; Gutiérrez-Vázquez, C.; Hewson, P.; Staszewski, O.; et al. Microglial control of astrocytes in response to microbial metabolites. Nature 2018, 557, 724–728. [Google Scholar] [CrossRef]
- Bhargava, P.; Smith, M.D.; Mische, L.; Harrington, E.; Fitzgerald, K.C.; Martin, K.; Kim, S.; Reyes, A.A.; Gonzalez-Cardona, J.; Volsko, C.; et al. Bile acid metabolism is altered in multiple sclerosis and supplementation ameliorates neuroinflammation. J. Clin. Investig. 2020, 130, 3467–3482. [Google Scholar] [CrossRef]
- Sarkar, A.; Lehto, S.M.; Harty, S.; Dinan, T.G.; Cryan, J.F.; Burnet, P.W.J. Psychobiotics and the Manipulation of Bacteria–Gut–Brain Signals. Trends Neurosci. 2016, 39, 763–781. [Google Scholar] [CrossRef]
- Zhou, L.; Foster, J.A. Psychobiotics and the gut-brain axis: In the pursuit of happiness. Neuropsychiatr. Dis. Treat. 2015, 11, 715–723. [Google Scholar] [CrossRef]
- Barrio, C.; Arias-Sánchez, S.; Martín-Monzón, I. The gut microbiota-brain axis, psychobiotics and its influence on brain and behaviour: A systematic review. Psychoneuroendocrinology 2022, 137, 105640. [Google Scholar] [CrossRef]
- Mayorgas, A.; Dotti, I.; Salas, A. Microbial Metabolites, Postbiotics, and Intestinal Epithelial Function. Mol. Nutr. Food Res. 2021, 65, 2000188. [Google Scholar] [CrossRef]
- Żółkiewicz, J.; Marzec, A.; Ruszczyński, M.; Feleszko, W. Postbiotics—A Step Beyond Pre- and Probiotics. Nutrients 2020, 12, 2189. [Google Scholar] [CrossRef] [PubMed]
- Irving, A.T.; Mimuro, H.; Kufer, T.A.; Lo, C.; Wheeler, R.; Turner, L.J.; Thomas, B.J.; Malosse, C.; Gantier, M.P.; Casillas, L.N.; et al. The Immune Receptor NOD1 and Kinase RIP2 Interact with Bacterial Peptidoglycan on Early Endosomes to Promote Autophagy and Inflammatory Signaling. Cell Host Microbe 2014, 15, 623–635. [Google Scholar] [CrossRef]
- Dinić, M.; Lukić, J.; Djokić, J.; Milenković, M.; Strahinić, I.; Golić, N.; Begović, J. Lactobacillus fermentum Postbiotic-induced Autophagy as Potential Approach for Treatment of Acetaminophen Hepatotoxicity. Front. Microbiol. 2017, 8, 594. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.D. Diet and Gut Microbiota in Health and Disease. Nestle Nutr. Inst. Workshop Ser. 2017, 88, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Bagherniya, M.; Butler, A.E.; Barreto, G.E.; Sahebkar, A. The effect of fasting or calorie restriction on autophagy induction: A review of the literature. Ageing Res. Rev. 2018, 47, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, C.; Zhao, Y.; Zhang, X.; Li, B.; Cui, R. The Effects of Calorie Restriction in Depression and Potential Mechanisms. Curr. Neuropharmacol. 2015, 13, 536–542. [Google Scholar] [CrossRef]
- Ghosh, T.S.; Rampelli, S.; Jeffery, I.B.; Santoro, A.; Neto, M.; Capri, M.; Giampieri, E.; Jennings, A.; Candela, M.; Turroni, S.; et al. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: The NU-AGE 1-year dietary intervention across five European countries. Gut 2020, 69, 1218–1228. [Google Scholar] [CrossRef]
- Staley, C.; Khoruts, A.; Sadowsky, M.J. Contemporary Applications of Fecal Microbiota Transplantation to Treat Intestinal Diseases in Humans. Arch. Med. Res. 2017, 48, 766–773. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Garge, N.; Zhang, X.; Sun, W.; O’Connell, T.M.; Bunger, M.K.; Bultman, S.J. The Microbiome and Butyrate Regulate Energy Metabolism and Autophagy in the Mammalian Colon. Cell Metab. 2011, 13, 517–526. [Google Scholar] [CrossRef]
- Lai, C.H.; Tsai, C.C.; Kuo, W.W.; Ho, T.J.; Day, C.H.; Pai, P.Y.; Chung, L.C.; Huang, C.C.; Wang, H.F.; Liao, P.H.; et al. Multi-Strain Probiotics Inhibit Cardiac Myopathies and Autophagy to Prevent Heart Injury in High-Fat Diet-Fed Rats. Int. J. Med. Sci. 2016, 13, 277–285. [Google Scholar] [CrossRef]
- Qi, R.; Sun, J.; Qiu, X.; Zhang, Y.; Wang, J.; Wang, Q.; Huang, J.; Ge, L.; Liu, Z. The intestinal microbiota contributes to the growth and physiological state of muscle tissue in piglets. Sci. Rep. 2021, 11, 11237. [Google Scholar] [CrossRef]

| Study Model | Study Type | Modulators | Mechanism | Reference | |
|---|---|---|---|---|---|
| Disease/GD | Autophagy | ||||
| Parkinson’s disease | In vivo | MPTP (Neurotoxic model for PD) |
| [104] | |
| Parkinson’s disease | In vitro (PC12 cell) | Rotenone (Neurotoxic model for PD) | Sodium butyrate |
| [105] |
| Multiple sclerosis | In vivo (Experimental autoimmune encephalomyelitis) | MCC950 and Rapamycin |
| [110] | |
| Alzheimer’s disease | In vivo (3× Tg mice) | Probiotics (SLAB51) |
| [117] | |
| Alzheimer’s disease | In vivo (3× Tg mice) | Probiotics (SLAB51) |
| [116] | |
| Ageing mice model/oxidative damage model on cell | In vivo/in vitro (PC12 cell) | Urolithin A |
| [115] | |
| Parkinson’s disease | In vivo | Manganese-induced neurotoxicity resembles PD | Fecal microbiota transplant (FMT) |
| [111] |
| Parkinson’s disease | In vitro (Enteroendocrine cells) | Sodium butyrate |
| [106] | |
| Neurological disorder mice model | In vivo | Microbial metabolite NAMO |
| [107] | |
| Alzheimer’s disease rodent model | In vivo | Aβ1-42-induced rats | Fructooligosaccharide |
| [118] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitra, S.; Munni, Y.A.; Dash, R.; Sadhu, T.; Barua, L.; Islam, M.A.; Chowdhury, D.; Bhattacharjee, D.; Mazumder, K.; Moon, I.S. Gut Microbiota in Autophagy Regulation: New Therapeutic Perspective in Neurodegeneration. Life 2023, 13, 957. https://doi.org/10.3390/life13040957
Mitra S, Munni YA, Dash R, Sadhu T, Barua L, Islam MA, Chowdhury D, Bhattacharjee D, Mazumder K, Moon IS. Gut Microbiota in Autophagy Regulation: New Therapeutic Perspective in Neurodegeneration. Life. 2023; 13(4):957. https://doi.org/10.3390/life13040957
Chicago/Turabian StyleMitra, Sarmistha, Yeasmin Akter Munni, Raju Dash, Toma Sadhu, Largess Barua, Md. Ariful Islam, Dipannita Chowdhury, Debpriya Bhattacharjee, Kishor Mazumder, and Il Soo Moon. 2023. "Gut Microbiota in Autophagy Regulation: New Therapeutic Perspective in Neurodegeneration" Life 13, no. 4: 957. https://doi.org/10.3390/life13040957
APA StyleMitra, S., Munni, Y. A., Dash, R., Sadhu, T., Barua, L., Islam, M. A., Chowdhury, D., Bhattacharjee, D., Mazumder, K., & Moon, I. S. (2023). Gut Microbiota in Autophagy Regulation: New Therapeutic Perspective in Neurodegeneration. Life, 13(4), 957. https://doi.org/10.3390/life13040957









