Abstract
This study was conducted to examine the associations between genome-wide association studies (GWAS)-important single nucleotide polymorphisms (SNPs) and knee osteoarthritis (KOA) among Europeans of Russia. The present replicative study (“patient-control” design has been used) was carried out on 1000 DNA samples from KOA (n = 500) and KOA-free (n = 500) participants. Ten GWAS-important for KOA SNPs of eight candidate genes (LYPLAL1, GNL3, GLT8D1, SBNO1, WWP2, NFAT5, TGFA, GDF5) were studied. To assess the link between SNPs and KOA susceptibility, logistic regression (to establish independent SNP effects) and MB-MDR (to identify SNP–SNP interactions) were used. As a result of this genetic analysis, the associations of individual SNPs with KOA have not been proven. Eight loci out of ten tested SNPs interacted with each other (within twelve genetic models) and determined susceptibility to KOA. The greatest contribution to the disease development were made by three polymorphisms/genes such as rs6976 (C>T) GLT8D1, rs56116847 (G>A) SBNO1, rs6499244 (T>A) NFAT5 (each was included in 2/3 [8 out 12] KOA-responsible genetic interaction models). A two-locus epistatic interaction of rs56116847 (G >A) SBNO1 × rs6499244 (T>A) NFAT5 determined the maximum percentage (0.86%) of KOA entropy. KOA-associated SNPs are regulatory polymorphisms that affect the expression/splicing level, epigenetic modification of 72 genes in KOA-pathogenetically significant organs such as skeletal muscles, tibial arteries/nerves, thyroid, adipose tissue, etc. These putative KOA-effector genes are mainly involved in the organization/activity of the exoribonuclease complex and antigen processing/presentation pathways. In conclusion, KOA susceptibility among Europeans of Russia is mediated by intergenic interactions (but not the main effects) of GWAS-important SNPs.
1. Introduction
Osteoarthritis (OA) is a common chronic degenerative joint disease [1], resulting in pathological changes in all joint tissues (cartilage/subchondral bone/ligaments/menisci/articular capsule/synovial membrane) [2]. More than 500 million people worldwide suffer from this disease; an especially high incidence of OA is observed among people over 65 years of age [3]. The prevalence of large-joint OA (knee and hip) worldwide is 3754.2 per 100 thousand population [4]. At the same time, knee joints are affected more often among the large joints [5,6]. The economic costs associated with OA are high, starting with the direct costs of care and treatment, and ending with the costs of total endoprosthetics [7]. Patients with knee OA (KOA) spend on average about USD 15,000 during their lifetime on direct medical expenses related to the disease [8]. At the same time, expensive endoprosthesis is not always effective; about 20% of patients report dissatisfaction with the result after total knee arthroplasty [9]. It is worth noting that OA leads to a decrease in the quality of life of patients through restriction of daily activity [10,11] and disability [12].
The genetic architecture of OA (including KOA) is complex and affects the individual’s total risk of developing the disorder, its clinical course, progression and prognosis [13,14,15]. The share of the hereditary component in OA is about 50% [13,14]. As a result of 24 GWAS conducted so far, 124 SNPs of 95 candidate genes associated with OA at various locations (KOA, hip joint OA, arm joint OA) have been established [15], among which 87 SNPs are linked with KOA at the genome-wide level (p < 5 × 10−8) [16]. At the same time, despite the large array of GWAS data for KOA, the number of replication studies performed is quite limited [17,18,19,20,21,22,23]. As a result of these replicative studies, associations with KOA were confirmed only for single GWAS-important SNPs: rs143383 GDF5, rs4867568 LOC105374701, rs3884606 FGF18, rs10947262 BTNL2/TSBP1-AS1, and rs7639618 COL6A4P1. Notably, the link with KOA in Asian populations was confirmed for three SNPs (rs143383 GDF5, rs4867568 LOC105374701, and rs3884606 FGF18) [22,23], and in European populations or mixed samples of Europeans and Asians for only two SNPs (rs10947262 BTNL2/TSBP1-AS1, rs7639618 COL6A4P1) [17,18].
Thus, there is an evident shortage of replicative studies of KOA. This dictates the necessity for further genetic studies of KOA in different ethnic groups of the world, including the Russian region. Therefore, we conducted this replication study of the involvement of GWAS-important SNPs for KOA disease susceptibility among Europeans of Russia.
2. Materials and Methods
2.1. Study Participants
The present replicative study (“patient-control” design has been used) was carried out on 1000 participants with KOA (n = 500) and KOA-free (n = 500). All participants (patients and control) included in the study met the following criteria (inclusion criteria): (1) individuals of European origin (all subjects had Russian nationality); (2) born and living in the central part of Russia [24,25]; (3) not related to each other; (4) aged 40 years and older; (5) the consent of all subjects (signed personally); (6) underwent the necessary clinical/laboratory/instrumental examinations in the “Orthopedics and Traumatology Department” at “Belgorod City Hospital №2” by competent orthopedic traumatologists. The following criteria were used to exclude subjects from this study (exclusion criteria): the presence of severe diabetes mellitus, coronary heart disease, hypertension, systemic connective tissue disorder, oncological diseases, renal-hepatic insufficiency, joint injuries/inflammatory, or musculoskeletal congenital malformations [26]. The sample was formed in the period between February 2016–December 2018 under the control of the hospital ethical commission.
The KOA diagnosis was established in accordance with the criteria of the ACR (American College of Rheumatology) [27,28]. The KOA group included patients with primary knee joint OA, who had a radiological stage of disease (Kellgren–Lawrence, K/L parameter) ≥ 2 [29] and pain symptoms (Visual Analog Scale [VAS] parameter was >40 points) [30]. The diagnostics of KOA was carried out by certified orthopedic traumatologists based on the results of all necessary tests (clinical/laboratory/instrumental [27,28,29,30]) conducted for all participants in the “Orthopedics and Traumatology Department” at “Belgorod City Hospital №2”. The analysis of all X-ray images was carried out by the orthopedic traumatologist Vitaly Novakov together (in consultation) with another qualified orthopedic traumatologist, which minimized the effect of bias among different observers and improved the quality of verification of the diagnosis of KOA (with r = 0.83 when reading X-ray images of patients with KOA stages 2–4 by one doctor or different doctors to r = 0.86–0.87 when reading the same X-rays by two doctors, one of whom is a permanent observer) [29]. The KOA-free group included subjects without musculoskeletal disorders. Hereditary burden (the presence of KOA and OA of other locations in relatives of 1–2 degrees of kinship, such as parents, siblings, uncles/aunts, grandparents) was assessed during a survey of the subjects (cases and controls).
2.2. GWAS SNPs Selection and Detection
For the present replicative study, GWAS SNPs for KOA (data from previously performed GWAS were taken into account [31,32,33,34,35,36]) (Supplementary Materials Table S1) possessing a distinct regulatory potential [37,38] (Haploreg data from the Roadmap Epigenomics and ENCODE projects were taken into account [39]) (Supplementary Table S2) were selected. As a result, we investigated ten KOA candidate SNPs: rs1060105 (C>T) and rs56116847 (G>A) SBNO1, rs2820436 (C>A) and rs2820443 (T>C) LYPLAL1, rs11177 (G>A) GNL3, rs6976 (C>T) GLT8D1, rs34195470 (G>A) WWP2, rs6499244 (T>A) NFAT5, rs3771501 (G>A) TGFA and rs143384 (A>G) GDF5.
The blood of KOA/KOA-free individuals was collected invasively from the elbow vein of the subjects and subsequently used for DNA allocation (the protocol for receiving DNA was set out earlier [40]). The NanoDrop spectrophotometer was employed to verify the DNA concentration/cleanliness [41]. DNA was stored in the freezer at −60 °C and subsequently utilized for the individual’s genotypic detection by real-time PCR (using CFX96 apparatus) [42,43]. Genotyping of DNA specimens was executed blindly by laboratory staff, and its accuracy was checked based on re-genotypic of several samples (≈7–10% case/control) [44,45], which showed a degree of 99% agreement.
2.3. Statistical Analysis
There was calculated the agreement of SNP genotype observed frequencies (KOA/KOA-free cohorts) to HWE [46,47]. To investigate the statistical dependence between individual SNP variants (allelic, additive, recessive, and dominant models have been employed [48]) and SNP interactions (simulation epistatic models were executed [49] with KOA using gPLINK [50] and MB-MDR [51,52] programs taking into account a number of confounders such as age, BMI, sex, hereditary, occupation-associated physical workload, leisure time physical activity, and the concomitant pathology availability (the musculoskeletal, cardiovascular, and endocrine systems) (Table 1). Parameters OR and 95% CI (odd ratio and confidence intervals of OR, respectively) have been utilized for the SNP-KOA dependence strength estimates [53,54]. Permutation-corrected p values were used [55] such as pperm ≤ 0.0125 for individual SNPs (Bonferroni-corrected p value based on four calculated models was calculated additionally, 0.05/4 [56]) and pperm ≤ 0.05 for models of SNP interactions [57]. Importantly, amongst the SNP epistatic models for further permutation assessment were the several models appropriate to the Bonferroni-corrected p value (the recombination of ten reported loci was taken into consideration) such as ≤1.11 × 10−3 (0.05/45 in 2–locus models), ≤4.17 × 10−4 (0.05/120 in 3–locus) and ≤2.38 × 10−4 (0.05/210 in 4–locus) [58]. The interaction map of high-confidence KOA effector SNPs was made with the MDR program [59].

Table 1.
Phenotypic characteristics of the study participants.
2.4. SNPs/Genes Predict Functions
The probable KOA effector SNPs/genes (taking into account proxy SNPs, r2 ≥ 0.80 [60,61]) have been chosen for their functionality (the in silico approach was applied [62,63] based on online integrated resources of bioinformatics information [64,65,66] including the GTEx [67], PolyPhen2 [68], HaploReg [39], Blood eQTL [69], SIFT [70], GeneMANIA [71]).
3. Results
The phenotypic characteristics (common biological parameters, occupation-related/leisure time physical activity, alcohol/smoking habits, hereditary burden, and concomitant pathology) of the examined KOA/control groups are presented in Table 1. The participants’ ages ranged from 40 to 70 years in the KOA group and 40–68 years in the KOA-free (control) group. The KOA patients did not differ from the control group in age, sex, height, smoking or alcohol status (p > 0.05). At the same time, KOA patients had a higher BMI (p < 1 × 10−6), hereditary burden (p = 0.0005), obesity rate (p = 0.0005), and high incidence of the cardiovascular (p = 0.0005), endocrine (p = 0.02) and musculoskeletal (p = 0.0005) diseases compared to the control group (Table 1). In the KOA cohort, the individuals number with a high level of occupational physical activity, as well as with low physical activity in their free time, is significantly higher compared to the control group (p = 0.0005 and p = 0.0005, respectively). In turn, the control subjects registered a greater number of individuals with irregular (1.24 times) and regular (2.33 times) physical activity in their free time compared with the KOA patients (p = 0.04 and p = 0.0007, respectively) (Table 1). Additionally, in the control group, the percentage of individuals with low occupational physical activity was significantly higher (2.12 times) than in the KOA subjects (p = 0.0005). The above parameters were used as factor-confounders in the evaluation of SNP-disease associations.
The statistical calculations showed that all examined GWAS loci in KOA/KOA-free participants were HWE-concordant (pHWE ≥ 0.117 in the KOA group and pHWE ≥ 0.059 in the KOA-free cohort) (Supplementary Table S3). As a result of the genetic analysis, associations of individual SNPs with KOA have not been proven (pbonf > 0.0125) (Table 2).

Table 2.
Associations of the studied gene polymorphisms with knee osteoarthritis.
The present MB-MDR analysis indicated intergenic interaction of the eight GWAS loci of 10 examined SNPs (with the exclusion of rs3771501 (G>A) TGFA and rs143384 (A>G) GDF5) within twelve genetic (epistatic) models as KOA-risk/protective factors (Table 3). The greatest contribution to the disease development has been made by three polymorphisms/genes: rs6976 (C>T) GLT8D1, rs56116847 (G>A) SBNO1, and rs6499244 (T>A) NFAT5 (each was included in 2/3 [8 out 12] KOA-responsible genetic interaction models). The highest Wald index (this statistic for high risk category equaled 47.56) was in a four–level genetic model: rs6499244 (T>A) NFAT5 × rs56116847 (G>A) SBNO1 × rs6976 (C>T) GLT8D1 × rs2820443 (T>C) LYPLAL1 (Table 3). Very important protective potential (−1.07 < beta value < −1.60 and p = 0.00003–0.00009) was found for some SNPs genotypic combinations such as TT(rs6499244)NFAT5 × AG(rs56116847)SBNO1 × CT(rs6976)GLT8D1, TT(rs6499244)NFAT5 x AG(rs56116847)SBNO1 × AG(rs11177)GNL3, TT(rs6499244)NFAT5 × AG(rs56116847)SBNO1 × CT(rs6976)GLT8D1 xTT(rs2820443)LYPLAL1, TT(rs6499244)NFAT5 × AG(rs56116847)SBNO1 × AG(rs11177)GNL3 × TT(rs2820443)LYPLAL1, TT(rs6499244)NFAT5 × AG(rs56116847)SBNO1 × AG(rs11177)GNL3 × CT(rs6976)GLT8D1 (Supplementary Table S4).

Table 3.
SNP × SNP interactions significantly associated with knee osteoarthritis.
Figure 1 portrays the interaction map for eight high-confidence KOA effector SNPs. The two-locus synergetic interaction (plotted in red) of rs56116847 (G>A) SBNO1 and rs6499244 (T>A) NFAT5 possessed a strong KOA effect (registering the maximum KOA entropy percentage-0.86%).
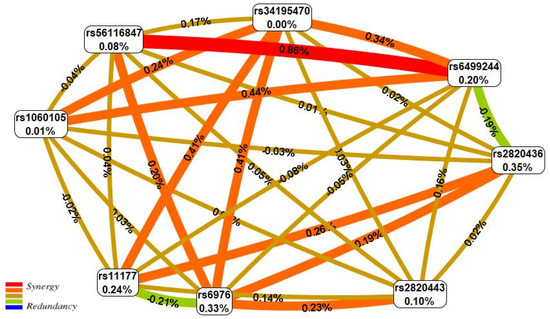
Figure 1.
The entropy graph of the SNP × SNP interactions with knee osteoarthritis based on the MDR analysis. Positive values of entropy indicate synergistic interactions while the negative values indicate redundancy. The red and orange colors denote strong and moderate synergism, respectively, brown color denotes the independent effect, green and blue colors denote moderate and strong antagonism, respectively.
3.1. Functional Genomics Data for KOA-Involved SNPs
3.1.1. SNPs Correlations with Amino Acid Replacements and Epigenetic Changes
Five of the investigated 315 SNPs (8 KOA-causal variants and 309 proxy SNPs) were missense loci leading to amino acid substitutions in the GNL3 (Arg39Gln, rs11177 and Val367Met, rs2289247), SPCS1 (Pro41Ala, rs6617), NEC4 (Pro225Ala, rs1029871) and SBNO1 (Ser728Asn, rs1060105) proteins. The above changes were predicted to be tolerated (SIFT tools information) and benign (PolyPhen-2 resource material), and only Pro>Ala replacement at position 225 in NEC4 protein (rs1029871) was possibly damaging (prediction effect revealed by PolyPhen-2)) (Supplementary Table S5).
Epigenetic modification of DNA regions near eighteen genes (C3orf78, CDK2AP1, CTD-2033A16.3, GLT8D1, GNL3, ITIH1, NEK4, NFAT5, NQO1, PBRM1, RN5S375, RP11-95P13.1, RP11-95P13.2, RP13-942N8.1, SBNO1, SETD8, SPCS1, WWP2) can be caused by eight KOA-involved SNPs and 292 loci out of 309 in LD SNPs (94.50%) due to the regulation of the activity of enhancers (139 SNPs, 44.13%) and promoters (24 SNPs, 7.62%), communication of specific DNA sites with protein-regulators (25 SNPs, 7.94%) and factors that promote the transcription process (266 SNPs, 86.08%) by changing the structure of chromatin (transformations of “open-closed” chromatin sites, methylation and acetylation of some histone protein fractions [H3K4me1,H3K27ac, etc.], etc.) (Supplementary Table S6).
It is important to underline that almost all KOA-associated polymorphic variants (with the exception of rs1060105) and a majority of strongly-linked loci exhibit their outstanding epigenetic effects in KOA-impact cell cultures and tissues such as primary osteoblast cells, chondrocytes, adipocytes, etc. For example, SNPs of genes such as LYPLAL1 (rs2820436 and rs2820443), GNL3 (rs11177), GLT8D1 (rs6976), WWP2 (rs34195470) and NFAT5 (rs6499244) were localized in the enhancer DNA regions at chondrocyte cell culture (E049(Epigenome ID)/STRM.CHON.MRW.DR.MSC(Mnemonic)), primary osteoblast cells (E129(Epigenome ID)/BONE.OSTEO(Mnemonic)); loci of LYPLAL1 (rs2820443), GNL3 (rs11177) and GLT8D1 (rs6976) genes have been positioned in the adipocytes promoter sites (E023(Epigenome ID)/FAT.MSC.DR.ADIP(Mnemonic)); specific nucleotide changes of GNL3 (rs11177), GLT8D1 (rs6976) and SBNO1 (rs56116847) genes were situated in the enhancer positions of peripheral blood T helper cells (E043(Epigenome ID)/BLD.CD4.CD25M.TPC(Mnemonic)).
3.1.2. KOA-Associated SNPs as Gene Quantitative Traits (eQTL and sQTL) Potential Predictors
The four KOA-associated loci (rs1060105 SBNO1, rs11177 GNL3, rs6499244 NFAT5, rs6976 GLT8D1) (Supplementary Table S7) and 70 highly-coupled SNPs with them (Supplementary Table S8) were modulators of blood transcribing six genes mRNA level (DNAH1, ITIH4, NT5DC2, RILPL2, SPCS1, WWP2).
Changes in the gene quantitative traits (transcription and splicing) in different organs (including KOA pathophysiology-involved organs such as skeletal muscles, tibial arteries and nerves, adipose tissue, thyroid, etc.) due to the allele-specific effects of the eight studied SNPs and high-LD loci (283 out 309 SNPs [91.59%]) have been detected as potential predictors of KOA. In total, the 61 genes transcription (ABCB9, ALAS1, ARL6IP4, C12orf65, CCDC62, CDK2AP1, CLEC18A, CLEC18C, COG4, DNAH1, EXOSC6, GLT8D1, GLYCTK, GLYCTK-AS1, GNL3, IL34, ITIH1, ITIH4, KMT5A, LYPLAL1-AS1, MPHOSPH9, MUSTN1, NEK4, NFAT5, NOB1, NPIPB14P, NQO1, NT5DC2, OGFOD2, PBRM1, PDXDC2P, PITPNM2, POC1A, PPM1M, PRKCD, RFT1, RILPL2, RIMKLBP2, RP11-168J18.6, RP11-282O18.3, RP11-296I10.3, RP11-392O17.1, RP11-394B2.1, RP11-394B2.5, RP11-546D6.3, RP11-894J14.2, RP11-972P1.11, RP5-1157M23.2, RP5-966M1.5, RP5-966M1.7, SBNO1, SERBP1P3, SFMBT1, SLC30A10, SMG1P7, SMIM4, SPCS1, TMEM110, WDR82, WWP2, ZC3H11B) (Supplementary Tables S9 and S10) and 24 genes splicing (ABCB9, C12orf65, GLT8D1, GLYCTK, GNL3, ITIH1, ITIH3, ITIH4, KMT5A, MPHOSPH9, MUSTN1, NOB1, NPIPB14P, NQO1, NT5DC2, PBRM1, PDXDC2P, PHF7, RILPL1, RP11-392O17.1, SMIM4, STAB1, TMEM110, WWP2) (Supplementary Tables S11 and S12) may be changed due to the apparent eQTL and sQTL influences of KOA-correlated SNPs.
3.1.3. Potential Interactions and Biological Pathways of KOA Putative Target Genes
The 72 putative KOA-effector genes (according to the functional genomics data given above and presented in the Supplementary Tables S5–S12) have been mainly involved in the organization/activity of the exoribonuclease complex and antigen processing/presentation pathways (Supplementary Table S13), and interact among themselves through joint transcription (55.16%), predictor communications (23.26%), overall domains of proteins molecules (10.76%), joint localization (6.08%), and physical mutual influences (4.74%) (Figure 2) with a predominant potential in these relationships of prioritized interactions between the follow pairs of genes: STIMATE-MUSTN1, STIMATE–TMEM110, STIMATE–MUSTN1, MRC1–STAB1, ARID2–PBRM1, NQO2–NQO1 (indicators of weight were 0.71–0.86) (Supplementary Table S14).
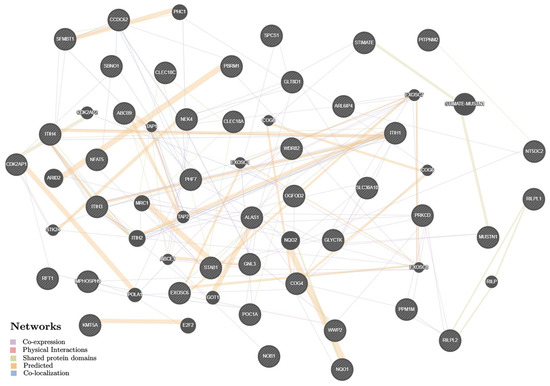
Figure 2.
The interaction networks of 72 KOA-associated genes in various tissues/organs inferred using GeneMANIA (http://genemania.org (accessed on 15 August 2022)). The candidate genes are cross-shaded.
4. Discussion
The present “patient-control” study demonstrated that KOA susceptibility among Europeans of Russia can be caused by intergenic interactions of eight GWAS-important SNPs (rs11177 GNL3, rs1060105 SBNO1, rs2820436 and rs2820443 LYPLAL1, rs56116847 SBNO1, rs6976 GLT8D1, rs6499244 NFAT5 and rs34195470 WWP2). The greatest contribution to the KOA susceptibility was made by the three polymorphisms/genes rs6499244 (T>A) NFAT5, rs6976 (C>T) GLT8D1, and rs56116847 (G>A) SBNO1 (each was included in 2/3 [8 out 12] KOA-responsible genetic interaction models). The associations between individual SNPs and KOA predisposition have not been supported.
The outcome of this work demonstrated involvement in the KOA genetic architecture (within gene-gene mutual influence) of rs6499244 (T>A) NFAT5. For rs6499244 NFAT5 (located on chromosome 16q22.1), in silico data indicated its pronounced functional actions by influences on enhancer sequences in chondrocytes (E049(Epigenome ID)/STRM.CHON.MRW.DR.MSC(Mnemonic)), primary osteoblast cells (E129(Epigenome ID)/BONE.OSTEO(Mnemonic)), primary T-regulatory and T-helper cells of peripheral blood (E044(Epigenome ID)/BLD.CD4.CD25.CD127M.TREGPC(Mnemonic) and E043(Epigenome ID)/BLD.CD4.CD25M.TPC(Mnemonic), respectively), fat nuclei (E063(Epigenome ID)/FAT.ADIP.NUC(Mnemonic)), promoter sites in fat nuclei (E063(Epigenome ID)/FAT.ADIP.NUC(Mnemonic)), transcriptional levels of 10 genes (CLEC18A, COG4, EXOSC6, IL34, NFAT5, NOB1, NPIPB14P, NQO1, PDXDC2P, SMG1P7), and transcript alternative splicing of four genes (NOB1, NPIPB14P, NQO1, WWP2) in the skeletal muscles, fatty tissue, thyroid gland, and other KOA-related organs. In a previous GWAS, the causal link between rs6499244 (T>A) NFAT5 and KOA risk was shown [35]. The potential epigenetic “authority” of this genome region was also known [15,72,73]. Rice et al. presented data on associations of the rs7359336 locus, which is in LD with rs6499244 NFAT5 (r2 = 0.91), with an increased level of DNA methylation in the WWP2 gene in patients with OA [72]. Additionally, the authors noted the connection of this functionally-active genome sequence with five cartilage-expressed genes such as PDXDC2P, CLEC18A, NOB1, NFAT5, IL34 [72]. The nuclear factor gene of activated T cells 5 (NFAT5) encodes a transcription factor that manages the quantitative expression traits of genes implicated in the regulation of osmoprotective and inflammatory reactions [73,74,75,76,77]. There is evidence of the role of NFAT5 in the formation of innate immunity by activating gene expression in macrophages during TLR (Toll-like receptor) ligation [75,78,79]. TLR2 and TLR4 are known to be highly expressed in synovial fluid macrophages and are responsible for macrophage activation [80]. NFAT5 also plays an important role in the proliferation of synoviocytes themselves [81]. A higher NFAT5 expression was found in the synovial membrane [81]. NFAT5 indirectly affects the migration of myoblasts during skeletal muscle myogenesis via the CYR61-dependent pathway [82]. Transcription factors NFAT 1, NFAT2, NFAT3 have been an essential function in the OA-pathobiology [83,84,85].
The data for rs56116847 (G>A) SBNO1 as a KOA risk locus were first established by Tachmazidou et al. in GWAS on samples of European origin from UK Biobank and Arthritis Research UK Osteoarthritis Genetics (arcOGEN) [35]. According to our data, this locus, interacting with rs6499244 (T>A) NFAT5, makes the greatest contribution to KOA susceptibility (0.86% of the disease entropy) among Europeans of Russia. The literature data indicate a serious epigenetic potential (relationship with the level of methylation in cartilage) of the locus rs56116847 (G>A) SBNO1 [15]. The information obtained by us in silico also confirms the position of this polymorphism in enhancers in primary T-regulatory and T-helper cells of peripheral blood (E044(Epigenome ID)/BLD.CD4.CD25.CD127M.TREGPC(Mnemonic) and E043(Epigenome ID)/BLD.CD4.CD25M.TPC(Mnemonic), respectively), primary peripheral blood monocytes (E029(Epigenome ID)/BLD.CD14.PC(Mnemonic)), its correlation with eQTL of eight genes (ABCB9, ARL6IP4, C12orf65, CDK2AP1, KMT5A, MPHOSPH9, OGFOD2 and RILPL2) and sQTL of four genes (ABCB9, KMT5A, MPHOSPH9 and RILPL1) in fibroblasts cell culture, tibial nerve, skeletal muscles, whole blood, etc. The KMT5A gene (also called PR-SET7, SET8) is closely related to the regulation of various biological processes such as DNA replication, chromosome condensation and activation of DNA replication checkpoints, cell proliferation, etc. [86]. KMT5A regulates the transcription of Ras, p53 and Wnt genes [86], whose protein products of the same name are involved in the OA-pathophysiology [87,88,89]. For instance, Wnt is well known for its participation in osteogenesis due to activation of the signaling pathways such as Wnt/calcium, Wnt/cyclic adenosine monophosphate (cAMP), Wnt/c-Jun NH(2)-terminal protein kinase (JNK) and Wnt/β-catenin [87]. ERa plays responsible role in cartilage degradation [89]. The inhibitory importance of KMT5A in the proliferation and metastasis of osteosarcoma cells through the transmission of β-catenin signals is also known [90].
We have identified the most substantial contribution of the rs6976 (C>T) GLT8D1 locus in the KOA-important intergenic interactions among Europeans of Russia. The rs6976 GLT8D1 association with OA was shown in three previously-published GWAS in Europeans [31,32,35]. However, in two other GWAS [91,92] and one meta-analysis of nine GWAS datasets [93] of significant associations of rs6976 (C>T) GLT8D1 with KOA in Europeans [93], African Americans [91] and Europeans of North America [92] has not been established. In the study of Lindner et al. the link between rs6976 GLT8D1 and one of the statistical models of the shape of the proximal femur in patients with hip OA of European origin was shown [94]. In accordance with some studies, a certain shape of the proximal femur can determine the occurrence and progression of OA [95,96]. According to our in silico materials, SNP rs6976 (C>T) GLT8D1 is functionally significant when associated with active enhancers in chondrocyte cell culture (E049(Epigenome ID)/STRM.CHON.MRW.DR.MSC(Mnemonic)), primary osteoblast cells (E129(Epigenome ID)/BONE.OSTEO(Mnemonic)), in skeletal muscle microtubule cells (E121(Epigenome ID)/MUS.HSMMT(Mnemonic)), and active promoters in adipocytes (E023(Epigenome ID)/FAT.MSC.DR.ADIP(Mnemonic)) and chondrocytes (E049(Epigenome ID)/STRM.CHON.MRW.DR.MSC(Mnemonic)). The rs6976 (C>T) GLT8D1 locus is linked to four non-synonymous SNPs (rs11177,rs2289247,rs6617,rs1029871), which determine amino acid modification in three proteins such as GNL3 (Arg39Gln and Val367Met), SPCS1 (Pro41Ala) and NEC4 (Pro225Ala). The SNP rs6976 studied by us is associated with the expression/splicing parameters of 28 genes (DNAH1, GLT8D1, GLYCTK, GLYCTK-AS1, GNL3, ITIH1, ITIH3, ITIH4, MUSTN1, NEK4, NT5DC2, PBRM1, PHF7, POC1A, PPM1M, RFT1, RP11-168J18.6, RP11-894J14.2, RP5-1157M23.2, RP5-966M1.5, RP5-966M1.7, SERBP1P3, SFMBT1, SMIM4, SPCS1, STAB1, TMEM110 and WDR82) in the tibial artery/nerve, skeletal muscles, thyroid gland, adipose tissue, etc. The data about rs6976 as a functional-impact locus were previously presented in other sources [14,97,98]. In a study by Rushton et al., rs6976 GLT8D1 was found to be associated with a lower level of methylation in the STAB1 gene in the cartilage of OA patients [98]. In Gee et al., the correlations of two polymorphic loci of the genes SPCS1 (rs6617) and GNL3 (rs11177), which are strongly coupled with rs6976 GLT8D1, with a significant imbalance of gene expression (SPCS1 and GNL3, respectively) in cartilage and other joint tissues such as ligaments, meniscus, adipose tissue, synovial membrane, were shown [97]. A number of studies have established the association of the rs11177 (G>A) GNL3 with the OA risk in both European and Asian populations [1,31,32,99], including on the GWAS level (p ≤ 5 × 10−8) [31,32].
The GLT8D1 gene, located in the 3p21.1 region, encodes a protein from the glycosyltransferase family. Glycosyltransferases are a family of enzymes that catalyze the biosynthesis of oligosaccharides, polysaccharides and glycoconjugates [100]. To date, very little is known about the physiological and pathological functions of the GLT8D1 gene and its corresponding protein. The GNL3 gene encodes a guanine-nucleotide-binding protein (nucleostemin), which plays an important role in many processes occurring in the cell, including participation in stem cell proliferation, regulation of the cell cycle, maintenance of telomerase activity [101,102,103]. GNL3 is expressed in mesenchymal stem cells, from which chondrocytes originate [101], shows association with chondrogenic differentiation and may participate in genomic regulation as an RNA-binding protein [104]. In a study by Louka et al., it was found that the GNL3 gene expression in synovial tissue/fluid samples was significantly higher in the group of patients with primary OA compared with the control cohort [105]. The nucleostemin level was also increased in chondrocytes in patients with OA [31].
This study has certain limitations, which include the following: (a) only ten SNP of KOA candidate genes have been studied in the work and the inclusion of more loci in the analysis may cause a “shift” in the estimates of the intergenic interactions role of the examined loci in the formation of KOA; (b) the results of this work were not replicated on another sample from the same population or samples of another ethnic group; (c) this study did not take into account all possible risk factors for KOA (immune factors, bone metabolism, diet features, etc.) and, accordingly, their effects as covariates were not taken into account when evaluating associations; and (d) much broader experimental evidence of the functional effects of SNPs determining susceptibility to KOA is required (only in silico data were used in this work).
5. Conclusions
In the current study it has been revealed that the KOA susceptibility among Europeans of Russia is mediated by intergenic interactions (but not the independent effects) of GWAS-important SNPs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/life13020405/s1, Table S1: The literature data about associations of the studied polymorphisms of the candidate genes with osteoarthritis; Table S2: The regulatory potential of the studied SNPs; Table S3: The allele and genotype frequencies of the studied SNPs in the knee osteoarthritis and control groups; Table S4: Genotype combinations associated with knee osteoarthritis; Table S5: Non-synonymous KOA-associated SNPs and SNPs in high LD (r2 ≥ 0.80); Table S6: Regulatory effects of the eight KOA-associated SNPs of the candidate genes and SNPs in high LD (r2 ≥ 0.80); Table S7: Effect of the KOA-associated candidate genes polymorphisms on affinity of the DNA regulatory motifs; Table S8: Cis-eQTL values of the KOA-associated SNPs of the candidate genes in blood; Table S9: Cis-eQTL values of the SNPs in high LD (r2 ≥ 0.80) with KOA-associated SNPs of the candidate genes in blood; Table S10: eQTL values of the KOA-associated SNPs of the candidate genes; Table S11: Effect of SNPs in high LD (r2 ≥ 0.80) with the KOA-associated polymorphisms of the candidate genes on gene expression level; Table S12: sQTL values of the KOA-associated SNPs of the candidate genes; Table S13: Effect of SNPs in high LD (r2 ≥ 0.80) with the KOA-associated polymorphisms of the candidate genes on alternative splicing (cis-sQTL); Table S14: Gene set enrichment analysis of biological pathways with 72 KOA-associated genes (KOA-associated polymorphisms functionality in various tissue/organs).
Author Contributions
Conceptualization, V.N., O.N. and M.C. (Maria Churnosova); Data curation, V.N., I.S. and A.P.; Formal analysis, M.C. (Mikhail Churnosov), E.R., I.A. and O.N.; Project administration, M.C. (Mikhail Churnosov); Writing—original draft, V.N. and M.C. (Maria Churnosova); Writing—review and editing, M.C. (Mikhail Churnosov), E.R. and I.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Local Ethical Committee of the Belgorod State University (28 January 2016, No. 4).
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
The data generated in the present study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, X.; Xiao, L.; Wang, Z.; Zhi, L.; Li, Q. Common variants in GNL3 gene contributed the susceptibility of hand osteoarthritis in Han Chinese population. Sci. Rep. 2022, 12, 16110. [Google Scholar] [CrossRef]
- Madry, H.; Luyten, F.P.; Facchini, A. Biological aspects of early osteoarthritis. Knee Surg. Sports Traumatol. Arthrosc. 2012, 20, 407–422. [Google Scholar] [CrossRef]
- Institute for Health Metrics and Evaluation. Available online: https://www.healthdata.org/results/gbd_summaries/2019/osteoarthritis-level-3-cause (accessed on 22 November 2022).
- Safiri, S.; Kolahi, A.A.; Smith, E.; Hill, C.; Bettampadi, D.; Mansournia, M.A.; Hoy, D.; Ashrafi-Asgarabad, A.; Sepidarkish, M.; Almasi-Hashiani, A.; et al. Global, regional and national burden of osteoarthritis 1990–2017: A systematic analysis of the Global Burden of Disease Study 2017. Ann. Rheum. Dis. 2020, 79, 819–828. [Google Scholar] [CrossRef]
- Michael, J.W.; Schlüter-Brust, K.U.; Eysel, P. The epidemiology, etiology, diagnosis, and treatment of osteoarthritis of the knee. Dtsch. Arztebl. Int. 2010, 107, 152–162, Erratum in Dtsch. Arztebl. Int. 2010, 107, 294. [Google Scholar] [CrossRef]
- Favero, M.; Belluzzi, E.; Ortolan, A.; Lorenzin, M.; Oliviero, F.; Doria, A.; Scanzello, C.R.; Ramonda, R. Erosive hand osteoarthritis: Latest findings and outlook. Nat. Rev. Rheumatol. 2022, 18, 171–183. [Google Scholar] [CrossRef]
- GBD 2016 DALYs and HALE Collaborators. Global, regional, and national disability-adjusted life-years (DALYs) for 333 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990-2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1260–1344, Erratum in Lancet 2017, 390, e38.. [Google Scholar] [CrossRef]
- Losina, E.; Paltiel, A.D.; Weinstein, A.M.; Yelin, E.; Hunter, D.J.; Chen, S.P.; Klara, K.; Suter, L.G.; Solomon, D.H.; Burbine, S.A.; et al. Lifetime medical costs of knee osteoarthritis management in the United States: Impact of extending indications for total knee arthroplasty. Arthritis Care Res. 2015, 67, 203–215. [Google Scholar] [CrossRef]
- Gunaratne, R.; Pratt, D.N.; Banda, J.; Fick, D.P.; Khan, R.J.K.; Robertson, B.W. Patient Dissatisfaction Following Total Knee Arthroplasty: A Systematic Review of the Literature. J. Arthroplasty 2017, 32, 3854–3860. [Google Scholar] [CrossRef]
- Barbour, K.E.; Helmick, C.G.; Boring, M.; Brady, T.J. Vital Signs: Prevalence of Doctor-Diagnosed Arthritis and Arthritis-Attributable Activity Limitation—United States, 2013-2015. MMWR Morb. Mortal. Wkly Rep. 2017, 66, 246–253. [Google Scholar] [CrossRef]
- Tan, J.S.; Tikoft, E.; O’Sullivan, P.; Smith, A.; Campbell, A.; Caneiro, J.P.; Kent, P. The Relationship Between Changes in Movement and Activity Limitation or Pain in People with Knee Osteoarthritis: A Systematic Review. J. Orthop. Sports Phys. Ther. 2021, 51, 492–502. [Google Scholar] [CrossRef]
- Gaudreault, N.; Maillette, P.; Coutu, M.F.; Durand, M.J.; Hagemeister, N.; Hébert, L.J. Work disability among workers with osteoarthritis of the knee: Risks factors, assessment scales, and interventions. Int. J. Rehabil. Res. 2014, 37, 290–296. [Google Scholar] [CrossRef]
- Zengini, E.; Finan, C.; Wilkinson, J.M. The Genetic Epidemiological Landscape of Hip and Knee Osteoarthritis: Where Are We Now and Where Are We Going? J. Rheumatol. 2016, 43, 260–266. [Google Scholar] [CrossRef]
- Klein, J.C.; Keith, A.; Rice, S.J.; Shepherd, C.; Agarwal, V.; Loughlin, J.; Shendure, J. Functional testing of thousands of osteoarthritis-associated variants for regulatory activity. Nat. Commun. 2019, 10, 2434. [Google Scholar] [CrossRef]
- Aubourg, G.; Rice, S.J.; Bruce-Wootton, P.; Loughlin, J. Genetics of osteoarthritis. Osteoarthr. Cartil. 2022, 30, 636–649. [Google Scholar] [CrossRef]
- GWAS Catalog. Available online: https://www.ebi.ac.uk/gwas/search?query=osteoarthritis,%20knee (accessed on 22 November 2022).
- Meulenbelt, I.; Chapman, K.; Dieguez-Gonzalez, R.; Shi, D.; Tsezou, A.; Dai, J.; Malizos, K.N.; Kloppenburg, M.; Carr, A.; Nakajima, M.; et al. Large replication study and meta-analyses of DVWA as an osteoarthritis susceptibility locus in European and Asian populations. Hum. Mol. Genet. 2009, 18, 1518–1523. [Google Scholar] [CrossRef]
- Shi, D.; Zheng, Q.; Chen, D.; Zhu, L.; Qin, A.; Fan, J.; Liao, J.; Xu, Z.; Lin, Z.; Norman, P.; et al. Association of single-nucleotide polymorphisms in HLA class II/III region with knee osteoarthritis. Osteoarthr. Cartil. 2010, 18, 1454–1457. [Google Scholar] [CrossRef][Green Version]
- Valdes, A.M.; Styrkarsdottir, U.; Doherty, M.; Morris, D.L.; Mangino, M.; Tamm, A.; Doherty, S.A.; Kisand, K.; Kerna, I.; Tamm, A.; et al. Large scale replication study of the association between HLA class II/BTNL2 variants and osteoarthritis of the knee in European-descent populations. PLoS ONE 2011, 6, e23371. [Google Scholar] [CrossRef]
- Nakajima, M.; Shi, D.; Dai, J.; Tsezou, A.; Zheng, M.; Norman, P.E.; Chou, C.H.; Lee, M.T.; Hwang, J.Y.; Kim, D.H.; et al. A large-scale replication study for the association of rs17039192 in HIF-2α with knee osteoarthritis. J. Orthop. Res. 2012, 30, 1244–1248. [Google Scholar] [CrossRef]
- Dai, J.; Ying, P.; Shi, D.; Hou, H.; Sun, Y.; Xu, Z.; Chen, D.; Zhang, G.; Ni, M.; Teng, H.; et al. FTO variant is not associated with osteoarthritis in the Chinese Han population: Replication study for a genome-wide association study identified risk loci. J. Orthop. Surg. Res. 2018, 13, 65. [Google Scholar] [CrossRef]
- Zhao, T.; Zhao, J.; Ma, C.; Wei, J.; Wei, B.; Liu, J. Evaluation of Relationship Between Common Variants in FGF18 Gene and Knee Osteoarthritis Susceptibility. Arch. Med. Res. 2020, 51, 76–81. [Google Scholar] [CrossRef]
- Li, Y.; Liu, F.; Xu, X.; Zhang, H.; Lu, M.; Gao, W.; Yin, L.; Yin, Z. A novel variant near LSP1P3 is associated with knee osteoarthritis in the Chinese population. Clin. Rheumatol. 2020, 39, 2393–2398. [Google Scholar] [CrossRef]
- Litovkina, O.; Nekipelova, E.; Dvornyk, V.; Polonikov, A.; Efremova, O.; Zhernakova, N.; Reshetnikov, E.; Churnosov, M. Genes involved in the regulation of vascular homeostasis determine renal survival rate in patients with chronic glomerulo-nephritis. Gene 2014, 546, 112–116. [Google Scholar] [CrossRef]
- Reshetnikov, E.A.; Akulova, L.Y.; Dobrodomova, I.S.; Dvornyk, V.Y.; Polonikov, A.V.; Churnosov, M.I. The insertion-deletion polymorphism of the ACE gene is associated with increased blood pressure in women at the end of pregnancy. J. Renin Angiotensin Aldosterone Syst. 2015, 16, 623–632. [Google Scholar] [CrossRef]
- Novakov, V.; Novakova, O.; Churnosova, M.; Aristova, I.; Sorokina, I.; Polonikov, A.; Reshetnikov, E.; Churnosov, M. The pronounced effect of obesity on the association of the rs143384 GDF5 with knee osteoarthritis. Life 2022, in press. [Google Scholar]
- Altman, R.; Asch, E.; Bloch, D.; Bole, G.; Borenstein, D.; Brandt, K.; Christy, W.; Cooke, T.D.; Greenwald, R.; Hochberg, M.; et al. Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Association. Arthritis Rheum. 1986, 29, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Altman, R.D. Criteria for classification of clinical osteoarthritis. J. Rheumatol. Suppl. 1991, 27, 10–12. [Google Scholar]
- Kellgren, J.H.; Lawrence, J.S. Radiological assessment of osteo-arthrosis. Ann. Rheum. Dis. 1957, 16, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Hawker, G.A.; Mian, S.; Kendzerska, T.; French, M. Measures of adult pain: Visual Analog Scale for Pain (VAS Pain), Numeric Rating Scale for Pain (NRS Pain), McGill Pain Questionnaire (MPQ), Short-Form McGill Pain Questionnaire (SF-MPQ), Chronic Pain Grade Scale (CPGS), Short Form-36 Bodily Pain Scale (SF-36 BPS), and Measure of Intermittent and Constant Osteoarthritis Pain (ICOAP). Arthritis Care Res. 2011, 63, S240–S252. [Google Scholar] [CrossRef]
- arcOGEN Consortium; arcOGEN Collaborators; Zeggini, E.; Panoutsopoulou, K.; Southam, L.; Rayner, N.W.; Day-Williams, A.G.; Lopes, M.C.; Boraska, V.; Esko, T.; et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): A genome-wide association study. Lancet 2012, 380, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Styrkarsdottir, U.; Lund, S.H.; Thorleifsson, G.; Zink, F.; Stefansson, O.A.; Sigurdsson, J.K.; Juliusson, K.; Bjarnadottir, K.; Sigurbjornsdottir, S.; Jonsson, S.; et al. Meta-analysis of Icelandic and UK data sets identifies missense variants in SMO, IL11, COL11A1 and 13 more new loci associated with osteoarthritis. Nat. Genet. 2018, 50, 1681–1687. [Google Scholar] [CrossRef]
- Zengini, E.; Hatzikotoulas, K.; Tachmazidou, I.; Steinberg, J.; Hartwig, F.P.; Southam, L.; Hackinger, S.; Boer, C.G.; Styrkarsdottir, U.; Gilly, A.; et al. Genome-wide analyses using UK Biobank data provide insights into the genetic architecture of osteoarthritis. Nat. Genet. 2018, 50, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Styrkarsdottir, U.; Stefansson, O.A.; Gunnarsdottir, K.; Thorleifsson, G.; Lund, S.H.; Stefansdottir, L.; Juliusson, K.; Agustsdottir, A.B.; Zink, F.; Halldorsson, G.H.; et al. GWAS of bone size yields twelve loci that also affect height, BMD, osteoarthritis or fractures. Nat. Commun. 2019, 10, 2054, Erratum in Nat. Commun. 2019, 10, 2358. [Google Scholar] [CrossRef] [PubMed]
- Tachmazidou, I.; Hatzikotoulas, K.; Southam, L.; Esparza-Gordillo, J.; Haberland, V.; Zheng, J.; Johnson, T.; Koprulu, M.; Zengini, E.; Steinberg, J.; et al. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat Genet. 2019, 51, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Boer, C.G.; Hatzikotoulas, K.; Southam, L.; Stefánsdóttir, L.; Zhang, Y.; Coutinho de Almeida, R.; Wu, T.T.; Zheng, J.; Hartley, A.; Teder-Laving, M.; et al. Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations. Cell 2021, 184, 4784–4818.e17, Erratum in Cell 2021, 184, 6003–6005. [Google Scholar] [CrossRef]
- Polonikov, A.V.; Bushueva, O.Y.; Bulgakova, I.V.; Freidin, M.B.; Churnosov, M.I.; Solodilova, M.A.; Shvetsov, Y.D.; Ivanov, V.P. A comprehensive contribution of genes for aryl hydrocarbon receptor signaling pathway to hypertension susceptibility. Pharm. Genom. 2017, 27, 57–69. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ponomarenko, I.; Reshetnikov, E.; Polonikov, A.; Verzilina, I.; Sorokina, I.; Yermachenko, A.; Dvornyk, V.; Churnosov, M. Candidate genes for age at menarche are associated with uterine leiomyoma. Front. Genet. 2021, 11, 512940. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016, 44, D877–D881. [Google Scholar] [CrossRef]
- Eliseeva, N.; Ponomarenko, I.; Reshetnikov, E.; Dvornyk, V.; Churnosov, M. LOXL1 gene polymorphism candidates for exfoliation glaucoma are also associated with a risk for primary open-angle glaucoma in a Caucasian population from central Russia. Mol. Vis. 2021, 27, 262–269. [Google Scholar]
- Ponomarenko, I.; Reshetnikov, E.; Altuchova, O.; Polonikov, A.; Sorokina, I.; Yermachenko, A.; Dvornyk, V.; Golovchenko, O.; Churnosov, M. Association of genetic polymorphisms with age at menarche in Russian women. Gene 2019, 686, 228–236. [Google Scholar] [CrossRef]
- Reshetnikov, E.; Zarudskaya, O.; Polonikov, A.; Bushueva, O.; Orlova, V.; Krikun, E.; Dvornyk, V.; Churnosov, M. Genetic markers for inherited thrombophilia are associated with fetal growth retardation in the population of Central Russia. J. Obstet. Gynaecol. Res. 2017, 43, 1139–1144. [Google Scholar] [CrossRef]
- Moskalenko, M.I.; Milanova, S.N.; Ponomarenko, I.V.; Polonikov, A.V.; Churnosov, M.I. Study of associations of polymorphism of matrix metalloproteinases genes with the development of arterial hypertension in men. Kardiologiia 2019, 59, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Golovchenko, O.; Abramova, M.; Ponomarenko, I.; Reshetnikov, E.; Aristova, I.; Polonikov, A.; Dvornyk, V.; Churnosov, M. Functionally significant polymorphisms of ESR1and PGR and risk of intrauterine growth restriction in population of Central Russia. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 253, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Starikova, D.; Ponomarenko, I.; Reshetnikov, E.; Dvornyk, V.; Churnosov, M. Novel data about association of the functionally significant polymorphisms of the MMP9 gene with exfoliation glaucoma in the caucasian population of Central Russia. Ophthalmic Res. 2021, 64, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Bushueva, O.; Solodilova, M.; Churnosov, M.; Ivanov, V.; Polonikov, A. The Flavin-Containing Monooxygenase 3 Gene and Essential Hypertension: The Joint Effect of Polymorphism E158K and Cigarette Smoking on Disease Susceptibility. Int. J. Hypertens. 2014, 2014, 712169. [Google Scholar] [CrossRef]
- Tikunova, E.; Ovtcharova, V.; Reshetnikov, E.; Dvornyk, V.; Polonikov, A.; Bushueva, O.; Churnosov, M. Genes of tumor necrosis factors and their receptors and the primary open angle glaucoma in the population of Central Russia. Int. J. Ophthalmol. 2017, 10, 1490–1494. [Google Scholar] [CrossRef]
- Pavlova, N.; Demin, S.; Churnosov, M.; Reshetnikov, E.; Aristova, I.; Churnosova, M.; Ponomarenko, I. The Modifying Effect of Obesity on the Association of Matrix Metalloproteinase Gene Polymorphisms with Breast Cancer Risk. Biomedicines 2022, 10, 2617. [Google Scholar] [CrossRef]
- Ponomarenko, I.; Reshetnikov, E.; Polonikov, A.; Sorokina, I.; Yermachenko, A.; Dvornyk, V.; Churnosov, M. Candidate genes for age at menarche are associated with endometriosis. Reprod. Biomed. Online 2020, 41, 943–956. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Calle, M.L.; Urrea, V.; Malats, N.; Van Steen, K. Mbmdr: An R package for exploring gene-gene interactions associated with binary or quantitative traits. Bioinformatics 2010, 26, 2198–2199. [Google Scholar] [CrossRef][Green Version]
- Ponomarenko, I.V. Using the method of Multifactor Dimensionality Reduction (MDR) and its modifications for analysis of gene-gene and gene-environment interactions in genetic-epidemiological studies (review). Res. Results Biomed. 2019, 5, 4–21. [Google Scholar] [CrossRef]
- Reshetnikov, E.; Ponomarenko, I.; Golovchenko, O.; Sorokina, I.; Batlutskaya, I.; Yakunchenko, T.; Dvornyk, V.; Polonikov, A.; Churnosov, M. The VNTR polymorphism of the endothelial nitric oxide synthase gene and blood pressure in women at the end of pregnancy. Taiwan J. Obstet. Gynecol. 2019, 58, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Minyaylo, O.; Ponomarenko, I.; Reshetnikov, E.; Dvornyk, V.; Churnosov, M. Functionally significant polymorphisms of the MMP-9 gene are associated with peptic ulcer disease in the Caucasian population of Central Russia. Sci. Rep. 2021, 11, 13515. [Google Scholar] [CrossRef] [PubMed]
- Che, R.; Jack, J.R.; Motsinger-Reif, A.A.; Brown, C.C. An adaptive permutation approach for genome-wide association study: Evaluation and recommendations for use. BioData Min. 2014, 7, 9. [Google Scholar] [CrossRef]
- Ponomarenko, I.; Reshetnikov, E.; Polonikov, A.; Sorokina, I.; Yermachenko, A.; Dvornyk, V.; Churnosov, M. Candidate genes for age at menarche are associated with endometrial hyperplasia. Gene 2020, 757, 144933. [Google Scholar] [CrossRef] [PubMed]
- Moskalenko, M.; Ponomarenko, I.; Reshetnikov, E.; Dvornyk, V.; Churnosov, M. Polymorphisms of the matrix metalloproteinase genes are associated with essential hypertension in a Caucasian population of Central Russia. Sci. Rep. 2021, 11, 5224. [Google Scholar] [CrossRef]
- Pavlova, N.; Demin, S.; Churnosov, M.; Reshetnikov, E.; Aristova, I.; Churnosova, M.; Ponomarenko, I. Matrix Metalloproteinase Gene Polymorphisms Are Associated with Breast Cancer in the Caucasian Women of Russia. Int. J. Mol. Sci. 2022, 23, 12638. [Google Scholar] [CrossRef] [PubMed]
- SOURCEFORGE. Available online: http://sourceforge.net/projects/mdr (accessed on 22 November 2022).
- Churnosov, M.; Abramova, M.; Reshetnikov, E.; Lyashenko, I.V.; Efremova, O.; Churnosova, M.; Ponomarenko, I. Polymorphisms of hypertension susceptibility genes as a risk factors of preeclampsia in the Caucasian population of central Russia. Placenta 2022, 129, 51–61. [Google Scholar] [CrossRef]
- Golovchenko, I.; Aizikovich, B.; Golovchenko, O.; Reshetnikov, E.; Churnosova, M.; Aristova, I.; Ponomarenko, I.; Churnosov, M. Sex Hormone Candidate Gene Polymorphisms Are Associated with Endometriosis. Int. J. Mol. Sci. 2022, 23, 13691. [Google Scholar] [CrossRef]
- Abramova, M.; Churnosova, M.; Efremova, O.; Aristova, I.; Reshetnikov, E.; Polonikov, A.; Churnosov, M.; Ponomarenko, I. Effects of pre-pregnancy over-weight/obesity on the pattern of association of hypertension susceptibility genes with preeclampsia. Life 2022, 12, 2018. [Google Scholar] [CrossRef]
- Zhabin, S.N. Association of the rs10508336 polymorphism of the TAF3 transcription factor gene with the risk of lower limb arterial disease in the Russian population. Res. Results Biomed. 2022, 8, 439–447. [Google Scholar] [CrossRef]
- Polonikov, A.; Kharchenko, A.; Bykanova, M.; Sirotina, S.; Ponomarenko, I.; Bocharova, A.; Vagaytseva, K.; Stepanov, V.; Bushueva, O.; Churnosov, M.; et al. Polymorphisms of CYP2C8, CYP2C9 and CYP2C19 and risk of coronary heart disease in Russian population. Gene 2017, 627, 451–459. [Google Scholar] [CrossRef]
- Polonikov, A.; Rymarova, L.; Klyosova, E.; Volkova, A.; Azarova, I.; Bushueva, O.; Bykanova, M.; Bocharova, I.; Zhabin, S.; Churnosov, M.; et al. Matrix metalloproteinases as target genes for gene regulatory networks driving molecular and cellular pathways related to a multistep pathogenesis of cerebrovascular disease. J. Cell Biochem. 2019, 120, 16467–16482. [Google Scholar] [CrossRef]
- Sirotina, S.; Ponomarenko, I.; Kharchenko, A.; Bykanova, M.; Bocharova, A.; Vagaytseva, K.; Stepanov, V.; Churnosov, M.; Solodilova, M.; Polonikov, A. A Novel Polymorphism in the Promoter of the CYP4A11 Gene Is Associated with Susceptibility to Coronary Artery Disease. Dis. Markers. 2018, 2018, 5812802. [Google Scholar] [CrossRef]
- GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef]
- Westra, H.J.; Peters, M.J.; Esko, T.; Yaghootkar, H.; Schurmann, C.; Kettunen, J.; Christiansen, M.W.; Fairfax, B.P.; Schramm, K.; Powell, J.E.; et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013, 45, 1238–1243. [Google Scholar] [CrossRef]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 7, 1073–1081. [Google Scholar] [CrossRef]
- Franz, M.; Rodriguez, H.; Lopes, C.; Zuberi, K.; Montojo, J.; Bader, G.D.; Morris, Q. GeneMANIA update 2018. Nucleic Acids Res. 2018, 46, W60–W64. [Google Scholar] [CrossRef]
- Rice, S.J.; Cheung, K.; Reynard, L.N.; Loughlin, J. Discovery and analysis of methylation quantitative trait loci (mQTLs) mapping to novel osteoarthritis genetic risk signals. Osteoarthr. Cartil. 2019, 27, 1545–1556. [Google Scholar] [CrossRef]
- Rice, S.J.; Beier, F.; Young, D.A.; Loughlin, J. Interplay between genetics and epigenetics in osteoarthritis. Nat. Rev. Rheumatol. 2020, 16, 268–281. [Google Scholar] [CrossRef]
- Lopez-Rodríguez, C.; Aramburu, J.; Rakeman, A.S.; Rao, A. NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc. Natl. Acad. Sci. USA 1999, 96, 7214–7219. [Google Scholar] [CrossRef]
- Kim, N.H.; Choi, S.; Han, E.J.; Hong, B.K.; Choi, S.Y.; Kwon, H.M.; Hwang, S.Y.; Cho, C.S.; Kim, W.U. The xanthine oxidase-NFAT5 pathway regulates macrophage activation and TLR-induced inflammatory arthritis. Eur. J. Immunol. 2014, 44, 2721–2736. [Google Scholar] [CrossRef]
- Cen, L.; Xing, F.; Xu, L.; Cao, Y. Potential Role of Gene Regulator NFAT5 in the Pathogenesis of Diabetes Mellitus. J. Diabetes Res. 2020, 2020, 6927429. [Google Scholar] [CrossRef]
- Gwon, D.H.; Kim, S.I.; Lee, S.H.; Noh, C.; Kim, Y.; Yun, S.; Lee, W.H.; Oh, J.Y.; Kim, D.W.; Hong, J.; et al. NFAT5 Deficiency Alleviates Formalin-Induced Inflammatory Pain Through mTOR. Int. J. Mol. Sci. 2021, 22, 2587. [Google Scholar] [CrossRef]
- Buxadé, M.; Lunazzi, G.; Minguillón, J.; Iborra, S.; Berga-Bolaños, R.; Del Val, M.; Aramburu, J.; López-Rodríguez, C. Gene expression induced by Toll-like receptors in macrophages requires the transcription factor NFAT5. J. Exp. Med. 2012, 209, 379–393. [Google Scholar] [CrossRef]
- Lee, N.; Kim, D.; Kim, W.U. Role of NFAT5 in the Immune System and Pathogenesis of Autoimmune Diseases. Front. Immunol. 2019, 10, 270. [Google Scholar] [CrossRef]
- Huang, Q.Q.; Pope, R.M. The role of toll-like receptors in rheumatoid arthritis. Curr. Rheumatol. Rep. 2009, 11, 357–364. [Google Scholar] [CrossRef]
- Yoon, H.J.; You, S.; Yoo, S.A.; Kim, N.H.; Kwon, H.M.; Yoon, C.H.; Cho, C.S.; Hwang, D.; Kim, W.U. NF-AT5 is a critical regulator of inflammatory arthritis. Arthritis Rheum. 2011, 63, 1843–1852. [Google Scholar] [CrossRef]
- O’Connor, R.S.; Mills, S.T.; Jones, K.A.; Ho, S.N.; Pavlath, G.K. A combinatorial role for NFAT5 in both myoblast migration and differentiation during skeletal muscle myogenesis. J. Cell Sci. 2007, 120, 149–159. [Google Scholar] [CrossRef]
- Greenblatt, M.B.; Ritter, S.Y.; Wright, J.; Tsang, K.; Hu, D.; Glimcher, L.H.; Aliprantis, A.O. NFATc1 and NFATc2 repress spontaneous osteoarthritis. Proc. Natl. Acad. Sci. USA 2013, 110, 19914–19919. [Google Scholar] [CrossRef]
- Sun, R.; Zhang, B.; Chen, L.; Sun, J. Role of nuclear factor of activated T cells 1 in the pathogenesis of osteoarthritis. Exp. Ther. Med. 2014, 7, 195–198. [Google Scholar] [CrossRef][Green Version]
- Zhang, M.; Lu, Q.; Budden, T.; Wang, J. NFAT1 protects articular cartilage against osteoarthritic degradation by directly regulating transcription of specific anabolic and catabolic genes. Bone Joint Res. 2019, 8, 90–100. [Google Scholar] [CrossRef]
- Liang, X.Q.; Du, Y.P.; Wang, D.L.; Yang, Y. The biological functions of lysine methyltransferase PR-SET7. Yi Chuan 2013, 35, 241–254. (In Chinese) [Google Scholar] [CrossRef]
- Kumarasinghe, D.D.; Hopwood, B.; Kuliwaba, J.S.; Atkins, G.J.; Fazzalari, N.L. An update on primary hip osteoarthritis including altered Wnt and TGF-β associated gene expression from the bony component of the disease. Rheumatology 2011, 50, 2166–2175. [Google Scholar] [CrossRef]
- Zhu, X.; Yang, S.; Lin, W.; Wang, L.; Ying, J.; Ding, Y.; Chen, X. Roles of Cell Cyle Regulators Cyclin D1, CDK4, and p53 in Knee Osteoarthritis. Genet. Test. Mol. Biomark. 2016, 20, 529–534. [Google Scholar] [CrossRef]
- Madzuki, I.N.; Lau, S.F.; Mohamad Shalan, N.A.A.; Mohd Ishak, N.I.; Mohamed, S. Does cartilage ERα overexpression correlate with osteoarthritic chondrosenescence? Indications from Labisia pumila OA mitigation. J. Biosci. 2019, 44, 100. [Google Scholar] [CrossRef]
- An, J.; Zhang, J.; Wang, Z.; Wang, K.; Liang, W.; Liu, X.; Zhou, H.; Zhang, H.; Wang, S. KMT5A Knockdown Suppresses Osteosarcoma Cell Proliferation and Metastasis Through Ꞵ-Catenin Signalling. Clin. Investig. Med. 2022, 45, E23–E31. [Google Scholar] [CrossRef]
- Liu, Y.; Yau, M.S.; Yerges-Armstrong, L.M.; Duggan, D.J.; Renner, J.B.; Hochberg, M.C.; Mitchell, B.D.; Jackson, R.D.; Jordan, J.M. Genetic Determinants of Radiographic Knee Osteoarthritis in African Americans. J. Rheumatol. 2017, 44, 1652–1658. [Google Scholar] [CrossRef]
- Yau, M.S.; Yerges-Armstrong, L.M.; Liu, Y.; Lewis, C.E.; Duggan, D.J.; Renner, J.B.; Torner, J.; Felson, D.T.; McCulloch, C.E.; Kwoh, C.K.; et al. Genome-Wide Association Study of Radiographic Knee Osteoarthritis in North American Caucasians. Arthritis Rheumatol. 2017, 69, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Fontenla, C.; Calaza, M.; Evangelou, E.; Valdes, A.M.; Arden, N.; Blanco, F.J.; Carr, A.; Chapman, K.; Deloukas, P.; Doherty, M.; et al. Assessment of osteoarthritis candidate genes in a meta-analysis of nine genome-wide association studies. Arthritis Rheumatol. 2014, 66, 940–949. [Google Scholar] [CrossRef]
- Lindner, C.; Thiagarajah, S.; Wilkinson, J.M.; Panoutsopoulou, K.; Day-Williams, A.G.; arcOGEN Consortium; Cootes, T.F.; Wallis, G.A. Investigation of association between hip osteoarthritis susceptibility loci and radiographic proximal femur shape. Arthritis Rheumatol. 2015, 67, 2076–2084. [Google Scholar] [CrossRef]
- Barr, R.J.; Gregory, J.S.; Reid, D.M.; Aspden, R.M.; Yoshida, K.; Hosie, G.; Silman, A.J.; Alesci, S.; Macfarlane, G.J. Predicting OA progression to total hip replacement: Can we do better than risk factors alone using active shape modelling as an imaging biomarker? Rheumatology 2012, 51, 562–570. [Google Scholar] [CrossRef]
- Agricola, R.; Reijman, M.; Bierma-Zeinstra, S.M.; Verhaar, J.A.; Weinans, H.; Waarsing, J.H. Total hip replacement but not clinical osteoarthritis can be predicted by the shape of the hip: A prospective cohort study (CHECK). Osteoarthr. Cartil. 2013, 21, 559–564. [Google Scholar] [CrossRef]
- Gee, F.; Clubbs, C.F.; Raine, E.V.; Reynard, L.N.; Loughlin, J. Allelic expression analysis of the osteoarthritis susceptibility locus that maps to chromosome 3p21 reveals cis-acting eQTLs at GNL3 and SPCS1. BMC Med. Genet. 2014, 15, 53. [Google Scholar] [CrossRef]
- Rushton, M.D.; Reynard, L.N.; Young, D.A.; Shepherd, C.; Aubourg, G.; Gee, F.; Darlay, R.; Deehan, D.; Cordell, H.J.; Loughlin, J. Methylation quantitative trait locus analysis of osteoarthritis links epigenetics with genetic risk. Hum. Mol. Genet. 2015, 24, 7432–7444. [Google Scholar] [CrossRef]
- Liu, B.; Cheng, H.; Ma, W.; Gong, F.; Wang, X.; Duan, N.; Dang, X. Common variants in the GNL3 contribute to the increasing risk of knee osteoarthritis in Han Chinese population. Sci. Rep. 2018, 8, 9610. [Google Scholar] [CrossRef]
- Moll, T.; Shaw, P.J.; Cooper-Knock, J. Disrupted glycosylation of lipids and proteins is a cause of neurodegeneration. Brain 2020, 143, 1332–1340. [Google Scholar] [CrossRef]
- Baddoo, M.; Hill, K.; Wilkinson, R.; Gaupp, D.; Hughes, C.; Kopen, G.C.; Phinney, D.G. Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection. J. Cell Biochem. 2003, 89, 1235–1249. [Google Scholar] [CrossRef] [PubMed]
- Romanova, L.; Kellner, S.; Katoku-Kikyo, N.; Kikyo, N. Novel role of nucleostemin in the maintenance of nucleolar architecture and integrity of small nucleolar ribonucleoproteins and the telomerase complex. J. Biol. Chem. 2009, 284, 26685–26694. [Google Scholar] [CrossRef] [PubMed]
- GeneCards: The Human Gene Database. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=GNL3 (accessed on 22 November 2022).
- Zhu, Z.; Xie, J.; Manandhar, U.; Yao, X.; Bian, Y.; Zhang, B. RNA binding protein GNL3 up-regulates IL24 and PTN to promote the development of osteoarthritis. Life Sci. 2021, 267, 118926. [Google Scholar] [CrossRef]
- Louka, M.L.; Zakaria, Z.M.; Nagaty, M.M.; Elsebaie, M.A.; Nabil, L.M. Expression of nucleostemin gene in primary osteoarthritis. Gene 2016, 587, 27–32. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).