Molecular Identification of Bacteria Isolated from Marketed Sparus aurata and Penaeus indicus Sea Products: Antibiotic Resistance Profiling and Evaluation of Biofilm Formation
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Material and Bacterial Isolation
2.2. Bacterial Identification and Phylogenetic Analyzes
2.3. Exoenzyme Production
2.4. Antibiotic Susceptibility Test
2.5. Adhesion Properties and Biofilm Formation Screening
2.5.1. Exopolysaccharide (Slime) Production
2.5.2. Wolfe Test
2.5.3. Biofilm Formation on Polystyrene Microtiter Plates
2.5.4. Biofilm Formation on Glass and Plastic Surfaces
2.6. Statistical Analysis
3. Results
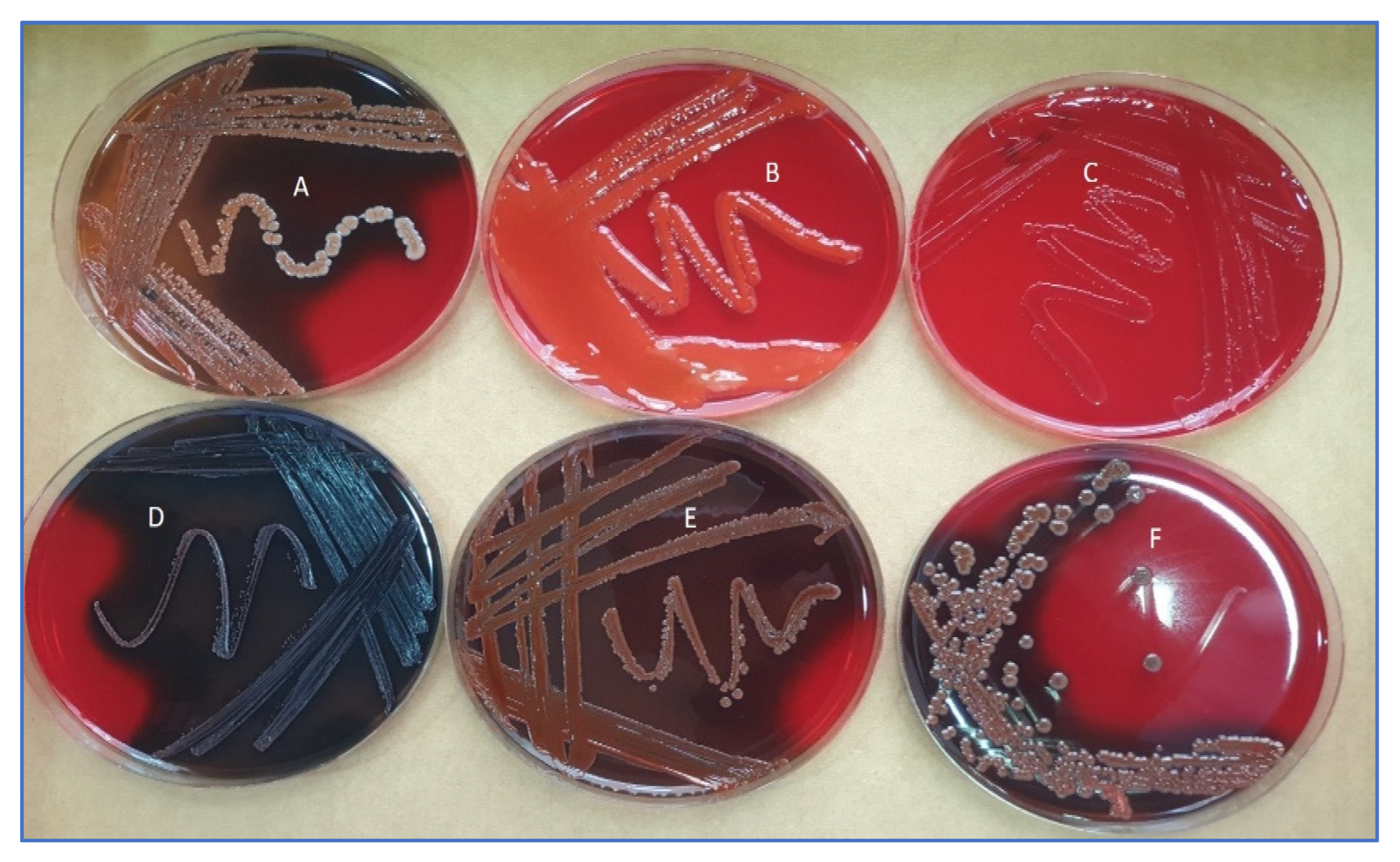
3.1. Morphological Characterization and 16SRNA Identification of Bacterial Isolates
3.2. Exoenzyme Production
3.3. Antibiotic Susceptibility Test
3.4. Slime Production on CRA Plates and Glass Tubes (Wolfe Test)
3.5. Quantitative Estimation of Biofilm Formation by Tasted Bacteria on Abiotic Surfaces
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Okocha, R.C.; Olatoye, I.O.; Adedeji, O.B. Food safety impacts of antimicrobial use and their residues in aquaculture. Public Health Rev. 2018, 39, 21. [Google Scholar] [CrossRef] [PubMed]
- Sheng, L.; Wang, L. The microbial safety of fish and fish products: Recent advances in understanding its significance, contamination sources, and control strategies. Compr. Rev. Food Sci. Food Saf. 2021, 20, 738–786. [Google Scholar] [CrossRef]
- Bolivar, A.; Costa, J.; Posada-Izquierdo, G.; Pérez-Rodríguez, F.; Bascón, I.; Zurera, G.; Valero, A. Characterization of foodborne pathogens and spoilage bacteria in mediterranean fish species and seafood products. Foodborne Pathog. Antibiot. Resist. 2016, 21–39. [Google Scholar] [CrossRef]
- Mokrani, D.; Oumouna, M.; Cuesta, A. Fish farming conditions affect to European sea bass (Dicentrarchus labrax L.) quality and shelf life during storage in ice. Aquaculture 2018, 490, 120–124. [Google Scholar] [CrossRef]
- Boziaris, I.S.; Parlapani, F.F. Specific spoilage organisms (SSOs) in fish. In The Microbiological Quality of Food; Elsevier: Amsterdam, The Netherlands, 2017; pp. 61–98. [Google Scholar]
- Baker-Austin, C.; Oliver, J.D. Vibrio vulnificus: New insights into a deadly opportunistic pathogen. Environ. Microbiol. 2018, 20, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, R.H.; Peng, T.L.; Ong, B.L.; Suhana, M.Y.S.; Hamid, N.H.; Afifah, M.N.F.; Raina, M.S. Antibiotics resistance of Vibrio spp. isolated from diseased seabass and tilapia in cage culture. In Proceedings of the International Seminar on Livestock Production and Veterinary Technology, Denpasar, Indonesia, 10–12 August 2016. [Google Scholar]
- Rippen, T.; Skonberg, D. Handling of Fresh Fish. In The Seafood Industry; Wiley: Hoboken, NJ, USA, 2012; pp. 249–260. [Google Scholar]
- Colombo, F.M.; Cattaneo, P.; Confalonieri, E.; Bernardi, C. Histamine food poisonings: A systematic review and meta-analysis. Crit. Rev. Food Sci. Nutr. 2018, 58, 1131–1151. [Google Scholar] [CrossRef]
- Parlapani, F.F. Microbial diversity of seafood. Curr. Opin. Food Sci. 2021, 37, 45–51. [Google Scholar] [CrossRef]
- Dang, H.; Song, L.; Chen, M.; Chang, Y. Concurrence of cat and tet Genes in Multiple Antibiotic-Resistant Bacteria Isolated from a Sea Cucumber and Sea Urchin Mariculture Farm in China. Microb. Ecol. 2006, 52, 634–643. [Google Scholar] [CrossRef]
- Snoussi, M.; Noumi, E.; Messaoud, A.; Hajlaoui, H.; Bakhrouf, A. Biochemical characteristics and genetic diversity of Vibrio spp. and Aeromonas hydrophila strains isolated from the Lac of Bizerte (Tunisia). World J. Microbiol. Biotechnol. 2010, 26, 2037–2046. [Google Scholar]
- Lajnef, R.; Snoussi, M.; Balboa, S.; Bastardo, A.; Laabidi, H.; Chatti, A.; Abdennaceur, H.; Romalde, J.L. Molecular typing of V. alginolyticus strains isolated from Tunisian marine biotopes by two PCR-based methods (ERIC and REP). Afr. J. Microbiol. Res. 2012, 6, 4647–4654. [Google Scholar] [CrossRef]
- Khoudja, S.; Snoussi, M.; Saidi, N.; Bakhrouf, A. Phenotypic characterization and RAPD fingerprinting of Vibrio parahaemolyticus and Vibrio alginolyticus isolated during Tunisian fish farm outbreaks. Folia Microbiol. 2013, 58, 17–26. [Google Scholar]
- Snoussi, M.; Noumi, E.; Lajnef, R.; Bellila, A.; Yazidi, N.; Bakhrouf, A. Phenotypic characterization and enterobacterial repetitive intergenic consensus PCR of Aeromonas spp. and Vibrio spp. strains isolated from Tunisian sea bream (Sparus aurata) fish farm. Afr. J. Microbiol. Res. 2011, 5, 2920–2928. [Google Scholar] [CrossRef]
- Snoussi, M.; Trabelsi, N.; Ben Taleb, S.; Dehmeni, A.; Flamini, G.; De Feo, V. Laurus nobilis, Zingiber officinale and Anethum graveolens essential oils: Composition, antioxidant and antibacterial activities against bacteria isolated from fish and shellfish. Molecules 2016, 21, 1414. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.N.K.; Van, T.T.H.; Nguyen, H.T.; Smooker, P.M.; Shimeta, J.; Coloe, P.J. Molecular characterization of antibiotic resistance in Pseudomonas and Aeromonas isolates from catfish of the Mekong Delta, Vietnam. Vet. Microbiol. 2014, 171, 397–405. [Google Scholar] [CrossRef]
- Snoussi, M.; Chaieb, K.; Mahmoud, R.; Bakhrouf, A. Quantitative study, identification and antibiotics sensitivity of someVibrionaceae associated to a marine fish hatchery. Ann. Microbiol. 2006, 56, 289–293. [Google Scholar] [CrossRef]
- Muyzer, G.; Teske, A.; Wirsen, C.O.; Jannasch, H.W. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 1995, 164, 165–172. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symposium Series; Information Retrieval Ltd.: London, UK, 1999; pp. c1979–c2000. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Hörmansdorfer, S.; Wentges, H.; Neugebaur-Büchler, K.; Bauer, J. Isolation of Vibrio alginolyticus from seawater aquaria. Int. J. Hyg. Environ. Health 2000, 203, 169–175. [Google Scholar] [CrossRef]
- Snoussi, M.; Hajlaoui, H.; Noumi, E.; Zanetti, S.; Bakhrouf, A. Phenotypic and genetic diversity of Vibrio alginolyticus strains recovered from juveniles and older Sparus aurata reared in a Tunisian marine farm. Ann. Microbiol. 2008, 58, 141–146. [Google Scholar] [CrossRef]
- Lajnef, R.; Snoussi, M.; Romalde, J.L.; Nozha, C.; Hassen, A. Comparative study on the antibiotic susceptibility and plasmid profiles of Vibrio alginolyticus strains isolated from four Tunisian marine biotopes. World J. Microbiol. Biotechnol. 2012, 28, 3345–3363. [Google Scholar] [CrossRef]
- Jorgensen, J.H. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria; approved guideline; Clinical & Laboratory Standards Institute: Wayne, PA, USA, 2010. [Google Scholar]
- Performance, C. Standards for Antimicrobial Susceptibility Testing, CLSI Supplement M100S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Manjusha, S.S.G.; Elyas, K.K.; Chandrasekaran, M. Multiple antibiotic resistances of Vibrio isolates from coastal and brackish water areas. Am. J. Biochem. Biotechnol. 2005, 1, 201–206. [Google Scholar]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Snoussi, M.; Noumi, E.; Cheriaa, J.; Usai, D.; Sechi, L.A.; Zanetti, S.; Bakhrouf, A. Adhesive properties of environmental Vibrio alginolyticus strains to biotic and abiotic surfaces. New Microbiol. 2008, 31, 489–500. [Google Scholar]
- Snoussi, M.; Noumi, E.; Hajlaoui, H.; Usai, D.; Sechi, L.A.; Zanetti, S.; Bakhrouf, A. High potential of adhesion to abiotic and biotic materials in fish aquaculture facility by Vibrio alginolyticus strains. J. Appl. Microbiol. 2009, 106, 1591–1599. [Google Scholar] [CrossRef]
- Wolfe, A.J.; Millikan, D.S.; Campbell, J.M.; Visick, K.L. Vibrio fischeri σ54 controls motility, biofilm formation, luminescence, and colonization. Appl. Environ. Microbiol. 2004, 70, 2520–2524. [Google Scholar] [CrossRef]
- Toledo-Arana, A.; Valle, J.; Solano, C.; Arrizubieta, M.J.; Cucarella, C.; Lamata, M.; Amorena, B.; Leiva, J.; Penadés, J.R.; Lasa, I. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 2001, 67, 4538–4545. [Google Scholar] [CrossRef]
- Henriques, M.; Azeredo, J.; Oliveira, R. The Influence of Subinhibitory Concentrations of Fluconazole and Amphotericin b in the Biofilm Formation of CANDIDA Albicans and Candida Dubliniensis. Int. J. Artif. Organs 2005, 28, 1181–1185. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Dakić, I.; Savić, B.; Švabić-Vlahović, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Snoussi, M.; Noumi, E.; Usai, D.; Sechi, L.A.; Zanetti, S.; Bakhrouf, A. Distribution of some virulence related-properties of Vibrio alginolyticus strains isolated from Mediterranean seawater (Bay of Khenis, Tunisia): Investigation of eight Vibrio cholerae virulence genes. World J. Microbiol. Biotechnol. 2008, 24, 2133–2141. [Google Scholar] [CrossRef]
- Sorroza, L.; Padilla, D.; Acosta, F.; Román, L.; Grasso, V.; Vega, J.; Real, F. Characterization of the probiotic strain Vagococcus fluvialis in the protection of European sea bass (Dicentrarchus labrax) against vibriosis by Vibrio anguillarum. Vet. Microbiol. 2011, 155, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Elhadi, N.; Radu, S.; Chen, C.H.; Nishibuchi, M. Prevalence of potentially pathogenic Vibrio species in the seafood marketed in Malaysia. J. Food Prot. 2004, 67, 1469–1475. [Google Scholar] [CrossRef]
- Colakoglu, F.A.; Sarmasik, A.; Koseoglu, B. Occurrence of Vibrio spp. and Aeromonas spp. in shellfish harvested off Dardanelles cost of Turkey. Food Control 2006, 17, 648–652. [Google Scholar] [CrossRef]
- Al-Sunaiher, A.E.; Ibrahim, A.S.; Al-Salamah, A.A. Association of Vibrio species with disease incidence in some cultured fishes in the Kingdom of Saudi Arabia. World Appl. Sci. J. 2010, 8, 653–660. [Google Scholar]
- Alikunhi, N.M.; Batang, Z.B.; AlJahdali, H.A.; Aziz, M.A.; Al-Suwailem, A.M. Culture-dependent bacteria in commercial fishes: Qualitative assessment and molecular identification using 16S rRNA gene sequencing. Saudi J. Biol. Sci. 2017, 24, 1105–1116. [Google Scholar] [CrossRef]
- Elhadi, N.; Aljeldah, M.; Aljindan, R. Microbiological contamination of imported frozen fish marketed in Eastern Province of Saudi Arabia. Int. Food Res. J. 2016, 23, 2723. [Google Scholar]
- Hassan, M.A.; Soliman, W.S.; Mahmoud, M.A.; AI-Shabeeb, S.S.; Imran, P.M. Prevalence of bacterial infections among cage-cultured marine fishes at the eastern province of Saudi Arabia. Res. J. Pharm. Biol. Chem. Sci. 2015, 6, 1112–1126. [Google Scholar]
- Ibrahim, M.M.; Al Shabeeb, S.S.; Noureldin, E.; Al Ramadhan, G.H. Occurrence of Potentially Pathogenic Vibrio and related species in Seafoods obtained from the Eastern Province of Saudi Arabia. Int. J. Adv. Res. Biol. Sci. 2016, 3, 71–80. [Google Scholar]
- Salama, A.J.; Satheesh, S.; Balqadi, A.A.; Kitto, M.R. Identifying suitable fin fish cage farming sites in the eastern Red Sea Coast, Saudi Arabia. Thalass. Int. J. Mar. Sci. 2016, 32, 1–9. [Google Scholar] [CrossRef]
- Alkuraieef, A.N.; Alsuhaibani, A.M.; Alshawi, A.H.; Alfaris, N.A.; Aljabryn, D.H. Chemical and microbiological quality of imported chilled, frozen, and locally cultured fish in Saudi Arabian markets. Food Sci. Technol. 2021, 42. [Google Scholar] [CrossRef]
- Beyari, E.A.; Aly, M.M.; Jastaniah, S.D. Bacterial Diversity in Some Local Fishes at Central Fish Market of Jeddah, Saudi Arabia. Ann. Med. Health Sci. Res. Vol. 2021, 11 (Suppl. 4), 33–38. [Google Scholar]
- Al-Ghanayem, A.; Joseph, B.; Bin Mahdi, M.; Scaria, B.; Saadabi, A.M. Multidrug Resistance Pattern of Bacteria Isolated from Fish Samples Sold in Retail Market. J. Clin. Diagn. Res. 2020, 14, 13–16. [Google Scholar] [CrossRef]
- Sivertsvik, M.; Jeksrud, W.K.; Rosnes, J.T. A review of modified atmosphere packaging of fish and fishery products–significance of microbial growth, activities and safety. Int. J. Food Sci. Technol. 2002, 37, 107–127. [Google Scholar] [CrossRef]
- Cabral, J.P. Water Microbiology. Bacterial pathogens and water. Int. J. Environ. Res. Public Health 2010, 7, 3657–3703. [Google Scholar] [CrossRef]
- Kirov, S.M.; Barnett, T.C.; Pepe, C.M.; Strom, M.S.; Albert, M.J. Investigation of the role of type IV Aeromonas pilus (Tap) in the pathogenesis of Aeromonas gastrointestinal infection. Infect. Immun. 2000, 68, 4040–4048. [Google Scholar] [CrossRef]
- Pogorelova, N.P.; A Zhuravleva, L.; Ibragimov, F.K.; Iushchenko, G.V. Bacteria of the genus Aeromonas as the causative agents of saprophytic infection. Zhurnal Mikrobiol. Epidemiol. I Immunobiol. 1995, 4, 9–12. [Google Scholar]
- Janda, J.M.; Abbott, S.L. Evolving concepts regarding the genus Aeromonas: An expanding panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 1998, 27, 332–344. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S. Aeromonas gastrointestinal disease: A case study in causation. Genus Aeromonas 1996, 311–326. [Google Scholar]
- Parker, J.L.; Shaw, J.G. Aeromonas spp. clinical microbiology and disease. J. Infect. 2011, 62, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Novotny, L.; Dvorska, L.; Lorencova, A.; Beran, V.; Pavlik, I. Fish: A potential source of bacterial pathogens for human beings. Vet. Med. 2004, 49, 343–358. [Google Scholar] [CrossRef]
- Wellington, E.M.H.; Boxall, A.B.A.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W.; et al. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Dai, W.; Qiu, Q.; Dong, C.; Zhang, J.; Xiong, J. Contrasting Ecological Processes and Functional Compositions Between Intestinal Bacterial Community in Healthy and Diseased Shrimp. Microb. Ecol. 2016, 72, 975–985. [Google Scholar] [CrossRef] [PubMed]
- Long, M.; Nielsen, T.K.; Leisner, J.J.; Hansen, L.H.; Shen, Z.X.; Zhang, Q.Q.; Li, A. Aeromonas salmonicida subsp. salmonicida strains isolated from Chinese freshwater fish contain a novel genomic island and possible regional-specific mobile genetic elements profiles. FEMS Microbiol. Lett. 2016, 363, fnw190. [Google Scholar] [CrossRef]
- Chandrarathna, H.; Nikapitiya, C.; Dananjaya, S.; Wijerathne, C.; Wimalasena, S.; Kwun, H.J.; Heo, G.-J.; Lee, J.; De Zoysa, M. Outcome of co-infection with opportunistic and multidrug resistant Aeromonas hydrophila and A. veronii in zebrafish: Identification, characterization, pathogenicity and immune responses. Fish Shellfish. Immunol. 2018, 80, 573–581. [Google Scholar] [CrossRef]
- Hamed, S.B.; Ranzani-Paiva, M.J.T.; Tachibana, L.; de Carla Dias, D.; Ishikawa, C.M.; Esteban, M.A. Fish pathogen bacteria: Adhesion, parameters influencing virulence and interaction with host cells. Fish Shellfish. Immunol. 2018, 80, 550–562. [Google Scholar] [CrossRef]
- Lafisca, A.; Pereira, C.S.; Giaccone, V.; Rodrigues, D.D.P. Enzymatic characterization of Vibrio alginolyticus strains isolated from bivalves harvested at Venice Lagoon (Italy) and Guanabara Bay (Brazil). Rev. Do Inst. De Med. Trop. De São Paulo 2008, 50, 199–202. [Google Scholar] [CrossRef]
- Bidinost, C.; Saka, H.A.; Aliendro, O.; Sola, C.; Panzetta-Duttari, G.; Carranza, P.; Echenique, J.; Patrito, E.; Bocco, J.L. Virulence factors of non-01 non-O139 Vibrio cholerae isolated in Cordoba, Argentina. Rev. Argent. De Microbiol. 2004, 36, 158. [Google Scholar]
- Duarte, A.S.; Correia, A.; Esteves, A. Bacterial collagenases—A review. Crit. Rev. Microbiol. 2016, 42, 106–126. [Google Scholar] [CrossRef]
- Maluping, R. Potential Aquatic Bacterial Pathogens in the Philippines and Thailand; Swedish University of Agricultural Sciences: Uppsala, Sweden, 2011. [Google Scholar]
- Costa, R.A.; Amorim, L.M.C.; Araújo, R.L.; dos Fernandes Vieira, R.H.S. Multiple enzymatic profiles of Vibrio parahaemolyticus strains isolated from oysters. Rev. Argent. De Microbiol. 2013, 45, 267–270. [Google Scholar] [CrossRef]
- Kemper, M.A.; Urrutia, M.M.; Beveridge, T.J.; Koch, A.L.; Doyle, R.J. Proton motive force may regulate cell wall-associated enzymes of Bacillus subtilis. J. Bacteriol. 1993, 175, 5690–5696. [Google Scholar] [CrossRef][Green Version]
- Natrah, F.; Ruwandeepika, H.D.; Pawar, S.; Karunasagar, I.; Sorgeloos, P.; Bossier, P.; Defoirdt, T. Regulation of virulence factors by quorum sensing in Vibrio harveyi. Vet. Microbiol. 2011, 154, 124–129. [Google Scholar] [CrossRef]
- Liuxy, P.C.; Lee, K.K.; Chen, S.N. Pathogenicity of different isolates of Vibrio harveyi in tiger prawn, Penaeus monodon. Lett. Appl. Microbiol. 1996, 22, 413–416. [Google Scholar] [CrossRef]
- Zhang, X.H.; Austin, B. Pathogenicity of Vibrio harveyi to salmonids. J. Fish Dis. 2000, 23, 93–102. [Google Scholar] [CrossRef]
- Watts, J.E.M.; Schreier, H.J.; Lanska, L.; Hale, M.S. The rising tide of antimicrobial resistance in aquaculture: Sources, sinks and solutions. Mar. Drugs 2017, 15, 158. [Google Scholar] [CrossRef]
- Titilawo, Y.; Sibanda, T.; Obi, L.; Okoh, A. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of faecal contamination of water. Environ. Sci. Pollut. Res. 2015, 22, 10969–10980. [Google Scholar] [CrossRef]
- Sechi, L.; Deriu, A.; Falchi, M.; Fadda, G.; Zanetti, S. Distribution of virulence genes in Aeromonas spp. isolated from Sardinian waters and from patients with diarrhoea. J. Appl. Microbiol. 2002, 92, 221–227. [Google Scholar] [CrossRef]
- Rajkumar, H.; Devaki, R.; Kandi, V. Evaluation of different phenotypic techniques for the detection of slime produced by bacteria isolated from clinical specimens. Cureus 2016, 8, e505. [Google Scholar]
- Muller, E.; Hübner, J.; Gutierrez, N.; Takeda, S.; A Goldmann, D.; Pier, G.B. Isolation and characterization of transposon mutants of Staphylococcus epidermidis deficient in capsular polysaccharide/adhesin and slime. Infect. Immun. 1993, 61, 551–558. [Google Scholar] [CrossRef]
- Abdallah, F.B.; Chaieb, K.; Zmantar, T.; Kallel, H.; Bakhrouf, A. Adherence assays and slime production of Vibrio alginolyticus and Vibrio parahaemolyticus. Braz. J. Microbiol. 2009, 40, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Odeyemi, O.A.; Ahmad, A. Population dynamics, antibiotics resistance and biofilm formation of Aeromonas and Vibrio species isolated from aquatic sources in Northern Malaysia. Microb. Pathog. 2017, 103, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Mack, D.; Rohde, H.; Dobinsky, S.; Riedewald, J.; Nedelmann, M.; Knobloch, J.K.-M.; Elsner, H.-A.; Feucht, H.H. Identification of three essential regulatory gene loci governing expression of Staphylococcus epidermidis polysaccharide intercellular adhesin and biofilm formation. Infect. Immun. 2000, 68, 3799–3807. [Google Scholar] [CrossRef] [PubMed]
- Alreshidi, M.M.; Dunstan, R.H.; Macdonald, M.M.; Gottfries, J.; Roberts, T.K. The uptake and release of amino acids by Staphylococcus aureus at mid-exponential and stationary phases and their corresponding responses to changes in temperature, pH and osmolality. Front. Microbiol. 2020, 10, 3059. [Google Scholar] [CrossRef] [PubMed]
- Abebe, G.M. The role of bacterial biofilm in antibiotic resistance and food contamination. Int. J. Microbiol. 2020, 2020, 1705814. [Google Scholar] [CrossRef]
- Alreshidi, M.M.; Dunstan, R.; Macdonald, M.M.; Singh, V.K.; Roberts, T.K. Analysis of Cytoplasmic and Secreted Proteins of Staphylococcus aureus Revealed Adaptive Metabolic Homeostasis in Response to Changes in the Environmental Conditions Representative of the Human Wound Site. Microorganisms 2020, 8, 1082. [Google Scholar] [CrossRef]
- Xing, Y.; Luo, X.; Liu, S.; Wan, W.; Huang, Q.; Chen, W. A novel eco-friendly recycling of food waste for preparing biofilm-attached biochar to remove Cd and Pb in wastewater. J. Clean. Prod. 2021, 311, 127514. [Google Scholar] [CrossRef]




| Agar | Site of Isolation | Colony Color | Code | Bacteria Name | Accession |
|---|---|---|---|---|---|
| TCBS agar | P. indicus abdomen muscles | Yellow | P12 | Vibrio hyugaensis | OP703739.1 |
| P. indicus abdomen muscles | Green yellow | P13 | Shewanella indica | OP704022.1 | |
| P. indicus abdomen muscles | Yellow | P14 | Vibrio natriegens | OP703736.1 | |
| Vibrio ChromoSelect agar | P. indicus abdomen muscles | Blue | P1 | Vibrio natriegens | OP703737.1 |
| P. indicus abdomen muscles | Turquoise | P2 | Vibrio alginolyticus | OP703632.1 | |
| P. indicus abdomen muscles | Colorless | P5 | Morganella morganii | OP704015.1 | |
| P. indicus abdomen muscles | Purple | P9 | Vibrio harveyi | OP704026.1 | |
| TCBS agar | S. aurata intestines | Yellow | SA1 | Aeromonas veronii | OP704025.1 |
| S. aurata intestines | Yellow | SA3 | Photobacterium. Piscicida | OP704011.1 | |
| S. aurata muscles | Yellow | SA17 | Aeromonas veronii | OP704024.1 | |
| S. aurata muscles | Blue green | SA21 | Vagococcus fluvialis | OP704018.1 | |
| S. aurata gills | Yellow | SA27 | Vibrio natriegens | OP703612.1 | |
| Vibrio ChromoSelect agar | S. aurata intestines | Turquoise | SA5 | Photobacterium damselae | OP704023.1 |
| S. aurata intestines | Light green with a green center | SA7 | Staphylococcus epidermidis | OP704017.1 | |
| S. aurata intestines | Colorless | SA9 | Bacillus cereus | OP704016.1 | |
| S. aurata gills | Light green with a green center | SA11 | Vibrio natriegens | OP704027.1 | |
| S. aurata gills | Colorless | SA13 | Vibrio natriegens | OP704012.1 | |
| S. aurata gills | Pink | SA15 | Aeromonas veronii | OP703806.1 | |
| S. aurata intestines | Green | SA25 | Aeromonas veronii | OP704013.1 | |
| S. aurata muscles | Blue | SA26 | Vibrio harveyi | OP703738.1 | |
| S. aurata muscles | Colorless | SA31 | Aeromonas veronii | OP704014.1 |
| Code | Strains | DNase | Lipase | Lecithinase | Caseinase | Hemolysis | Amylase | Gelatinase |
|---|---|---|---|---|---|---|---|---|
| P1 | V.natriegens | + | + | + | + | − | + | − |
| P14 | V.natriegens | + | + | − | − | − | + | − |
| SA11 | V.natriegens | + | + | − | − | − | + | − |
| SA13 | V.natriegens | − | + | + | + | − | + | + |
| SA27 | V.natriegens | + | + | − | − | − | + | − |
| P9 | V.harveyi * | + | + | + | + | + | + | + |
| SA26 | V.harveyi | − | + | − | + | − | + | + |
| P2 | V.alginolyticus * | + | + | + | + | + | + | + |
| P12 | V.hyugaensis | + | + | − | − | − | + | − |
| SA1 | A.veronii | − | − | − | + | − | + | + |
| SA15 | A.veronii * | + | + | + | + | + | + | + |
| SA17 | A.veronii * | + | + | + | + | + | + | − |
| SA25 | A.veronii * | + | − | + | + | + | + | + |
| SA31 | A.veronii * | + | − | + | + | + | + | − |
| SA3 | P.piscicida * | − | + | − | − | + | + | − |
| SA5 | P.damselae * | − | + | − | − | + | + | − |
| P5 | M.morganii | + | − | − | − | − | + | − |
| P13 | S.indica * | + | + | + | + | + | + | + |
| SA9 | B.cereus * | + | + | + | + | + | + | − |
| SA7 | S.epidermidis | − | + | + | + | − | + | + |
| SA21 | V.fluvialis * | + | + | + | + | + | + | + |
| Microorganisms Tested | Antibiotic Resistance Index (ARI) |
|---|---|
| Vibrio spp. (n = 9) | 0.542 |
| Aeromonas spp. (n = 5) | 0.553 |
| All Gram-negative | 0.544 |
| All Gram-positive | 0.462 |
| Code | Bacteria Tested | Slime Production on CRA | Wolfe Test | |
|---|---|---|---|---|
| Morphotype | Interpretation | |||
| P1 | V. natriegens | Red | Non producer | ++ |
| P14 | V. natriegens | Black | Producer | + |
| SA11 | V. natriegens | Black | Producer | ++ |
| SA13 | V. natriegens | Orange | Non producer | ++ |
| SA27 | V. natriegens | Black | Producer | ++ |
| P9 | V. harveyi | Black | Producer | ++ |
| SA26 | V. harveyi | Red | Non producer | + |
| P2 | V. alginolyticus | Bordeaux | Non producer | ++ |
| P12 | V. hyugaensis | Bordeaux | Non producer | + |
| SA1 | A. veronii | Black | Producer | + |
| SA15 | A. veronii | Bordeaux | Non producer | ++ |
| SA17 | A. veronii | White with red center | Non producer | + |
| SA25 | A. veronii | Black | Producer | ++ |
| SA31 | A. veronii | Black gray | Non producer | + |
| SA3 | P. piscicida | Red | Non producer | + |
| SA5 | P. damselae | Red | Non producer | − |
| P5 | M. morganii | Red | Non producer | + |
| P13 | S. indica | Bordeaux | Non producer | ++ |
| SA9 | B. cereus | Bordeaux | Non producer | ++ |
| SA7 | S. epidermidis | Red | Non producer | + |
| SA21 | V. fluvialis | Bordeaux | Non producer | + |
| Code | Bacteria Tested | Polystyrene * | Glass ** | Plastic ** | |||
|---|---|---|---|---|---|---|---|
| OD595nm ± SD | Interpretation | OD595nm ± SD | Interpretation | OD595nm ± SD | Interpretation | ||
| P1 | V. natriegens | 0.360 ± 0.024 | (−); Non biofilm forming | 0.102 ± 0.002 | (+); Weakly adherent | 0.152 ± 0.036 | (++); Moderately adherent |
| P14 | V. natriegens | 0.746 ± 0.002 | (−); Non biofilm forming | 0.114 ± 0.006 | (+); Weakly adherent | 0.412 ± 0.022 | (+++), Strongly adherent |
| SA11 | V. natriegens | 2.029 ± 0.166 | (++); Medium biofilm forming | 0.070 ± 0.004 | (−); Non adherent | 0.081 ± 0.013 | (+); Weakly adherent |
| SA13 | V. natriegens | 0.976 ± 0.061 | (−); Non biofilm forming | 0.108 ± 0.009 | (+); Weakly adherent | 0.387 ± 0.514 | (+++), Strongly adherent |
| SA27 | V. natriegens | 0.599 ± 0.026 | (−); Non biofilm forming | 0.189 ± 0.010 | (++); Moderately adherent | 0.148 ± 0.008 | (++); Moderately adherent |
| P9 | V. harveyi | 1.492 ± 0.119 | (+); Weak biofilm forming | 0.101 ± 0.027 | (+); Weakly adherent | 0.081 ± 0.017 | (+); Weakly adherent |
| SA26 | V. harveyi | 0.981 ± 0.178 | (−); Non biofilm forming | 0.245 ± 0.023 | (++); Moderately adherent | 0.219 ± 0.061 | (++); Moderately adherent |
| P2 | V. alginolyticus | 2.070 ± 0.076 | (++); Medium biofilm forming | 0.188 ± 0.045 | (++); Moderately adherent | 0.116 ± 0.033 | (+); Weakly adherent |
| P12 | V. hyuganesis | 2.029 ± 0.206 | (++); Medium biofilm forming | 0.075 ± 0.008 | (−); Non adherent | 0.067 ± 0.009 | (−); Non adherent |
| SA1 | A. veronii | 1.505 ± 0.072 | (+); Weak biofilm forming | 0.112 ± 0.007 | (+); Weakly adherent | 0.099 ± 0.022 | (+); Weakly adherent |
| SA15 | A. veronii | 1.500 ± 0.118 | (+); Weak biofilm forming | 0.140 ± 0.013 | (+); Weakly adherent | 0.055 ± 0.005 | (−); Non adherent |
| SA17 | A. veronii | 0.835 ± 0.055 | (−); Non biofilm forming | 0.091 ± 0.016 | (+); Weakly adherent | 0.077 ± 0.005 | (+); Weakly adherent |
| SA25 | A. veronii | 0.351 ± 0.021 | (−); Non biofilm forming | 0.158 ± 0.021 | (+); Weakly adherent | 0.060 ± 0.004 | (−); Non adherent |
| SA31 | A. veronii | 1.214 ± 0.216 | (+); Weak biofilm forming | 0.142 ± 0.010 | (+); Weakly adherent | 0.099 ± 0.005 | (+); Weakly adherent |
| SA3 | P. piscicida | 2.792 ± 0.244 | (++); Medium biofilm forming | 0.246 ± 0.027 | (++); Moderately adherent | 0.077 ± 0.001 | (+); Weakly adherent |
| SA5 | P. damselae | 0.410 ± 0.028 | (−); Non biofilm forming | 0.109 ± 0.010 | (+); Weakly adherent | 0.118 ± 0.019 | (+); Weakly adherent |
| P13 | S. indica | 0.505 ± 0.078 | (−); Non biofilm forming | 0.110 ± 0.007 | (+); Weakly adherent | 0.418 ± 0.004 | (+++), Strongly adherent |
| P5 | M. morganii | 0.782 ± 0.053 | (−); Non biofilm forming | 0.114 ± 0.012 | (+); Weakly adherent | 0.133 ± 0.009 | (+); Weakly adherent |
| SA9 | B. cereus | 2.525 ± 0.210 | (++); Medium biofilm forming | 0.313 ± 0.061 | (++); Moderately adherent | 0.151 ± 0.025 | (++); Moderately adherent |
| SA7 | S. epidermidis | 0.781 ± 0.023 | (−); Non biofilm forming | 0.114 ± 0.009 | (+); Weakly adherent | 0.182 ± 0.010 | (++); Moderately adherent |
| SA21 | V. fluvialis | 0.307 ± 0.011 | (−); Non biofilm forming | 0.116 ± 0.012 | (+); Weakly adherent | 0.078 ± 0.018 | (+); Weakly adherent |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdulhakeem, M.A.; Alreshidi, M.; Bardakci, F.; Hamadou, W.S.; De Feo, V.; Noumi, E.; Snoussi, M. Molecular Identification of Bacteria Isolated from Marketed Sparus aurata and Penaeus indicus Sea Products: Antibiotic Resistance Profiling and Evaluation of Biofilm Formation. Life 2023, 13, 548. https://doi.org/10.3390/life13020548
Abdulhakeem MA, Alreshidi M, Bardakci F, Hamadou WS, De Feo V, Noumi E, Snoussi M. Molecular Identification of Bacteria Isolated from Marketed Sparus aurata and Penaeus indicus Sea Products: Antibiotic Resistance Profiling and Evaluation of Biofilm Formation. Life. 2023; 13(2):548. https://doi.org/10.3390/life13020548
Chicago/Turabian StyleAbdulhakeem, Mohammad A., Mousa Alreshidi, Fevzi Bardakci, Walid Sabri Hamadou, Vincenzo De Feo, Emira Noumi, and Mejdi Snoussi. 2023. "Molecular Identification of Bacteria Isolated from Marketed Sparus aurata and Penaeus indicus Sea Products: Antibiotic Resistance Profiling and Evaluation of Biofilm Formation" Life 13, no. 2: 548. https://doi.org/10.3390/life13020548
APA StyleAbdulhakeem, M. A., Alreshidi, M., Bardakci, F., Hamadou, W. S., De Feo, V., Noumi, E., & Snoussi, M. (2023). Molecular Identification of Bacteria Isolated from Marketed Sparus aurata and Penaeus indicus Sea Products: Antibiotic Resistance Profiling and Evaluation of Biofilm Formation. Life, 13(2), 548. https://doi.org/10.3390/life13020548










