Cannabichromene Induces Neuronal Differentiation in NSC-34 Cells: Insights from Transcriptomic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Extraction, Isolation, and Purification of CBC from C. sativa
2.2. Cells Culture and Treatment
2.3. MTT Test
2.4. Total RNA Extraction, Library Preparation and Transcriptomic Analysis
2.5. Bioinformatic Analysis
2.6. Western Blot Analysis
- -
- GFAP (GA5): 1:1000 (#3670), Cell Signaling Technology, Danvers, MA, USA;
- -
- AChRα1 (153): 1:500 (sc-65829) Santa Cruz Biotechnology, Inc., Dallas, TX, USA;
- -
- GAPDH (14C10): 1:1000 (#3683), Cell Signaling Technology, Danvers, MA, USA.
- -
- Chicken anti-rat IgG-HRP: 1:2000 (sc-2956) Santa Cruz Biotechnology, Inc., Dallas, TX, USA;
- -
- Chicken anti-mouse IgG-HRP: 1:2000 (SA1-72021) ThermoFisher Scientific, Rockford, IL, USA.
2.7. Statistical Data Analysis
3. Results
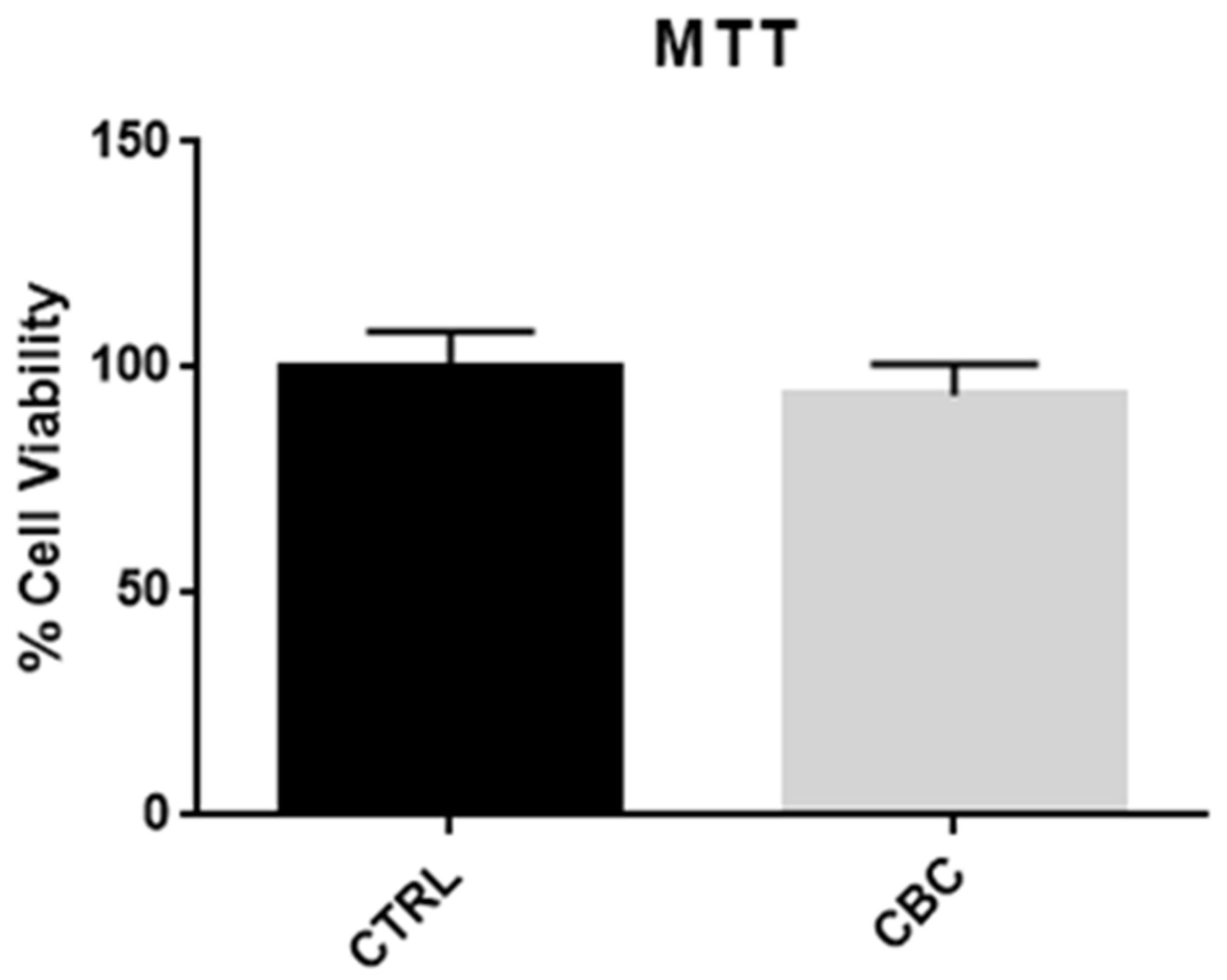
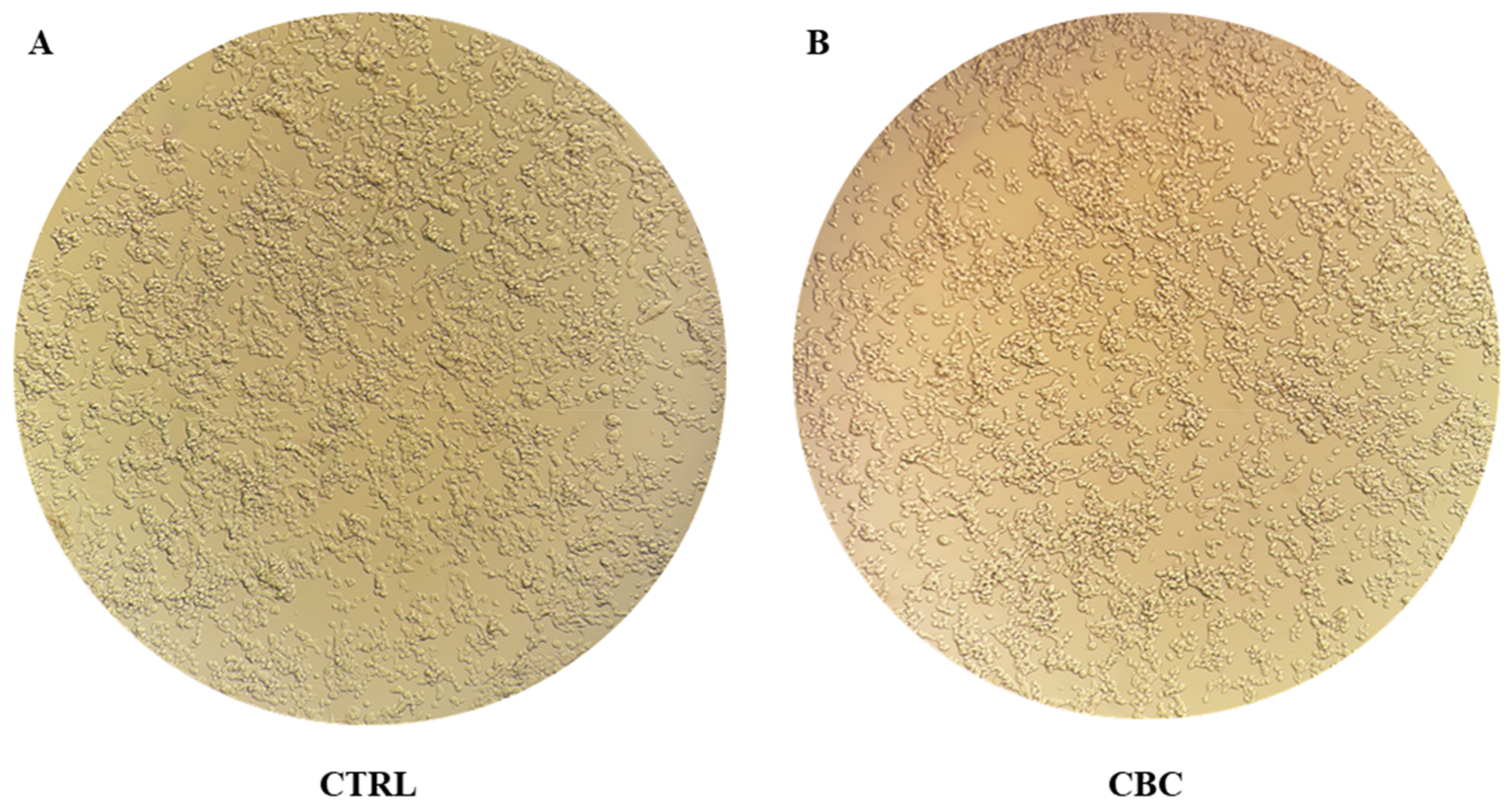
3.1. MTT Test and Evaluation of Cells Morphology
3.2. Inspection of the Transcriptomic Profile
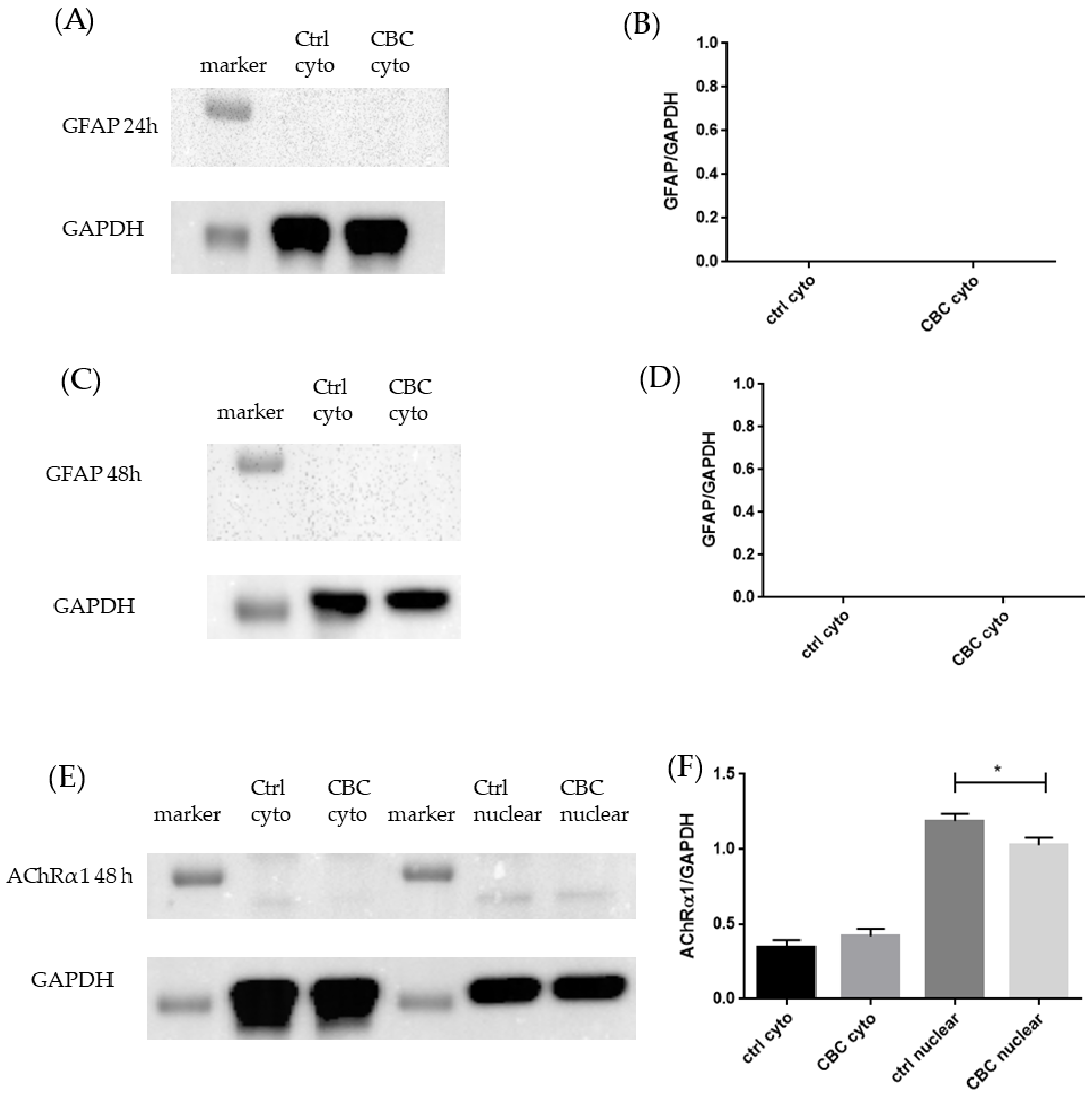
3.3. Western Blot Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, P.; Mahato, D.K.; Kamle, M.; Borah, R.; Sharma, B.; Pandhi, S.; Tripathi, V.; Yadav, H.S.; Devi, S.; Patil, U.; et al. Pharmacological properties, therapeutic potential, and legal status of Cannabis sativa L.: An overview. Phytother. Res. PTR 2021, 35, 6010–6029. [Google Scholar] [CrossRef]
- Blando, S.; Raffaele, I.; Chiricosta, L.; Valeri, A.; Gugliandolo, A.; Silvestro, S.; Pollastro, F.; Mazzon, E. Cannabidiol Promotes Neuronal Differentiation Using Akt and Erk Pathways Triggered by Cb1 Signaling. Molecules 2022, 27, 5644. [Google Scholar] [CrossRef]
- Cristino, L.; Bisogno, T.; Di Marzo, V. Cannabinoids and the expanded endocannabinoid system in neurological disorders. Nat. Rev. Neurol. 2020, 16, 9–29. [Google Scholar] [CrossRef]
- Compagnucci, C.; Di Siena, S.; Bustamante, M.B.; Di Giacomo, D.; Di Tommaso, M.; Maccarrone, M.; Grimaldi, P.; Sette, C. Type-1 (CB1) cannabinoid receptor promotes neuronal differentiation and maturation of neural stem cells. PLoS ONE 2013, 8, e54271. [Google Scholar] [CrossRef]
- Valeri, A.; Mazzon, E. Cannabinoids and Neurogenesis: The Promised Solution for Neurodegeneration? Molecules 2021, 26, 6313. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, P.S.; Perfilieva, E.; Björk-Eriksson, T.; Alborn, A.-M.; Nordborg, C.; Peterson, D.A.; Gage, F.H. Neurogenesis in the adult human hippocampus. Nat. Med. 1998, 4, 1313–1317. [Google Scholar] [CrossRef]
- Sailor, K.A.; Ming, G.L.; Song, H. Neurogenesis as a potential therapeutic strategy for neurodegenerative diseases. Expert Opin. Biol. Ther. 2006, 6, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Izzo, A.A.; Capasso, R.; Aviello, G.; Borrelli, F.; Romano, B.; Piscitelli, F.; Gallo, L.; Capasso, F.; Orlando, P.; Di Marzo, V. Inhibitory effect of cannabichromene, a major non-psychotropic cannabinoid extracted from Cannabis sativa, on inflammation-induced hypermotility in mice. Br. J. Pharmacol. 2012, 166, 1444–1460. [Google Scholar] [CrossRef]
- Pattnaik, F.; Nanda, S.; Mohanty, S.; Dalai, A.K.; Kumar, V.; Ponnusamy, S.K.; Naik, S. Cannabis: Chemistry, extraction and therapeutic applications. Chemosphere 2022, 289, 133012. [Google Scholar] [CrossRef] [PubMed]
- Shinjyo, N.; Di Marzo, V. The effect of cannabichromene on adult neural stem/progenitor cells. Neurochem. Int. 2013, 63, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Cashman, N.R.; Durham, H.D.; Blusztajn, J.K.; Oda, K.; Tabira, T.; Shaw, I.T.; Dahrouge, S.; Antel, J.P. Neuroblastoma x spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 1992, 194, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Maier, O.; Böhm, J.; Dahm, M.; Brück, S.; Beyer, C.; Johann, S. Differentiated NSC-34 motoneuron-like cells as experimental model for cholinergic neurodegeneration. Neurochem. Int. 2013, 62, 1029–1038. [Google Scholar] [CrossRef]
- Gaoni, Y.; Mechoulam, R. The isolation and structure of delta-1-tetrahydrocannabinol and other neutral cannabinoids from hashish. J. Am. Chem. Soc. 1971, 93, 217–224. [Google Scholar] [CrossRef]
- Silvestro, S.; Chiricosta, L.; Gugliandolo, A.; Pizzicannella, J.; Diomede, F.; Bramanti, P.; Trubiani, O.; Mazzon, E. Extracellular Vesicles Derived from Human Gingival Mesenchymal Stem Cells: A Transcriptomic Analysis. Genes 2020, 11, 118. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 22 January 2023).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Carbon, S.; Ireland, A.; Mungall, C.J.; Shu, S.; Marshall, B.; Lewis, S.; Ami, G.O.H.; Web Presence Working, G. AmiGO: Online access to ontology and annotation data. Bioinformatics 2009, 25, 288–289. [Google Scholar] [CrossRef]
- Silvestro, S.; Chiricosta, L.; Gugliandolo, A.; Iori, R.; Rollin, P.; Perenzoni, D.; Mattivi, F.; Bramanti, P.; Mazzon, E. The Moringin/α-CD Pretreatment Induces Neuroprotection in an In Vitro Model of Alzheimer’s Disease: A Transcriptomic Study. Curr. Issues Mol. Biol. 2021, 43, 197–214. [Google Scholar] [CrossRef] [PubMed]
- Gugliandolo, A.; Silvestro, S.; Chiricosta, L.; Pollastro, F.; Bramanti, P.; Mazzon, E. The Transcriptomic Analysis of NSC-34 Motor Neuron-Like Cells Reveals That Cannabigerol Influences Synaptic Pathways: A Comparative Study with Cannabidiol. Life 2020, 10, 227. [Google Scholar] [CrossRef]
- Nieto-Estévez, V.; Defterali, Ç.; Vicario-Abejón, C. IGF-I: A Key Growth Factor that Regulates Neurogenesis and Synaptogenesis from Embryonic to Adult Stages of the Brain. Front. Neurosci. 2016, 10, 52. [Google Scholar] [CrossRef]
- Otaegi, G.; Yusta-Boyo, M.J.; Vergaño-Vera, E.; Méndez-Gómez, H.R.; Carrera, A.C.; Abad, J.L.; González, M.; de la Rosa, E.J.; Vicario-Abejón, C.; de Pablo, F. Modulation of the PI 3-kinase-Akt signalling pathway by IGF-I and PTEN regulates the differentiation of neural stem/precursor cells. J. Cell Sci. 2006, 119, 2739–2748. [Google Scholar] [CrossRef] [PubMed]
- Dráberová, E.; Sulimenko, V.; Vinopal, S.; Sulimenko, T.; Sládková, V.; D’Agostino, L.; Sobol, M.; Hozák, P.; Křen, L.; Katsetos, C.D.; et al. Differential expression of human γ-tubulin isotypes during neuronal development and oxidative stress points to a γ-tubulin-2 prosurvival function. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2017, 31, 1828–1846. [Google Scholar] [CrossRef]
- Wang, J.-M.; Zeng, Y.-S.; Wu, J.-L.; Li, Y.; Teng, Y.D. Cograft of neural stem cells and schwann cells overexpressing TrkC and neurotrophin-3 respectively after rat spinal cord transection. Biomaterials 2011, 32, 7454–7468. [Google Scholar] [CrossRef]
- Bamburg, J.R.; Minamide, L.S.; Wiggan, O.; Tahtamouni, L.H.; Kuhn, T.B. Cofilin and Actin Dynamics: Multiple Modes of Regulation and Their Impacts in Neuronal Development and Degeneration. Cells 2021, 10, 2726. [Google Scholar] [CrossRef]
- Tedeschi, A.; Dupraz, S.; Curcio, M.; Laskowski, C.J.; Schaffran, B.; Flynn, K.C.; Santos, T.E.; Stern, S.; Hilton, B.J.; Larson, M.J.E.; et al. ADF/Cofilin-Mediated Actin Turnover Promotes Axon Regeneration in the Adult CNS. Neuron 2019, 103, 1073–1085.e1076. [Google Scholar] [CrossRef] [PubMed]
- Matusica, D.; Fenech, M.P.; Rogers, M.-L.; Rush, R.A. Characterization and use of the NSC-34 cell line for study of neurotrophin receptor trafficking. J. Neurosci. Res. 2008, 86, 553–565. [Google Scholar] [CrossRef] [PubMed]
- Chacon, J.; Rogers, C.D. Early expression of Tubulin Beta-III in avian cranial neural crest cells. Gene Expr. Patterns 2019, 34, 119067. [Google Scholar] [CrossRef]
- Mesman, S.; van Hooft, J.A.; Smidt, M.P. Mest/Peg1 Is Essential for the Development and Maintenance of a SNc Neuronal Subset. Front. Mol. Neurosci. 2016, 9, 166. [Google Scholar] [CrossRef]
- Prasad, R.; Jung, H.; Tan, A.; Song, Y.; Moon, S.; Shaker, M.R.; Sun, W.; Lee, J.; Ryu, H.; Lim, H.K.; et al. Hypermethylation of Mest promoter causes aberrant Wnt signaling in patients with Alzheimer’s disease. Sci. Rep. 2021, 11, 20075. [Google Scholar] [CrossRef]
- Cho, Y.; Cavalli, V. HDAC signaling in neuronal development and axon regeneration. Curr. Opin. Neurobiol. 2014, 27, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jin, J.; Yang, X.-J. Histone Deacetylase 3 Governs Perinatal Cerebral Development via Neural Stem and Progenitor Cells. iScience 2019, 20, 148–167. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, R.L.; Hsieh, J.; Barbosa, A.C.; Richardson, J.A.; Olson, E.N. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc. Natl. Acad. Sci. USA 2009, 106, 7876–7881. [Google Scholar] [CrossRef]
- Davenne, M.; Maconochie, M.K.; Neun, R.; Pattyn, A.; Chambon, P.; Krumlauf, R.; Rijli, F.M. Hoxa2 and Hoxb2 Control Dorsoventral Patterns of Neuronal Development in the Rostral Hindbrain. Neuron 1999, 22, 677–691. [Google Scholar] [CrossRef] [PubMed]
- Thakurela, S.; Tiwari, N.; Schick, S.; Garding, A.; Ivanek, R.; Berninger, B.; Tiwari, V.K. Mapping gene regulatory circuitry of Pax6 during neurogenesis. Cell Discov. 2016, 2, 15045. [Google Scholar] [CrossRef]
- Pattyn, A.; Hirsch, M.; Goridis, C.; Brunet, J.F. Control of hindbrain motor neuron differentiation by the homeobox gene Phox2b. Development 2000, 127, 1349–1358. [Google Scholar] [CrossRef]
- Ochi, S.; Imaizumi, Y.; Shimojo, H.; Miyachi, H.; Kageyama, R. Oscillatory expression of Hes1 regulates cell proliferation and neuronal differentiation in the embryonic brain. Development 2020, 147, dev182204. [Google Scholar] [CrossRef] [PubMed]
- See, K.; Yadav, P.; Giegerich, M.; Cheong, P.S.; Graf, M.; Vyas, H.; Lee, S.G.; Mathavan, S.; Fischer, U.; Sendtner, M.; et al. SMN deficiency alters Nrxn2 expression and splicing in zebrafish and mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2014, 23, 1754–1770. [Google Scholar] [CrossRef]
- Schieweck, R.; Schöneweiss, E.C.; Harner, M.; Rieger, D.; Illig, C.; Saccà, B.; Popper, B.; Kiebler, M.A. Pumilio2 Promotes Growth of Mature Neurons. Int. J. Mol. Sci. 2021, 22, 8998. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chi, T.; Xue, Y.; Zhou, S.; Kuo, A.; Crabtree, G.R. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc. Natl. Acad. Sci. USA 1998, 95, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.I.; Lessard, J.; Olave, I.A.; Qiu, Z.; Ghosh, A.; Graef, I.A.; Crabtree, G.R. Regulation of Dendritic Development by Neuron-Specific Chromatin Remodeling Complexes. Neuron 2007, 56, 94–108. [Google Scholar] [CrossRef]
- Wang, T.; Chen, J.; Tang, C.-X.; Zhou, X.-Y.; Gao, D.-S. Inverse Expression Levels of EphrinA3 and EphrinA5 Contribute to Dopaminergic Differentiation of Human SH-SY5Y Cells. J. Mol. Neurosci. 2016, 59, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.J.; Kangas, C.D.; Macklin, W.B. Neuronal expression of the proteolipid protein gene in the medulla of the mouse. J. Neurosci. Res. 2009, 87, 2842–2853. [Google Scholar] [CrossRef]
- Harlow, D.E.; Saul, K.E.; Culp, C.M.; Vesely, E.M.; Macklin, W.B. Expression of proteolipid protein gene in spinal cord stem cells and early oligodendrocyte progenitor cells is dispensable for normal cell migration and myelination. J. Neurosci. Off. J. Soc. Neurosci. 2014, 34, 1333–1343. [Google Scholar] [CrossRef]
- Pataskar, A.; Jung, J.; Smialowski, P.; Noack, F.; Calegari, F.; Straub, T.; Tiwari, V.K. NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. EMBO J. 2016, 35, 24–45. [Google Scholar] [CrossRef] [PubMed]
- Resende, R.R.; Adhikari, A. Cholinergic receptor pathways involved in apoptosis, cell proliferation and neuronal differentiation. Cell Commun. Signal. 2009, 7, 20. [Google Scholar] [CrossRef]
- Takarada, T.; Nakamichi, N.; Kitajima, S.; Fukumori, R.; Nakazato, R.; Le, N.Q.; Kim, Y.-H.; Fujikawa, K.; Kou, M.; Yoneda, Y. Promoted Neuronal Differentiation after Activation of Alpha4/Beta2 Nicotinic Acetylcholine Receptors in Undifferentiated Neural Progenitors. PLoS ONE 2012, 7, e46177. [Google Scholar] [CrossRef]
- Rotundo, R.L. Acetylcholinesterase at the neuromuscular junction. Neurosci. Lett. 2020, 735, 135157. [Google Scholar] [CrossRef]
- Coleman, B.A.; Taylor, P. Regulation of Acetylcholinesterase Expression during Neuronal Differentiation (∗). J. Biol. Chem. 1996, 271, 4410–4416. [Google Scholar] [CrossRef] [PubMed]
- Sperling, L.E.; Klaczinski, J.; Schütz, C.; Rudolph, L.; Layer, P.G. Mouse acetylcholinesterase enhances neurite outgrowth of rat R28 cells through interaction with laminin-1. PLoS ONE 2012, 7, e36683. [Google Scholar] [CrossRef] [PubMed]
- Reeves, S.A.; Helman, L.J.; Allison, A.; Israel, M.A. Molecular cloning and primary structure of human glial fibrillary acidic protein. Proc. Natl. Acad. Sci. USA 1989, 86, 5178–5182. [Google Scholar] [CrossRef] [PubMed]


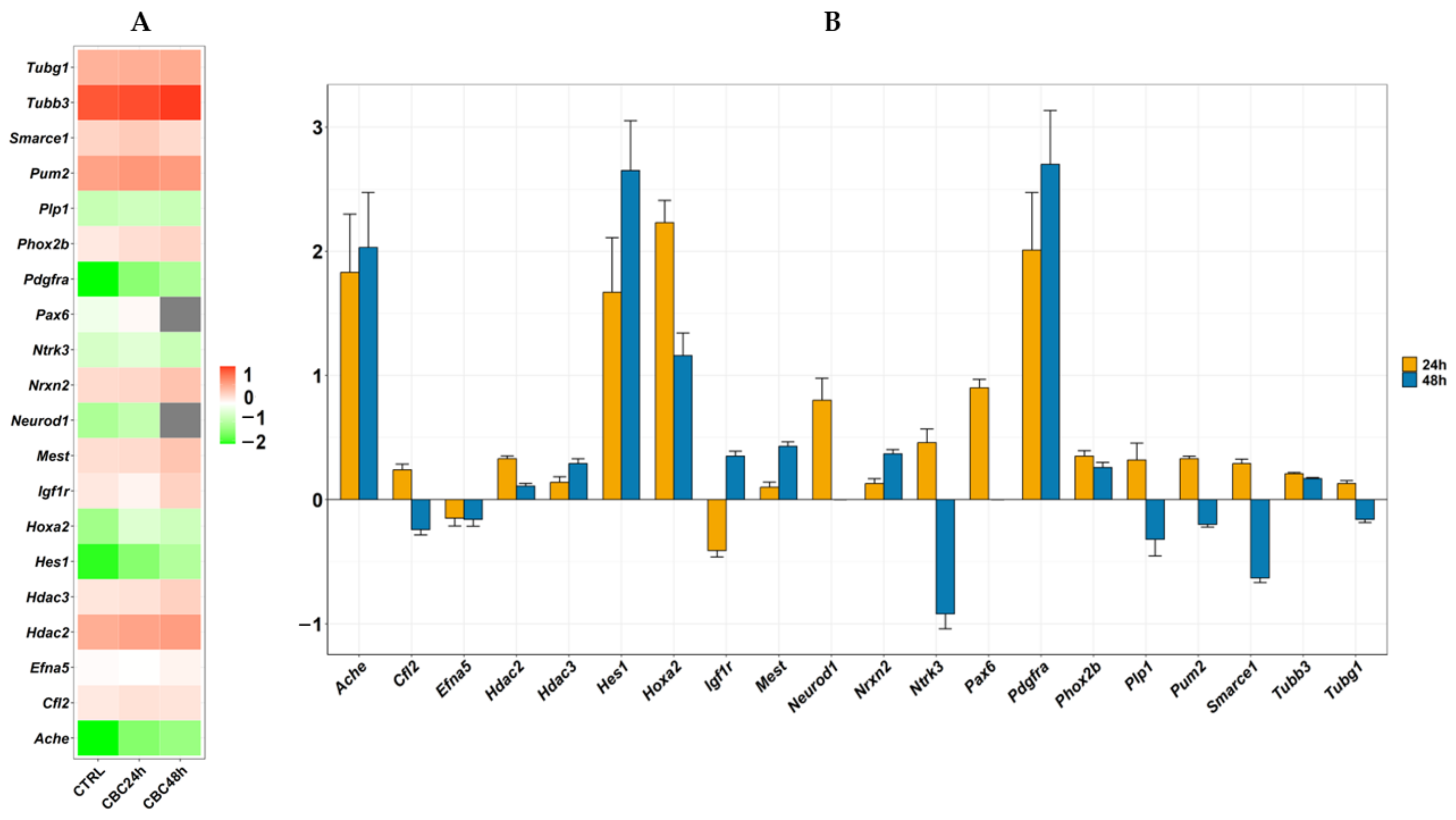

| Gene | CTRL vs. CBC Fold Change | p-Value | q-Value | CTRL vs. CBC-48 h Fold Change | p-Value | q-Value |
|---|---|---|---|---|---|---|
| Ache | 1.83 | 1.03 × 10−4 | 5.16 × 10−4 | 2.03 | 4.91 × 10−6 | 2.46 × 10−05 |
| Cfl2 | 0.24 | 1.52 × 10−7 | 1.44 × 10−6 | −0.24 | 4.99 × 10−8 | 3.30 × 10−07 |
| Efna5 | −0.15 | 1.75 × 10−2 | 4.05 × 10−2 | −0.16 | 3.59 × 10−3 | 9.33 × 10−03 |
| Hdac2 | 0.33 | 1.05 × 10−47 | 1.82 × 10−45 | 0.11 | 7.56 × 10−8 | 4.88 × 10−07 |
| Hdac3 | 0.14 | 2.21 × 10−3 | 7.08 × 10−3 | 0.29 | 2.32 × 10−13 | 2.54 × 10−12 |
| Hes1 | 1.67 | 1.54 × 10−4 | 7.33 × 10−4 | 2.65 | 4.21 × 10−11 | 3.90 × 10−10 |
| Hoxa2 | 2.23 | 2.18 × 10−35 | 2.38 × 10−33 | 1.16 | 1.61 × 10−10 | 1.41 × 10−09 |
| Igf1r | −0.41 | 1.95 × 10−15 | 5.58 × 10−14 | 0.35 | 1.06 × 10−17 | 1.51 × 10−16 |
| Mest | 0.10 | 1.27 × 10−2 | 3.11 × 10−2 | 0.43 | 2.79 × 10−33 | 7.69 × 10−32 |
| Neurod1 | 0.80 | 6.75 × 10−6 | 4.57 × 10−5 | - | >0.05 | >0.05 |
| Nrxn2 | 0.13 | 7.25 × 10−4 | 2.77 × 10−3 | 0.37 | 5.24 × 10−26 | 1.08 × 10−24 |
| Ntrk3 | 0.46 | 2.54 × 10−5 | 1.49 × 10−4 | −0.92 | 1.03 × 10−14 | 1.21 × 10−13 |
| Pax6 | 0.90 | 1.04 × 10−38 | 1.30 × 10−36 | - | >0.05 | >0.05 |
| Pdgfra | 2.01 | 1.47 × 10−5 | 9.09 × 10−5 | 2.7 | 4.46 × 10−10 | 3.74 × 10−09 |
| Phox2b | 0.35 | 4.68 × 10−15 | 1.29 × 10−13 | 0.26 | 3.29 × 10−10 | 2.80 × 10−09 |
| Plp1 | 0.32 | 1.87 × 10−2 | 4.28 × 10−2 | −0.32 | 1.57 × 10−2 | 3.29 × 10−02 |
| Pum2 | 0.33 | 2.01 × 10−61 | 5.46 × 10−59 | −0.2 | 1.57 × 10−25 | 3.16 × 10−24 |
| Smarce1 | 0.29 | 1.19 × 10−15 | 3.51 × 10−14 | −0.63 | 2.25 × 10−67 | 1.28 × 10−65 |
| Tubb3 | 0.21 | 8.27 × 10−109 | 3.83 × 10−106 | 0.17 | 1.46 × 10−88 | 1.11 × 10−86 |
| Tubg1 | 0.13 | 1.68 × 10−7 | 1.57 × 10−6 | −0.16 | 1.61 × 10−12 | 1.64 × 10−11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valeri, A.; Chiricosta, L.; D’Angiolini, S.; Pollastro, F.; Salamone, S.; Mazzon, E. Cannabichromene Induces Neuronal Differentiation in NSC-34 Cells: Insights from Transcriptomic Analysis. Life 2023, 13, 742. https://doi.org/10.3390/life13030742
Valeri A, Chiricosta L, D’Angiolini S, Pollastro F, Salamone S, Mazzon E. Cannabichromene Induces Neuronal Differentiation in NSC-34 Cells: Insights from Transcriptomic Analysis. Life. 2023; 13(3):742. https://doi.org/10.3390/life13030742
Chicago/Turabian StyleValeri, Andrea, Luigi Chiricosta, Simone D’Angiolini, Federica Pollastro, Stefano Salamone, and Emanuela Mazzon. 2023. "Cannabichromene Induces Neuronal Differentiation in NSC-34 Cells: Insights from Transcriptomic Analysis" Life 13, no. 3: 742. https://doi.org/10.3390/life13030742
APA StyleValeri, A., Chiricosta, L., D’Angiolini, S., Pollastro, F., Salamone, S., & Mazzon, E. (2023). Cannabichromene Induces Neuronal Differentiation in NSC-34 Cells: Insights from Transcriptomic Analysis. Life, 13(3), 742. https://doi.org/10.3390/life13030742







