Random and Natural Non-Coding RNA Have Similar Structural Motif Patterns but Differ in Bulge, Loop, and Bond Counts
Abstract
1. Introduction
2. Results
2.1. Abstract RNA Shapes
2.2. Nature Uses High-Frequency Shapes
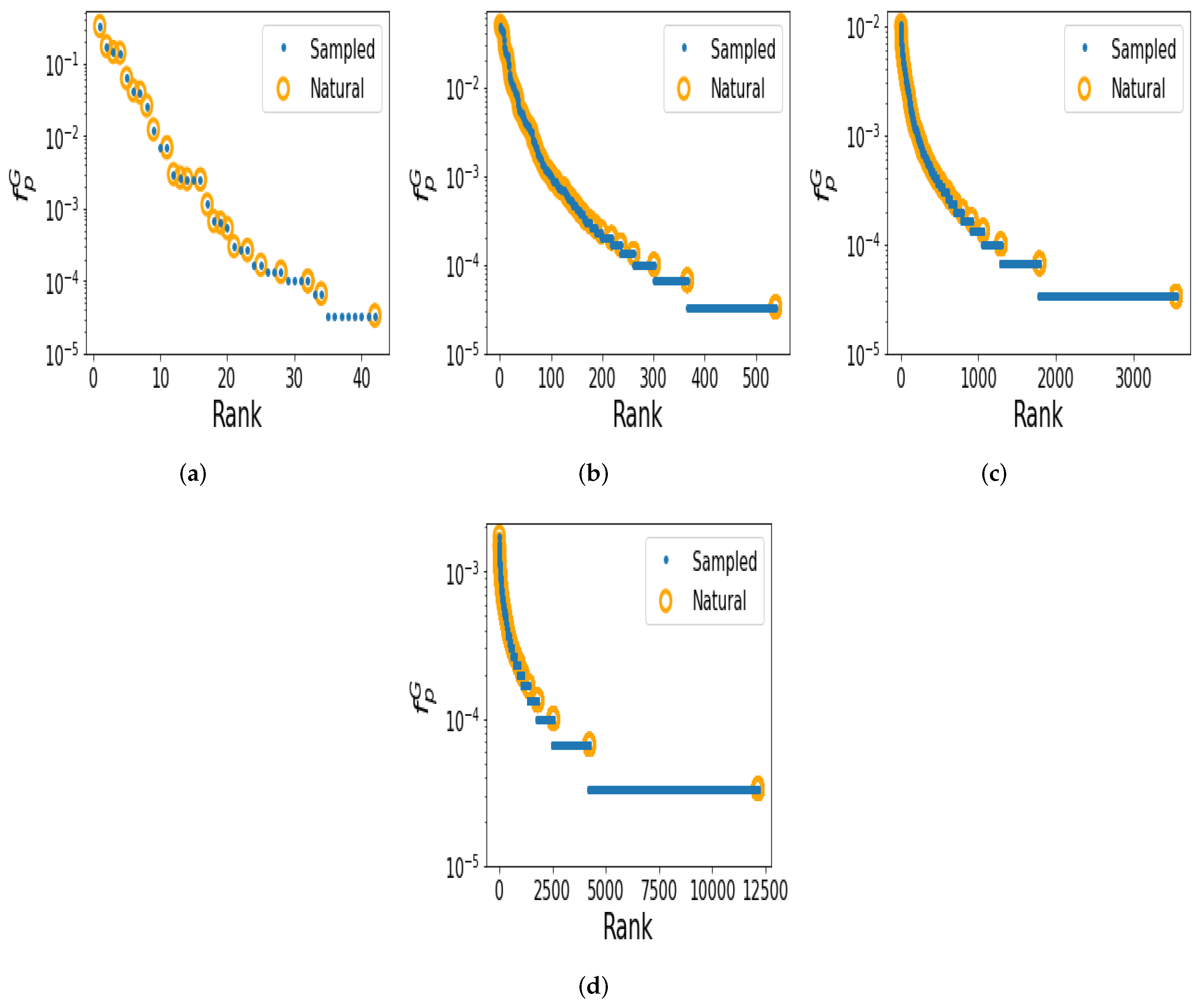
2.3. Shape Abundance Can Be Predicted from Random Sampling
2.4. Studying Structural Motif Frequencies for Larger RNA
2.5. Biological Functions of Some High and Low-Frequency Shapes
3. Classifying Natural and Random RNA Using Motif Counts
3.1. Can We Use Motif Frequency to Detect Functional RNA?
3.2. Classifying RNA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Methods
Appendix A.1. Code and Data Availability
Appendix A.2. Random RNA Sequences
Appendix A.3. Natural RNA Sequences
Appendix A.4. Folding RNA
Appendix A.5. Drawing RNA
Appendix A.6. Motif Counting
Appendix A.7. Abstract Shapes
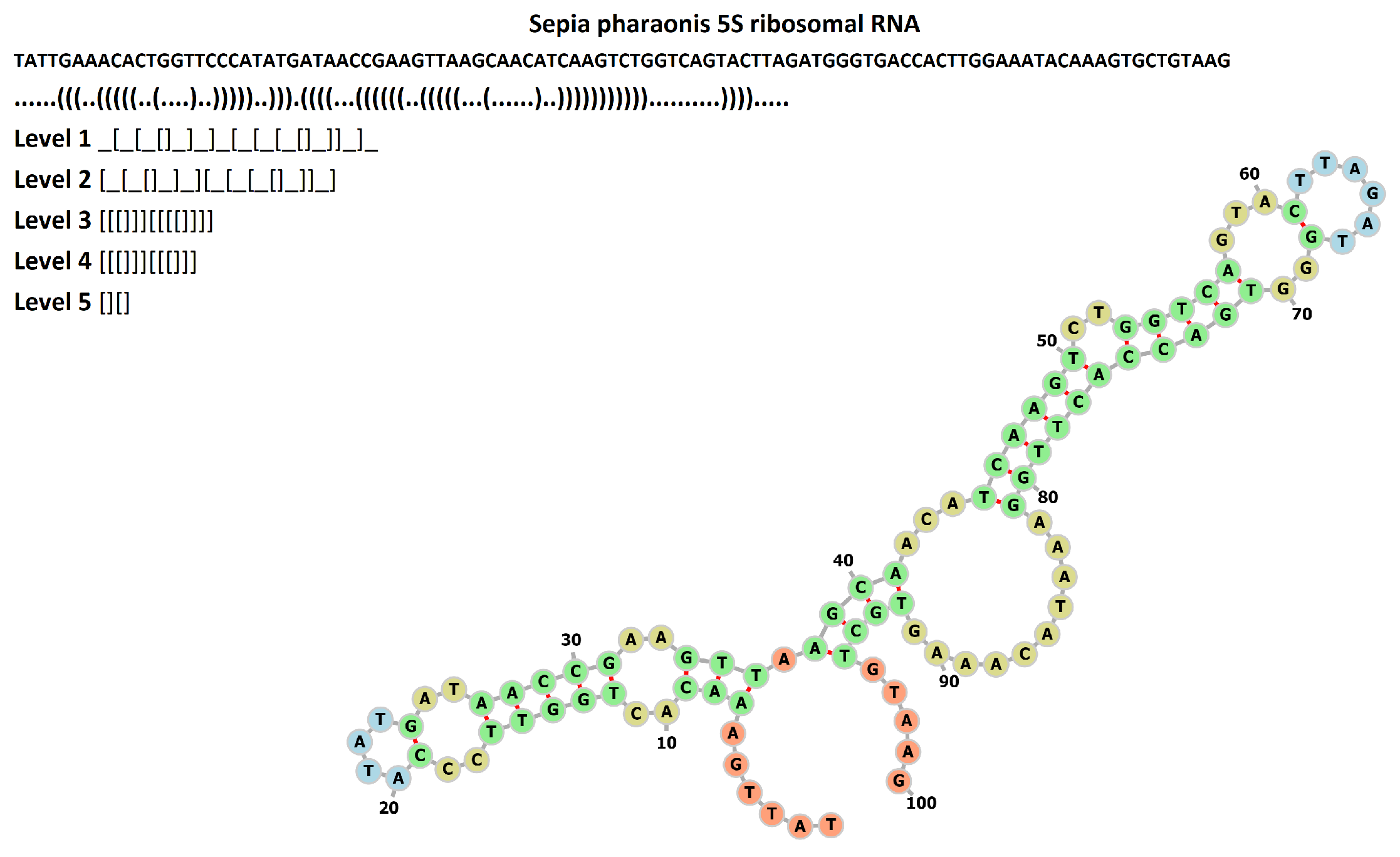
Appendix B. Motif Counts and Overall RNA Shape
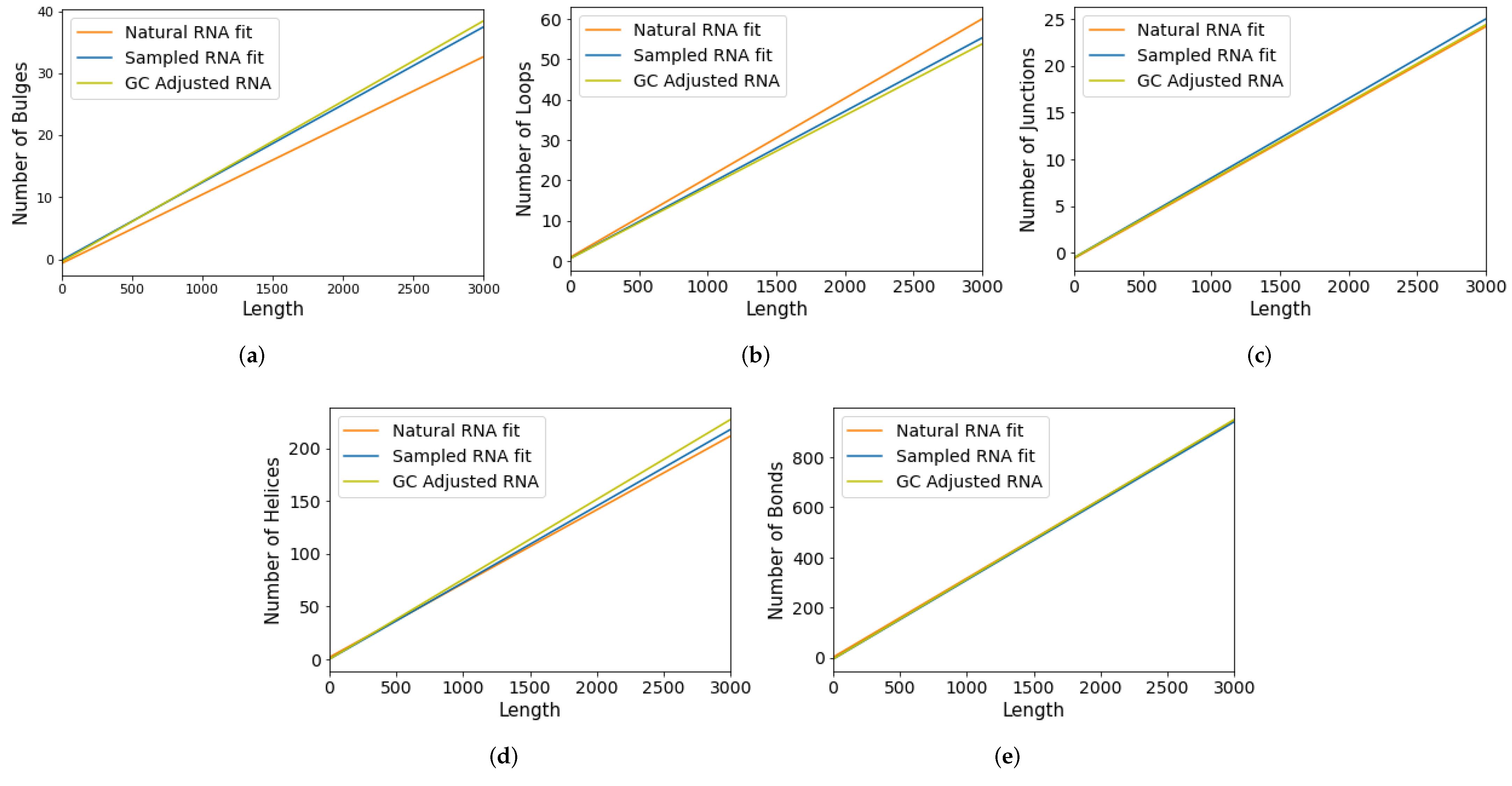
Appendix C. Adjusting for GC Content
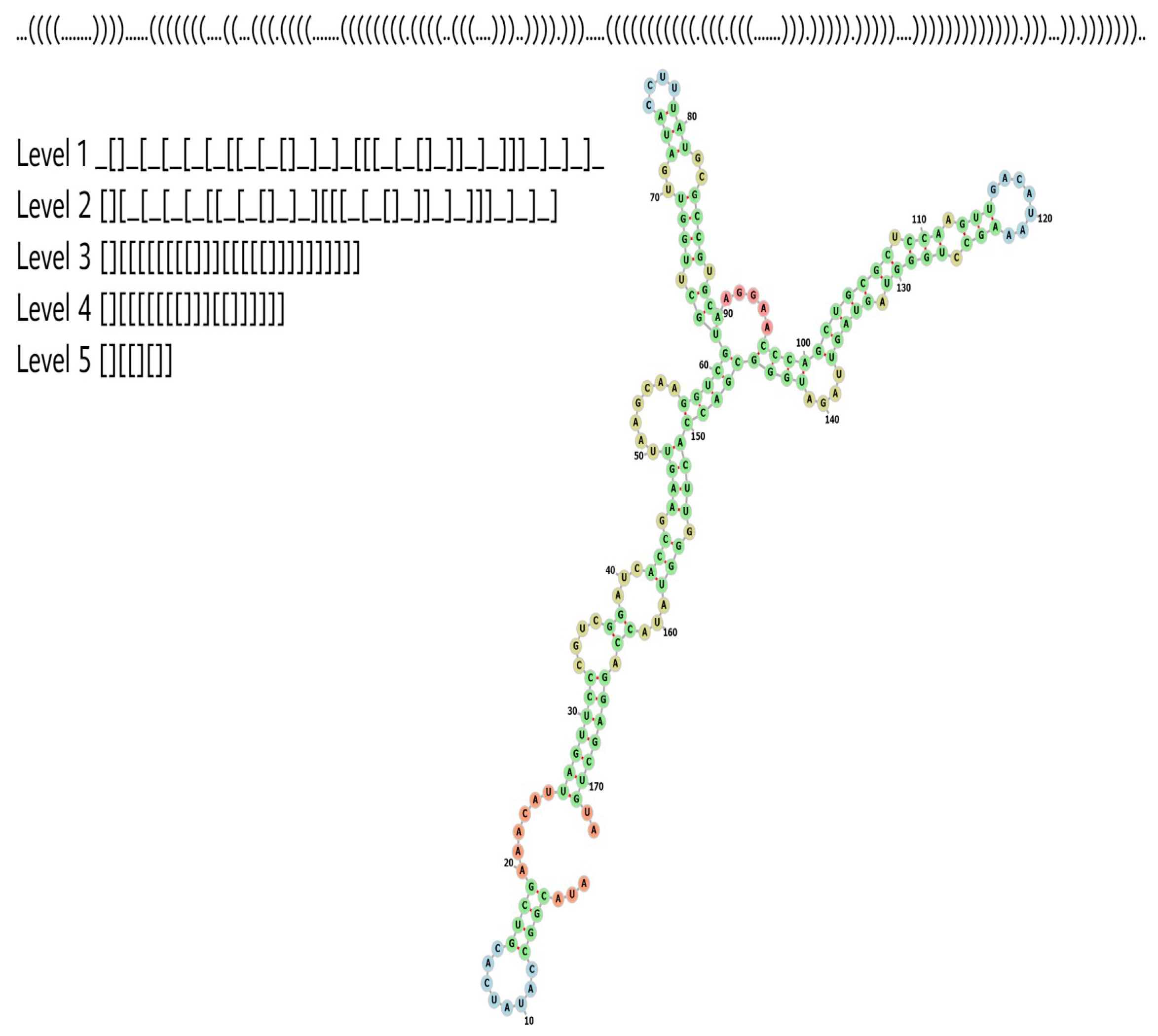
References
- Smith, J.M.; Burian, R.; Kauffman, S.; Alberch, P.; Campbell, J.; Goodwin, B.; Lande, R.; Raup, D.; Wolpert, L. Developmental constraints and evolution: A perspective from the mountain lake conference on development and evolution. Q. Rev. Biol. 1985, 60, 265–287. [Google Scholar] [CrossRef]
- Stoltzfus, A. Mutation, Randomness, and Evolution; Oxford University Press: Oxford, UK, 2021. [Google Scholar]
- Gould, S.J. Wonderful Life: The Burgess Shale and the Nature of History; WW Norton & Company: New York, NY, USA, 1990. [Google Scholar]
- Blount, Z.D.; Lenski, R.E.; Losos, J.B. Contingency and determinism in evolution: Replaying life’s tape. Science 2018, 362, eaam5979. [Google Scholar] [CrossRef]
- Arthur, W. Developmental drive: An important determinant of the direction of phenotypic evolution. Evol. Dev. 2001, 3, 271–278. [Google Scholar] [CrossRef]
- Uller, T.; Laland, K.N. Evolutionary Causation: Biological and Philosophical Reflections; MIT Press: Cambridge, MA, USA, 2019; Volume 23. [Google Scholar]
- Borenstein, E.; Krakauer, D.C. An end to endless forms: Epistasis, phenotype distribution bias, and nonuniform evolution. PLoS Comput. Biol. 2008, 4, e1000202. [Google Scholar] [CrossRef] [PubMed]
- Uller, T.; Moczek, A.P.; Watson, R.A.; Brakefield, P.M.; Laland, K.N. Developmental bias and evolution: A regulatory network perspective. Genetics 2018, 209, 949–966. [Google Scholar] [CrossRef] [PubMed]
- Jablonski, D. Developmental bias, macroevolution, and the fossil record. Evol. Dev. 2020, 22, 103–125. [Google Scholar] [CrossRef]
- Yampolsky, L.Y.; Stoltzfus, A. Bias in the introduction of variation as an orienting factor in evolution. Evol. Dev. 2001, 3, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Stoltzfus, A.; Yampolsky, L.Y. Climbing mount probable: Mutation as a cause of nonrandomness in evolution. J. Hered. 2009, 100, 637–647. [Google Scholar] [CrossRef]
- Cano, A.V.; Rozhoňová, H.; Stoltzfus, A.; McCandlish, D.M.; Payne, J.L. Mutation bias shapes the spectrum of adaptive substitutions. Proc. Natl. Acad. Sci. USA 2022, 119, e2119720119. [Google Scholar] [CrossRef]
- Zuker, M.; Mathews, D.H.; Turner, D.H. Algorithms and thermodynamics for RNA secondary structure prediction: A practical guide. RNA Biochem. Biotechnol. 1999, 70, 11–44. [Google Scholar]
- Hofacker, I.L.; Fontana, W.; Stadler, P.F.; Bonhoeffer, L.S.; Tacker, M.; Schuster, P. Fast folding and comparison of RNA secondary structures. MMon. Chem/Chem. Mon. 1994, 125, 167–188. [Google Scholar] [CrossRef]
- Contrant, M.; Fender, A.; Chane-Woon-Ming, B.; Randrianjafy, R.; Vivet-Boudou, V.; Richer, D.; Pfeffer, S. Importance of the RNA secondary structure for the relative accumulation of clustered viral microRNAs. Nucleic Acids Res. 2014, 42, 7981–7996. [Google Scholar] [CrossRef]
- Elliott, D.; Ladomery, M. Molecular Biology of RNA; Oxford University Press: Oxford, UK, 2017. [Google Scholar]
- Wang, X.-W.; Liu, C.-X.; Chen, L.-L.; Zhang, Q.C. RNA structure probing uncovers RNA structure-dependent biological functions. Nat. Chem. Biol. 2021, 17, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.N.; Gabay, J.; Débarbouillé, M.; Schwartz, M. A role for mRNA secondary structure in the control of translation initiation. Nature 1982, 295, 616–618. [Google Scholar] [CrossRef] [PubMed]
- Kramer, M.C.; Gregory, B.D. Does RNA secondary structure drive translation or vice versa? Nat. Struct. Mol. Biol. 2018, 25, 641–643. [Google Scholar] [CrossRef] [PubMed]
- Ermolenko, D.N.; Mathews, D.H. Making ends meet: New functions of mRNA secondary structure. Wiley Interdiscip. Rev. RNA 2021, 12, e1611. [Google Scholar] [CrossRef]
- Bailor, M.H.; Sun, X.; Al-Hashimi, H.M. Topology links RNA secondary structure with global conformation, dynamics, and adaptation. Science 2010, 327, 202. [Google Scholar] [CrossRef]
- Fontana, W.; Konings, D.A.M.; Stadler, P.F.; Schuster, P. Statistics of RNA secondary structures. Biopolym. Orig. Res. Biomol. 1993, 33, 1389–1404. [Google Scholar] [CrossRef]
- Schuster, P. Genotypes with phenotypes: Adventures in an RNA toy world. Biophys. Chem. 1997, 66, 75–110. [Google Scholar] [CrossRef]
- Fontana, W. Modelling ‘evo-devo’ with RNA. BioEssays 2002, 24, 1164–1177. [Google Scholar] [CrossRef]
- Schuster, P.; Fontana, W.; Stadler, P.F.; Hofacker, I.L. From sequences to shapes and back: A case study in RNA secondary structures. Proc. Biol. Sci. 1994, 255, 279–284. [Google Scholar] [PubMed]
- Carothers, J.M.; Oestreich, S.C.; Davis, J.H.; Szostak, J.W. Informational complexity and functional activity of RNA structures. J. Am. Chem. Soc. 2004, 126, 5130–5137. [Google Scholar] [CrossRef]
- Knight, R.; Sterck, H.D.; Markel, R.; Smit, S.; Oshmyansky, A.; Yarus, M. Abundance of correctly folded RNA motifs in sequence space, calculated on computational grids. Nucleic Acids Res. 2005, 33, 5924–5935. [Google Scholar] [CrossRef] [PubMed]
- Stich, M.; Briones, C.; Manrubia, S.C. On the structural repertoire of pools of short, random RNA sequences. J. Theor. Biol. 2008, 252, 750–763. [Google Scholar] [CrossRef] [PubMed]
- Cowperthwaite, M.C.; Economo, E.P.; Harcombe, W.R.; Miller, E.L.; Meyers, L.A. The ascent of the abundant: How mutational networks constrain evolution. PLoS Comput. Biol. 2008, 4, e1000110. [Google Scholar] [CrossRef]
- Ahnert, S.E. Structural properties of genotype–phenotype maps. J. R. Soc. Interface 2017, 14, 20170275. [Google Scholar] [CrossRef]
- Dingle, K.; Schaper, S.; Louis, A.A. The structure of the genotype–phenotype map strongly constrains the evolution of non-coding RNA. Interface Focus 2015, 5, 20150053. [Google Scholar] [CrossRef]
- Dingle, K.; Ghaddar, F.; Šulc, P.; Louis, A.A. Phenotype bias determines how natural RNA structures occupy the morphospace of all possible shapes. Mol. Biol. Evol. 2022, 39, msab280. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef]
- Farley, E.J.; Eggleston, H.; Riehle, M.M. Filtering the junk: Assigning function to the mosquito non-coding genome. Insects 2021, 12, 186. [Google Scholar] [CrossRef]
- Feingold, E.A.; Pachter, L. The encode (encyclopedia of DNA elements) project. Science 2004, 306, 636–640. [Google Scholar]
- Roulois, D.; Yau, H.L.; Singhania, R.; Wang, Y.; Danesh, A.; Shen, S.Y.; Han, H.; Liang, G.; Jones, P.A.; Pugh, T.J.; et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 2015, 162, 961–973. [Google Scholar] [CrossRef]
- Chung, H.; Calis, J.J.A.; Wu, X.; Sun, T.; Yu, Y.; Sarbanes, S.L.; Thi, V.L.D.; Shilvock, A.R.; Hoffmann, H.-H.; Rosenberg, B.R.; et al. Human adar1 prevents endogenous RNA from triggering translational shutdown. Cell 2018, 172, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Johnston, I.G.; Dingle, K.; Greenbury, S.F.; Camargo, C.Q.; Doye, J.P.K.; Ahnert, S.E.; Louis, A.A. Symmetry and simplicity spontaneously emerge from the algorithmic nature of evolution. Proc. Natl. Acad. Sci. USA 2022, 119, e2113883119. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.S.; Ahnert, S.E. Insertions and deletions in the RNA sequence–structure map. J. R. Soc. Interface 2021, 18, 20210380. [Google Scholar] [CrossRef] [PubMed]
- Giegerich, R.; Voß, B.; Rehmsmeier, M. Abstract shapes of RNA. Nucleic Acids Res. 2004, 32, 4843–4851. [Google Scholar] [CrossRef]
- Janssen, S.; Giegerich, R. The RNA shapes studio. Bioinformatics 2014, 31, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Siederdissen, C.H.Z.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. Viennarna package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- RNAcentral Consortium. RNAcentral 2021: Secondary structure integration, improved sequence search and new member databases. Nucleic Acids Res. 2021, 49, D212–D220. [Google Scholar] [CrossRef]
- Stich, M.; Manrubia, S.C. Motif frequency and evolutionary search times in RNA populations. J. Theor. Biol. 2011, 280, 117–126. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nebel, M.E.; Scheid, A. On quantitative effects of RNA shape abstraction. Theory Biosci. 2009, 128, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Hofacker, I.L.; Schuster, P.; Stadler, P.F. Combinatorics of RNA secondary structures. Discret. Appl. Math. 1998, 88, 207–237. [Google Scholar] [CrossRef]
- Jorg, T.; Martin, O.C.; Wagner, A. Neutral network sizes of biological RNA molecules can be computed and are not atypically small. BMC Bioinform. 2008, 9, 464. [Google Scholar] [CrossRef]
- Rivas, E.; Eddy, S.R. Secondary structure alone is generally not statistically significant for the detection of noncoding RNAs. Bioinformatics 2000, 16, 583–605. [Google Scholar] [CrossRef] [PubMed]
- Carter, R.J.; Dubchak, I.; Holbrook, S.R. A computational approach to identify genes for functional RNAs in genomic sequences. Nucleic Acids Res. 2001, 29, 3928–3938. [Google Scholar] [CrossRef]
- Bonnet, E.; Wuyts, J.; Rouzé, P.; de Peer, Y.V. Evidence that microrna precursors, unlike other non-coding rnas, have lower folding free energies than random sequences. Bioinformatics 2004, 20, 2911–2917. [Google Scholar] [CrossRef]
- Washietl, S.; Hofacker, I.L.; Stadler, P.F. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. USA 2005, 102, 2454–2459. [Google Scholar] [CrossRef]
- Wu, C.-Y.; Li, Q.-Z.; Feng, Z.-X. Non-coding RNA identification based on topology secondary structure and reading frame in organelle genome level. Genomics 2016, 107, 9–15. [Google Scholar] [CrossRef]
- Sutanto, K.; Turcotte, M. Assessing the use of secondary structure fingerprints and deep learning to classify RNA sequences. In Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Republic of Korea, 16–19 December 2020; pp. 42–49. [Google Scholar]
- Dingle, K. Probabilistic Bias in Genotype-Phenotype Maps. PhD Thesis, University of Oxford, Oxford, UK, 2014. [Google Scholar]
- Manrubia, S.; Cuesta, J.A.; Aguirre, J.; Ahnert, S.E.; Altenberg, L.; Cano, A.V.; Catalán, P.; Diaz-Uriarte, R.; Elena, S.F.; García-Martín, J.A.; et al. From genotypes to organisms: State-of-the-art and perspectives of a cornerstone in evolutionary dynamics. Phys. Life Rev. 2021, 38, 55–106. [Google Scholar] [CrossRef]
- Ekland, E.H.; Szostak, J.W.; Bartel, D.P. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 1995, 269, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Neme, R.; Amador, C.; Yildirim, B.; McConnell, E.; Tautz, D. Random sequences are an abundant source of bioactive RNAs or peptides. Nat. Ecol. Evol. 2017, 1, 0127. [Google Scholar] [CrossRef]
- Smit, S.; Yarus, M.; Knight, R. Natural selection is not required to explain universal compositional patterns in rRNA secondary structure categories. RNA 2006, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Schaper, S.; Louis, A.A. The arrival of the frequent: How bias in genotype-phenotype maps can steer populations to local optima. PLoS ONE 2014, 9, e86635. [Google Scholar] [CrossRef] [PubMed]
- Catalán, P.; Manrubia, S.; Cuesta, J.A. Populations of genetic circuits are unable to find the fittest solution in a multilevel genotype–phenotype map. J. R. Soc. Interface 2020, 17, 20190843. [Google Scholar] [CrossRef] [PubMed]
- Psujek, S.; Beer, R.D. Developmental bias in evolution: Evolutionary accessibility of phenotypes in a model evo-devo system. Evol. Dev. 2008, 10, 375–390. [Google Scholar] [CrossRef]
- Braendle, C.; Baer, C.F.; Félix, M.A. Bias and evolution of the mutationally accessible phenotypic space in a developmental system. PLoS Genetics 2010, 6, e1000877. [Google Scholar] [CrossRef]
- Arthur, W. Biased Embryos and Evolution; Cambridge University Press: Cambridge, UK, 2004. [Google Scholar]
- Atallah, J.; Liu, N.H.; Dennis, P.; Hon, A.; Godt, D.; Larsen, E.W. Cell dynamics and developmental bias in the ontogeny of a complex sexually dimorphic trait in Drosophila melanogaster. Evol. Dev. 2009, 11, 191–204. [Google Scholar] [CrossRef]
- Arthur, W. The interaction between developmental bias and natural selection: From centipede segments to a general hypothesis. Heredity 2002, 89, 239–246. [Google Scholar] [CrossRef]
- Johnston, I.G.; Ahnert, S.A.; Doye, J.P.K.; Louis, A.A. Evolutionary dynamics in a simple model of self-assembly. Phys. Rev. E 2011, 83, 066105. [Google Scholar] [CrossRef]
- Monroe, J.; Srikant, T.; Carbonell-Bejerano, P.; Becker, C.; Lensink, M.; Exposito-Alonso, M.; Klein, M.; Hildebrandt, J.; Neumann, M.; Kliebenstein, D.; et al. Mutation bias reflects natural selection in arabidopsis thaliana. Nature 2022, 602, 101–105. [Google Scholar] [CrossRef]
- Salazar-Ciudad, I. Why call it developmental bias when it is just development? Biol. Direct 2021, 16, 3. [Google Scholar] [CrossRef] [PubMed]
- Xiong, K.; Gerstein, M.; Masel, J. Differences in evolutionary accessibility determine which equally effective regulatory motif evolves to generate pulses. Genetics 2021, 219, iyab140. [Google Scholar] [CrossRef] [PubMed]
- Dingle, K. Optima and simplicity in nature. arXiv 2022, arXiv:2210.02564. [Google Scholar]
- Dingle, K. Fitness, optima, and simplicity. Preprints 2022, 2022080402. [Google Scholar] [CrossRef]
- Dingle, K.; Camargo, C.Q.; Louis, A.A. Input–output maps are strongly biased towards simple outputs. Nat. Commun. 2018, 9, 761. [Google Scholar] [CrossRef]
- Dingle, K.; Pérez, G.V.; Louis, A.A. Generic predictions of output probability based on complexities of inputs and outputs. Sci. Rep. 2020, 10, 4415. [Google Scholar] [CrossRef]
- García-Martín, J.A.; Catalán, P.; Manrubia, S.; Cuesta, J.A. Statistical theory of phenotype abundance distributions: A test through exact enumeration of genotype spaces (a). EPL (Europhys. Lett.) 2018, 123, 28001. [Google Scholar] [CrossRef]
- Schultes, E.A.; Bartel, D.P. One sequence, two ribozymes: Implications for the emergence of new ribozyme folds. Science 2000, 289, 448–452. [Google Scholar] [CrossRef]
- Ponty, Y. Efficient sampling of RNA secondary structures from the Boltzmann ensemble of low-energy. J. Math. Biol. 2008, 56, 107–127. [Google Scholar] [CrossRef]
- Morgan, S.R.; Higgs, P.G. Evidence for kinetic effects in the folding of large RNA molecules. J. Chem. Phys. 1996, 105, 7152–7157. [Google Scholar] [CrossRef]
- Govindarajan, S.; Recabarren, R.; Goldstein, R.A. Estimating the total number of protein folds. Proteins Struct. Funct. Bioinform. 1999, 35, 408–414. [Google Scholar] [CrossRef]
- Oberai, A.; Ihm, Y.; Kim, S.; Bowie, J.U. A limited universe of membrane protein families and folds. Protein Sci. 2006, 15, 1723–1734. [Google Scholar] [CrossRef]
- Liu, M.; Poppleton, E.; Pedrielli, G.; Šulc, P.; Bertsekas, D.P. Expertrna: A new framework for RNA secondary structure prediction. INFORMS J. Comput. 2022, 34, 2464–2484. [Google Scholar] [CrossRef]
- Pucci, F.; Schug, A. Shedding light on the dark matter of the biomolecular structural universe: Progress in RNA 3D structure prediction. Methods 2019, 162, 68–73. [Google Scholar] [CrossRef]
- Johnston, I.G.; Dingle, K.; Greenbury, S.F.; Camargo, C.Q.; Doye, J.P.K.; Ahnert, S.E.; Louis, A.A. Reply to Ocklenburg and Mundorf: The interplay of developmental bias and natural selection. Proc. Natl. Acad. Sci. USA 2022, 119, e2205299119. [Google Scholar] [CrossRef]
- Jiménez, J.I.; Xulvi-Brunet, R.; Campbell, G.W.; Turk-MacLeod, R.; Chen, I.A. Comprehensive experimental fitness landscape and evolutionary network for small RNA. Proc. Natl. Acad. Sci. USA 2013, 110, 14984–14989. [Google Scholar] [CrossRef]
- Kun, Á.; Szathmáry, E. Fitness landscapes of functional RNAs. Life 2015, 5, 1497–1517. [Google Scholar] [CrossRef]
- Gioacchino, A.D.; Procyk, J.; Molari, M.; Schreck, J.S.; Zhou, Y.; Liu, Y.; Monasson, R.; Cocco, S.; Šulc, P. Generative and interpretable machine learning for aptamer design and analysis of in vitro sequence selection. PLoS Comput. Biol. 2022, 18, e1010561. [Google Scholar] [CrossRef]
- Rotrattanadumrong, R.; Yokobayashi, Y. Experimental exploration of a ribozyme neutral network using evolutionary algorithm and deep learning. Nat. Commun. 2022, 13, 4847. [Google Scholar] [CrossRef]
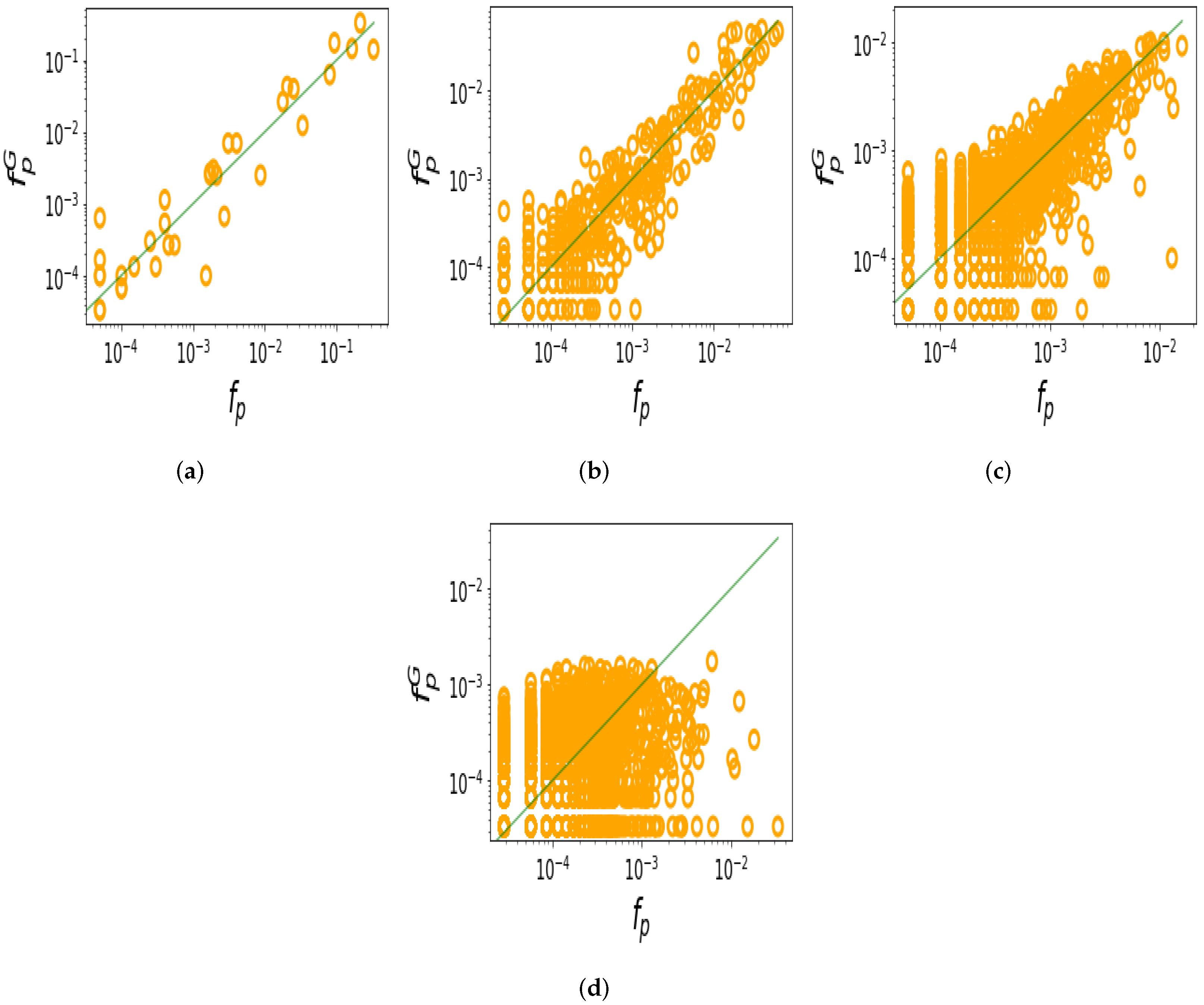
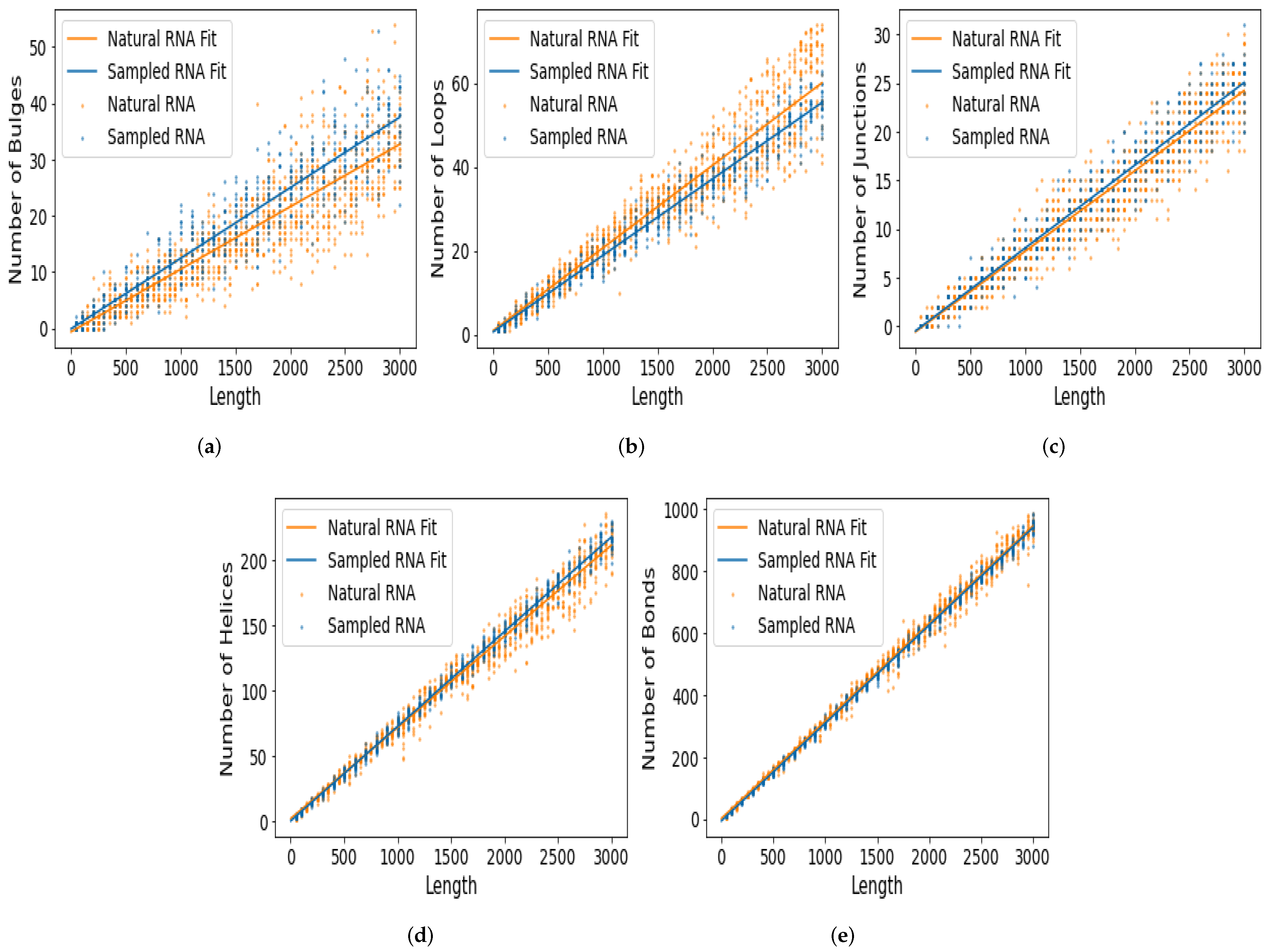
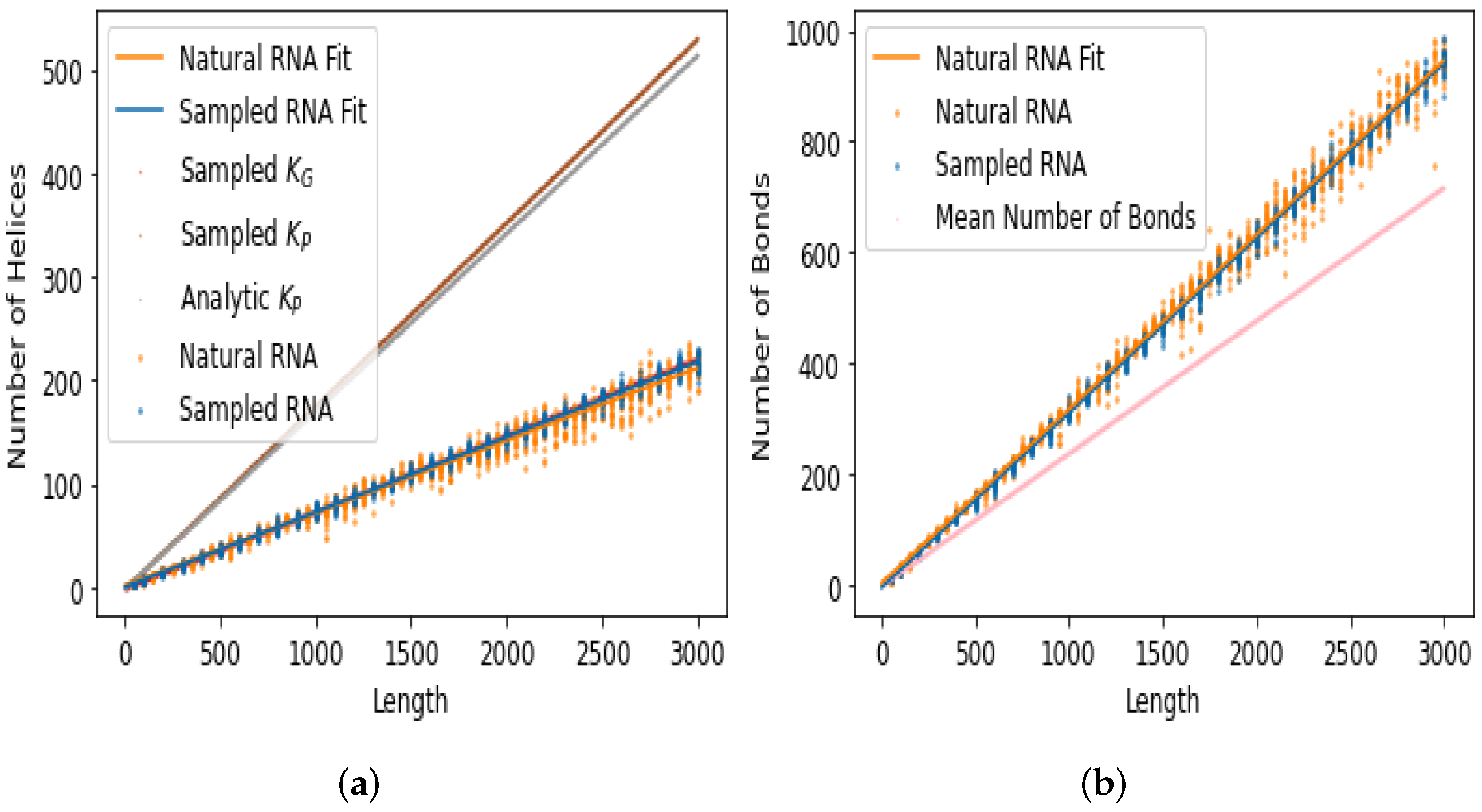
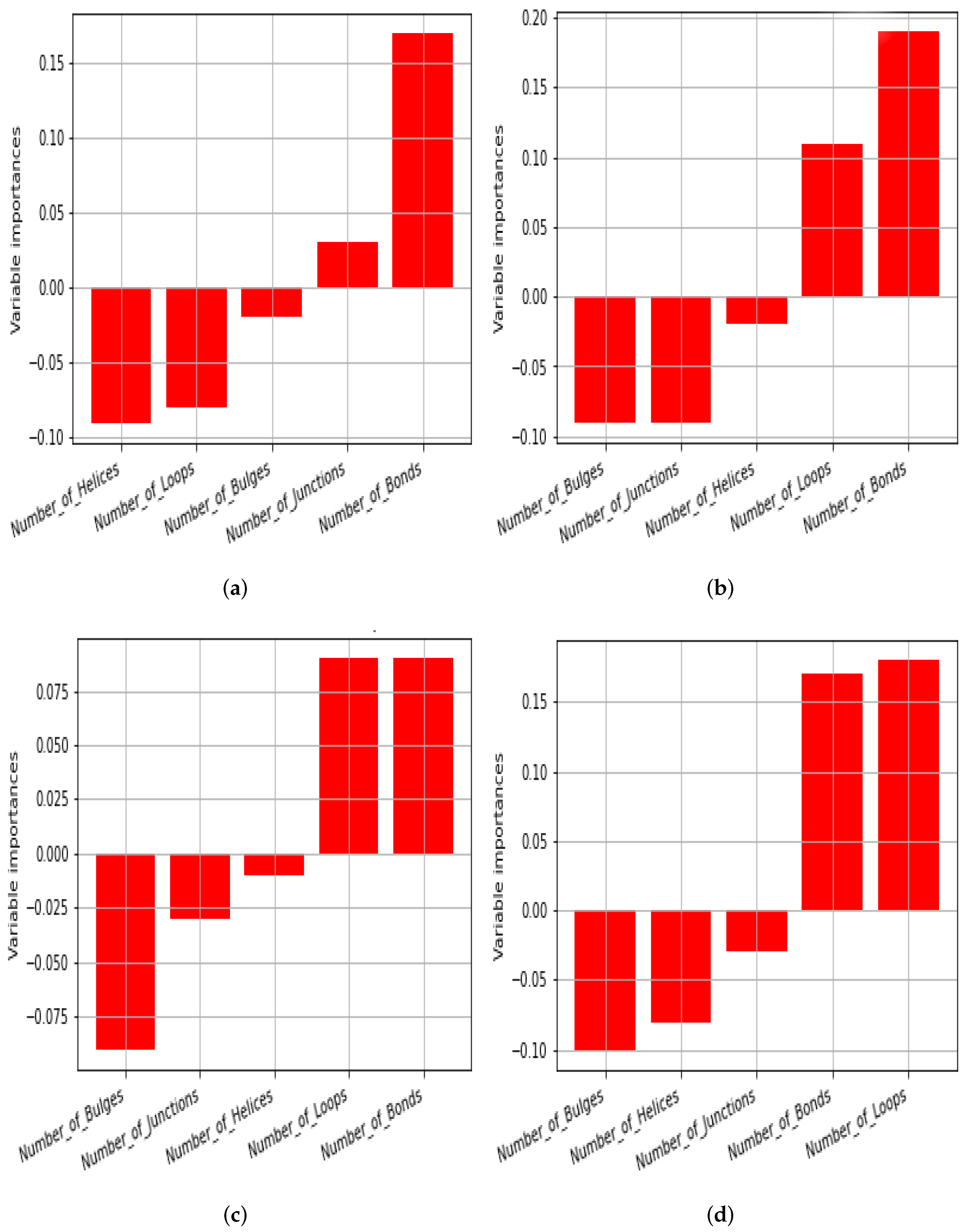
| Motif | Natural | Random Samples |
|---|---|---|
| Bulges | 0.010 | 0.013 |
| Loops | 0.020 | 0.018 |
| Junctions | 0.0083 | 0.0085 |
| Helices | 0.070 | 0.073 |
| Bonds | 0.31 | 0.32 |
| (a) Slopes | ||
|---|---|---|
| Motif | Natural | Random Samples |
| Bulges | [0.010, 0.011] | [0.012, 0.013] |
| Loops | [0.011, 0.020] | [0.012, 0.018] |
| Junctions | [0.0082, 0.020] | [0.0084, 0.018] |
| Helices | [0.0082, 0.070] | [0.0084, 0.073] |
| Bonds | [0.0082, 0.32] | [0.0085, 0.32] |
| (b) Intercepts | ||
| Motif | Natural | Random Samples |
| Bulges | [−0.97, −0.35] | [−0.50, 0.28] |
| Loops | [−0.90, 1.2] | [−0.42, 0.86] |
| Junctions | [−0.85, 1.2] | [−0.62, 0.82] |
| Helices | [−0.81, 2.1] | [−0.61, 0.79] |
| Bonds | [−0.79, 2.6] | [−6.4, 0.76]] |
| (a) kNN | ||
|---|---|---|
| Length (L) | Original ROC Area | 95% Confidence Interval |
| 100 | 0.72 | [0.72–0.73] |
| 400 | 0.81 | [0.81–0.82] |
| 1000 | 0.86 | [0.85–0.88] |
| 1000 GC adjusted | 0.86 | [0.85–0.87] |
| (b) PLSDA | ||
| Length (L) | Original ROC Area | 95% Confidence Interval |
| 100 | 0.70 | [0.70–0.71] |
| 400 | 0.78 | [0.78–0.78] |
| 1000 | 0.83 | [0.82–0.84] |
| 1000 GC adjusted | 0.83 | [0.82–0.84] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghaddar, F.; Dingle, K. Random and Natural Non-Coding RNA Have Similar Structural Motif Patterns but Differ in Bulge, Loop, and Bond Counts. Life 2023, 13, 708. https://doi.org/10.3390/life13030708
Ghaddar F, Dingle K. Random and Natural Non-Coding RNA Have Similar Structural Motif Patterns but Differ in Bulge, Loop, and Bond Counts. Life. 2023; 13(3):708. https://doi.org/10.3390/life13030708
Chicago/Turabian StyleGhaddar, Fatme, and Kamaludin Dingle. 2023. "Random and Natural Non-Coding RNA Have Similar Structural Motif Patterns but Differ in Bulge, Loop, and Bond Counts" Life 13, no. 3: 708. https://doi.org/10.3390/life13030708
APA StyleGhaddar, F., & Dingle, K. (2023). Random and Natural Non-Coding RNA Have Similar Structural Motif Patterns but Differ in Bulge, Loop, and Bond Counts. Life, 13(3), 708. https://doi.org/10.3390/life13030708







