Abstract
Although the importance of lipid homeostasis in neuronal function is undisputed, how they are regulated within neurons to support their unique function is an area of active study. NHR-49 is a nuclear hormone receptor functionally similar to PPARα, and a major lipid regulator in C. elegans. Although expressed in most tissues, little is known about its roles outside the intestine, the main metabolic organ of C. elegans. Here, using tissue- and neuron-type-specific transgenic strains, we examined the contribution of neuronal NHR-49 to cell-autonomous and non-autonomous nhr-49 mutant phenotypes. We examined lifespan, brood size, early egg-laying, and reduced locomotion on food. We found that lifespan and brood size could be rescued by neuronal NHR-49, and that NHR-49 in cholinergic and serotonergic neurons is sufficient to restore lifespan. For behavioral phenotypes, NHR-49 in serotonergic neurons was sufficient to control egg-laying, whereas no single tissue or neuron type was able to rescue the enhanced on-food slowing behavior. Our study shows that NHR-49 can function in single neuron types to regulate C. elegans physiology and behavior, and provides a platform to further investigate how lipid metabolism in neurons impact neuronal function and overall health of the organism.
1. Introduction
Many instances of neurological and neurodegenerative disorders involve various forms of underlying metabolic dysfunctions, including lipid dysregulation [1]. However, precise understanding of the upstream causes that lead to such effects remain unclear. In addition, studies of lipid metabolism in neurons are made difficult in most systems by the fact that they work closely with the glia, making it difficult to disentangle one’s function from another. The free-living nematode Caenorhabditis elegans provides an ideal in vivo model to study the effect of lipid metabolism with a simple nervous system composed of all major neuron subtypes, such as glutamatergic, GABAergic, cholinergic, serotonergic, and dopaminergic, and includes only a few glial cells.
NHR-49 is one of the many nuclear hormone receptors expressed in C. elegans. C. elegans possesses a total of 284 nuclear hormone receptors in its genome, but the function of most remains elusive [2]. NHR-49 is one of the well-studied nuclear hormone receptors, first discovered for its role in the C. elegans fasting response, where animals metabolize its lipid stores to produce energy [3]. NHR-49 regulates expression of genes involved in fatty acid β-oxidation and desaturation [4]. Lack of nhr-49 resulted in decreased survival during starvation [3]. Aside from the fasting response, nhr-49 mutants display additional pleiotropic defects, such as shorter lifespan, impaired proteostasis, and vulnerability to various stressors such as oxidative stress, pathogenic infection, and hypoxia [5,6,7,8,9,10]. Such defects demonstrate the importance of lipid homeostasis in a wide variety of processes.
Although the intestine is the main metabolic organ and site of lipid storage in C. elegans, NHR-49 is expressed in most tissues in the animal, including neurons. Neurons generally have limited lipid catabolic capabilities due to their vulnerability to ROS [11]. They do not store fats, and it has been shown in mammalian and fly nervous systems that they instead receive lipid-derived lactate from astrocytes or glia as an energy source [11,12]. Considering this, it is unclear what role a transcription factor that mainly regulates fatty acid β-oxidation genes would play in neurons. In light of recent studies that have shown contributions of neuronal NHR-49 to lifespan and immunity [8,13], further study of the role of the lipid regulator in neurons may improve our understanding of how neuronal lipid metabolism contributes to cell-autonomous and non-autonomous functions. Similar to the neuronal subtypes in the vertebrate nervous system, C. elegans neurons express neurotransmitters and neuromodulators such as acetylcholine, GABA, serotonin, and dopamine. Therefore, studies in contributions of specific neuronal subtypes to lipid-mediated inter-tissue signaling in C. elegans may be significant and applicable to mammalian systems. In addition, since phenotypes such as lifespan and immunity are complex phenotypes with many contributing factors, it is difficult to precisely define the mechanism of neuronal NHR-49. Therefore, identification of more specific neuronal and behavioral phenotypes of nhr-49 may provide a better system to study the precise roles and mechanism of lipid metabolism in neurons, which may in turn reveal how it supports the overall health of the animal.
Here, we used tissue- and neuron-type-specific transgenic rescue strains of nhr-49 to investigate the contribution of neuronal NHR-49 to various nhr-49 phenotypes. The phenotypes we observed are lifespan, a previously established phenotype, as well as brood size, egg-laying, and on-food slowing behavior, which are previously unreported phenotypes of nhr-49 described here for the first time. We found that neuronal NHR-49 can cell-non-autonomously contribute to both lifespan and brood size, and more specifically, NHR-49 in cholinergic and serotonergic neurons are sufficient to prolong lifespan in the mutants. For egg-laying, NHR-49 in serotonergic neurons was sufficient to restore the defect, likely demonstrating a direct cell-autonomous effect of NHR-49 in neuronal function. On the contrary, enhanced on-food slowing behavior could not be rescued by NHR-49 in any single tissue, suggesting it is likely mediated by signaling between multiple tissues with functional NHR-49.
2. Materials and Methods
2.1. C. elegans Growth and Maintenance
All strains used for the assay are listed in Table S2. Strains were grown in nematode growth media (NGM) plates seeded with OP50 E. coli bacteria as previously described. Strains were maintained at 20 °C.
2.2. Plasmid Constructs and Generation of Transgenic Lines
Transgenic strains generated for the study are listed in Table S2. All primers used in this study are listed in Table S3. All neuronal-promoter-driven nhr-49 plasmids were constructed using the pPD95.75 plasmid as backbone. nhr-49c was cloned from C. elegans cDNA. This particular isoform was chosen due to having the longest coding region and because it had been previously used by other labs for transgenic rescue strains [13]. Each segment of the plasmid, promoter, nhr-49, and the SL2 trans-splicing sequence was ligated into the plasmid backbone using the restriction enzyme sites indicated in the primer sequences in Table S3. To subclone tph-1 and dat-1 promoters, which were flanked by NotI and SmaI sites, we created a NotI site in the plasmid by inserting a custom-designed multiple cloning site using annealed single strand oligos with appropriate overhangs (MCS, Table S3). For muscle-specific expression of nhr-49, the same nhr-49 insert was inserted into the myo-3 promoter containing plasmid, which was a gift from Junho Lee. nhr-49(nr2041) mutant strains were injected with each promoter plasmid (20 ng/μL) and co-injection marker unc-122p::mCherry (30 ng/μL), unc-122p::gfp or myo-2p::mCherry (5 ng/μL) co-injection marker. Promoters used for tissue- and neuron-type-specific expression are as follows: 3.5 kb rgef-1 promoter for pan-neuronal expression [14], 3.2 kb unc-17 promoter for cholinergic neurons [15], 2.4 kb eat-4 promoter for glutamatergic neurons [16], 1.75 kb tph-1 promoter for serotonergic neurons [17], 716 bp dat-1 promoter for dopaminergic neurons [18], and 1.8 kb unc-25 promoter for GABAergic neurons [19].
2.3. Lifespan Assays
Worms were synchronized by bleach and grown until L4 larval stage. Approximately 20–25 worms were placed in each NGM plate containing 120 μM FuDR (5-Fluoro-2′-deoxyuridine) to prevent eggs from hatching. Worms were counted every 2–3 days until all worms were dead. Three trials were conducted for each strain.
2.4. Scoring Embryonic Stage of Freshly Laid Eggs
A previously established protocol was followed [20]. In short, 5 adult worms were placed on seeded NGM plates and left for an hour, after which they were removed. Immediately, eggs laid on plates were observed under a microscope to determine their embryonic stage. Brood size was determined by placing a single L4 worm in a seeded NGM plate. Worms were moved to a new plate daily until they stopped laying new eggs. Hatched worms were counted in each plate after 2–3 days to assess total number of eggs laid by the worm.
2.5. Measuring Worm Speed and Path Angle
Worm speed was measured following a previously established protocol with some modifications [21]. Between 7–9 worms were placed in a bacterial lawn, were left to acclimate for 4 min, then their movement was recorded for 4 min at two frames per second. Three trials were conducted for each strain, with the exception of the muscle-specific rescue strain. For measuring off-food speed, about 20 worms were picked and placed in an unseeded agar plate to remove any food adhered to the body, then moved to a fresh assay plate where movement was recorded for 4 min. Average speed of worms was determined using the wrmTrck plugin in Fiji (https://imagej.net/Fiji, accessed on 2 May 2023) [22]. Worm location within the plate was tracked using the “Analyze Particle” function in Fiji. The coordinates provided in the results table were used to plot its location relative to time using Matplotlib in Python. Path angle was measured by first finding the coordinates of each worm using the MTrack2 plugin in Fiji, then extracting coordinates at 10-s intervals to determine path angle with a Python script.
2.6. Statistical Analysis
Statistical analysis for worm lifespan was conducted using OASIS 2 Kaplan–Meier analysis and Log-Rank test [23]. Statistical analysis for worm velocity was compared using unpaired t-test. Brood size was compared using one-way ANOVA with Dunnett’s post hoc test comparison to nhr-49. Statistical analysis for embryonic stage distribution of fresh laid eggs was determined by Wilcoxon Mann–Whitney rank sum test in comparison to nhr-49. Statistical significance is indicated by asterisks as follows: p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), and p < 0.0001 (****).
3. Results
3.1. NHR-49 in Select Neuron Types Contribute to Lifespan
Mutants of nhr-49 display short lifespan compared to wild-type C. elegans. Interestingly, previous studies have shown that transgenic expression of NHR-49 in either neuronal or intestinal tissue restores most of the short lifespan defect in nhr-49 mutants [8,13]. We confirmed these results by generating a pan-neuronal rescue strain in nhr-49 mutants using the rgef-1 promoter, a pan-neuronal promoter with no observable intestinal leakiness often seen in other neuronal promoters [24,25,26] (Figure 1A,H and Figure S1; Table S1). We also generated a muscle-specific rescue transgenic strain and found that expression of NHR-49 in the muscle of nhr-49 mutants had no effect on lifespan (Figure 1B,H). The intestine-specific rescue showed restored lifespan as reported previously [13] (Figure 1C,H).
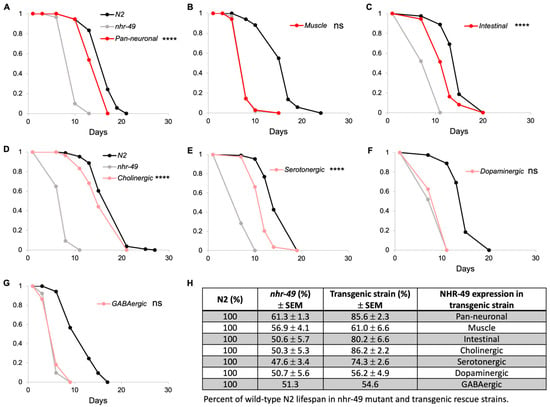
Figure 1.
NHR-49 in select neuron types contributes to lifespan. (A–C) Lifespan of pan-neuronal (A), muscle (B), and intestinal (C) transgenic rescue strains in comparison to wild-type N2 and nhr-49 mutant strains. (D–G) Lifespan of neuron-type specific rescue strains. Each graph represents one trial of lifespan assay, since lifespan can vary significantly between trials. (H) Summary of all trials indicated in percentages of mean lifespan compared to N2 wild-type. Mean lifespan of nhr-49 mutant and transgenic strains for each trial was normalized against that of wild-type N2. Values shown represent average of the normalized values and standard error of mean. Three trials were conducted with the exception of the GABAergic rescue strain. Complete data and statistical analyses for all trials are presented in Table S1. Asterisks indicate p value (**** p < 0.0001, ns: not significant).
If NHR-49 in neurons alone is sufficient to restore normal lifespan, would any specific neuron type be contributing to this? C. elegans neurons express neurotransmitters and neuromodulators such as acetylcholine, GABA, serotonin, and dopamine that regulate motor control, behaviors, and various neuronal functions (Table 1). To examine if NHR-49 expression in specific subsets of neurons is responsible for lifespan, we generated strains in which nhr-49 was restored in either cholinergic, GABAergic, serotonergic, or dopaminergic neurons. Interestingly, we found that there was indeed a neuron-type specificity to the contribution of NHR-49 to lifespan: whereas NHR-49 expression in dopaminergic and GABAergic had no effect (Figure 1F–H; Table S1), NHR-49 expression in cholinergic and serotonergic showed recovery of lifespan to 86.2% and 74.3%, respectively, that appeared similar to the lifespan recovery of 85.6% in the pan-neuronal rescue strain (Figure 1D,E,H; Table S1). Thus, we show that not all neurons equally contribute to lifespan, but NHR-49 in certain neuron types contributes more than others.

Table 1.
Neuron subtypes in C. elegans according to neurotransmitters or neuromodulators.
Table 1.
Neuron subtypes in C. elegans according to neurotransmitters or neuromodulators.
| Promoter | Neuron Type | # of Neurons | Associated Function |
|---|---|---|---|
| dat-1 | Dopaminergic | 8 | Touch sensation [27], locomotion [28], foraging behavior [27,29], learning [30] |
| tph-1 | Serotonergic | 3 | Egg-laying [31], foraging behavior [27], pharyngeal pumping [32] |
| unc-25 | GABAergic | 26 | Locomotion [33], immunity [34] |
| unc-17 | Cholinergic | 160 | Locomotion, male mating, egg-laying [35] |
| eat-4 | Glutamatergic | 79 | Sensory signaling [36] |
3.2. NHR-49 in Neurons Contributes to Egg Production
C. elegans hermaphrodites begin laying eggs a few hours after becoming adults and continue to lay at a regular rate for a few days, resulting in about 300 laid eggs. Egg-laying behavior in the hermaphrodite is tightly regulated by vulval and uterine muscles innervated by motor neurons that express the neurotransmitters acetylcholine and serotonin (Table 1) [37]. Accumulation of embryos in the uteri induces motor neuron activity resulting in egg-laying [38]. While working with the mutants, we noticed that nhr-49 mutants carried a smaller number of eggs in the uterus. Wild-type N2 worms carry 10–15 eggs in the body, but nhr-49 adults tend to carry less than 10 (Figure 2A). Although easily recognizable, the phenotype has never been reported or closely studied. The number of eggs carried by the adult worms may indicate a difference in the rate of input—oocyte maturation and fertilization, or the rate of output—how soon the eggs are laid. We investigated both possibilities to see which are affected by NHR-49.
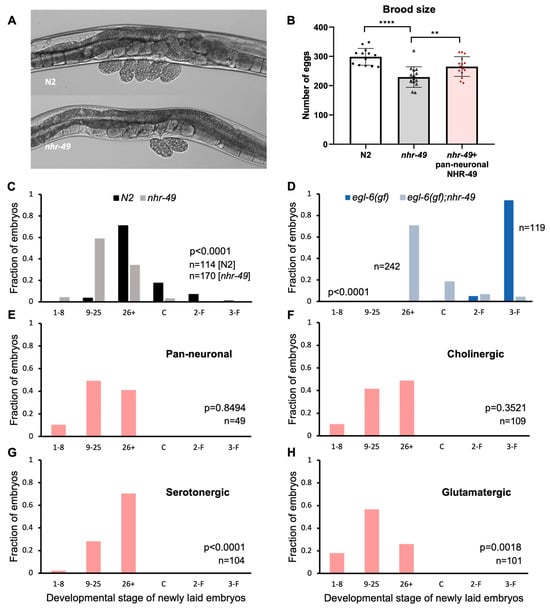
Figure 2.
NHR-49 in select neuron types contribute to egg-laying. (A) Images of eggs carried by N2 and nhr-49. Recently laid eggs collected around the vulva in N2 show embryos in the 9–25 and 26+ stages, and two 9–25 stage eggs for nhr-49 mutants. (B) Total number of eggs laid by N2, nhr-49, and pan-neuronal NHR-49 transgenic. Asterisks indicate p value (** p < 0.01, **** p < 0.0001) (C) Embryonic stages of freshly laid eggs in N2 and nhr-49 mutants. (D) Embryonic stages of freshly laid eggs in egl-6(gf) and nhr-49;egl-6(gf). (E–H) Embryonic stages of freshly laid eggs in pan-neuronal (E), cholinergic (F), serotonergic (G), and glutamatergic (H) transgenic rescue strains. p values determined by the Wilcoxon Mann–Whitney rank sum test are indicated for each graph (C–H).
To see if there is a difference in brood size between N2 and nhr-49 mutants, we counted the total number of eggs laid during the lifetime of the worms. We found that nhr-49 mutants did indeed lay fewer eggs—an average of 230 eggs compared to almost 300 laid by wild-type worms (Figure 2B). Interestingly, we found that pan-neuronal rescue of nhr-49 was sufficient to bring the total eggs closer to wild-type levels, laying an average of 265 eggs. This indicates that NHR-49 function in the neurons can contribute to brood size (Figure 2B).
3.3. NHR-49 in Serotonergic Neurons Contributes to Egg Laying
Next, to see if the absence of nhr-49 affected egg-laying behavior, we used a previously established protocol to score embryonic stages of freshly laid eggs [20]. The mitotic divisions and morphological changes of C. elegans embryos can be easily observed through its clear outer shell, which allowed us to sort the egg stages into six categories: 1–8 cell, 9–25 cell, 26+ cell, comma, 2-fold, and 3-fold stages. The first two stages are easily recognizable by the number of cells in the embryo. At 26+ cell stage, the embryo is a cluster of innumerable cells and undergoes gastrulation. Afterwards an indentation forms at the side of the embryo, which is called the comma stage, then the worm shape begins to take hold, with the elongated shape curled into 2-fold then 3-fold in the tight space within the egg shell. Wild-type N2 hermaphrodites laid eggs that are mostly in the 26+ embryonic stage, which is consistent with previous studies [20] (Figure 2C). On the other hand, more than half of the eggs laid by nhr-49 mutants were in the 9–25 stage, showing a clear shift to earlier stage embryos (Figure 2C).
Vulva muscles receive input from the egg-laying motor circuit, which includes VC cholinergic motor neurons and the HSN motor neuron that is both serotonergic and cholinergic [37]. Is NHR-49 acting through the neurons to control egg-laying? To strengthen the possibility that the egg laying phenotype of nhr-49 mutants is due to altered activity of the neuronal circuit, we took advantage of the egl-6 gain-of-function mutant strain, which exhibits high egg retention due to suppression of HSN motor neuron activity [20]. Genes that counteract the HSN inhibition can be tested through monitoring egg-laying in the double mutant with the egl-6(gf) allele.
When we observed egg-laying in the egl-6(gf) background, there was a much wider difference between nhr-49 wild-type and mutant allele (Figure 2D). Whereas eggs laid by egl-6(gf) mutants were mostly in the 3-fold stage, the last embryonic stage before hatching, eggs from nhr-49;egl-6(gf) double mutants were mostly in the 26+ stage. The fact that nhr-49 mutants relieved egg retention caused by the suppression of a very specific neuron, indicated that the egg-laying phenotype of nhr-49 mutants is likely due to a neuronal defect.
To see if neuronal NHR-49 can restore normal egg-laying, we conducted the same assay on tissue-specific transgenic rescue strains. When eggs were scored in the pan-neuronal rescue, very little difference was observed compared to the mutant (Figure 2E). NHR-49 expression in cholinergic neurons resulted in a slight shift toward later embryonic stages, but was not statistically significant (Figure 2F). However, NHR-49 expression in serotonergic neurons resulted in a much larger shift towards later stage eggs, bringing the distribution closer to which is seen in wild-type worms (Figure 2G). Interestingly, NHR-49 in glutamatergic neurons resulted in even earlier stage eggs than in the mutant, indicating that restoring NHR-49 in the glutamatergic neurons exacerbated the early egg-laying phenotype (Figure 2H). This may explain the lack of improvement in the pan-neuronal rescue strain, as different neuron types affect egg-laying in different directions. Our results show that functional NHR-49 in the serotonergic neurons contributes to their neuronal function to mediate egg-laying.
3.4. Reduced On-Food Speed in nhr-49 Mutants Is Not Due to NHR-49 in Any Single Tissue
Lastly, we noticed that while nhr-49 mutants display no overt movement defects, they traveled very little when placed in the bacterial lawn, a trait easily observable by the tracks left by the worms (Videos S1 and S2). C. elegans employ various foraging and exploratory behaviors to increase their chance of finding food and staying on food. The speed of movement, and size of the area explored, and distance traveled are dependent on factors such as time the animal has been away from food, internal nutrient levels, quality of food, as well as past nutritional experience of the worm [27,39,40,41]. When C. elegans approach a bacterial lawn, they slow down their speed as they enter the bacterial food, which is called the basal slowing response [27]. This is thought to allow worms to stay where food is plentiful, and requires dopaminergic signaling and mechanical sensing of bacteria [27]. We found that N2 animals display basal slowing response reducing their movement speed by approximately 50% on food (Figure 3A). nhr-49 mutants show slightly reduced speed off food compared to wild-type, although their motility and gait overall appeared normal (Figure 3A, Videos S3 and S4). However, on food, nhr-49 mutants displayed around 80% decreased velocity, a drastic reduction compared to basal slowing response in wild-type animals (Figure 3A).
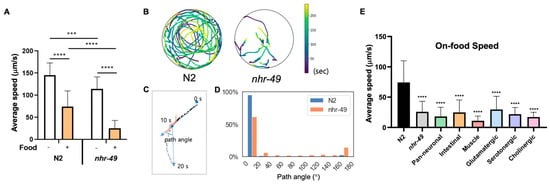
Figure 3.
Reduced on-food speed in nhr-49 mutants is not due to NHR-49 in any single tissue. (A) Average speed of N2 and nhr-49 worms with and without food. (B) Trajectories of nine N2 and eight nhr-49 worms on bacterial lawn. Color indicates the time during the 240 s when worms were found in the given location. Video recordings used for analysis can be seen in Videos S1 and S2, respectively. (C) Sample worm trajectory for 20 s and definition of path angle. Each dot indicates location of a worm in each frame (2 frame/s). Path angle was determined using worm locations at 10-s intervals. (D) Histogram of path angle frequency in each genotype. (E) Average speed of N2, nhr-49, and each tissue and neuron type transgenic rescue strains. Asterisks indicate p value (*** p < 0.001, **** p < 0.0001).
C. elegans displays two modes of movement behavior on food called roaming and dwelling. Roaming behavior is characterized by relatively high movement speed with little curvature, whereas dwelling is defined by much slower speed with curved trajectories and frequent reversal of movement [41,42,43]. Differences in on-food movement are seen particularly between well-fed and starved animals: well-fed C. elegans display both roaming and dwelling behaviors, whereas starved worms exclusively show dwelling behavior [43]. To characterize whether the acute change in on-food speed in nhr-49 is consistent with dwelling behavior, we recorded by video the movement trajectories of N2 and nhr-49 mutants on food (Figure 3B). Overall, we found that wild-type worms moved about, exploring the whole area of the lawn, whereas movement of nhr-49 mutants was much more subdued, with some animals hardly moving during the 4 min of recording (Figure 3B). In addition, when we analyzed the path angle of their movements, N2 worms mostly traveled with little curvature, whereas nhr-49 movement contained more angle bends and reversals, which are indicated by path angles close to 180° (Figure 3C,D). On the contrary, when off-food, very little difference was observed in the distribution of path angles (Figure S2A,B). This suggests that the enhanced on-food slowing observed in nhr-49 mutants is not only due to a general decrease in motility, but a heightened reaction to the presence of food. Thus, nhr-49 mutants display altered on-food behavior, with much decreased speed and increased curvature and changes in direction compared to wild-type.
To find out whether NHR-49 in a specific tissue was responsible for the enhanced slowing on food, we tested on-food speed of the tissue-specific transgenic rescue strains. We tested pan-neuronal as well as intestinal rescue strains, since nhr-49 could be involved in the internal nutrition signal originating from the intestine. Surprisingly, none of the tissue-specific strains tested—neuronal, intestinal, and muscle-specific—restored the on-food speed back to wild-type levels (Figure 3E). In fact, the pan-neuronal rescue and muscle-specific rescue strains showed a tendency to suppress on-food speed even further. Likewise, none of the neuron-type specific rescues of nhr-49 showed improvement in on-food speed (Figure 3E). This may indicate either the enhanced basal slowing is mediated by yet another tissue that we have not yet tested, such as the hypodermis, or that it cannot be attributed to NHR-49 function in any single tissue. In the latter case, it may indicate that the behavior likely involves NHR-49 function in multiple tissues.
4. Discussion
The importance of lipid regulation in metabolism, health, and lifespan has been well established. Indeed, the importance of the lipid regulator NHR-49 in fat regulation, lifespan, and health has been substantiated by numerous studies in C. elegans [3,5,6,7,8,9,10,44]. However, the role that lipid regulation plays in the nervous system and behavior is not as clear.
Studies of lipid regulation in C. elegans inevitably focus on the intestine, since it is the main metabolic organ that stores and processes fat. However, a few nhr-49 mutant phenotypes, such as reduced lifespan and immunity, have been shown to improve upon restoring NHR-49 in the neurons, suggesting that NHR-49 contributes to the overall health of the worm through its lipid regulator function in neurons [8,13]. Studies of how lipid utilization and metabolism within neurons contribute to these phenotypes, though, have been lacking. In light of reports that various neurodegenerative diseases are often found to occur together with metabolic dysfunction [45,46], understanding how metabolism in neurons affects the rest of the body and vice versa will help illuminate the complex interplay between the nervous system and the rest of the body. C. elegans provides an ideal model to study the role of lipid regulators in neurons, with its simple nervous system of 302 neurons and only a small number of glia. Because phenotypes such as lifespan and immunity are complex phenotypes with many contributing factors, identification of more specific neuronal and behavioral phenotypes of nhr-49 may provide a better system to study the immediate roles and mechanism of lipid metabolism in neurons, which in turn may help reveal the mechanism of its cell-non-autonomous role.
Here, we characterized various known, as well as newly described, phenotypes of nhr-49 mutants using tissue-specific transgenic strains. We confirmed previous reports that neuronal NHR-49 can restore lifespan, and further narrowed down the neurons responsible to cholinergic and serotonergic neurons. We also identified additional nhr-49 mutant phenotypes, such as reduced brood size, early egg-laying, and slow on-food speed. Reduced brood size was also restored closer to wild-type levels by expression of neuronal NHR-49. For early egg-laying, pan-neuronal NHR-49 had very little effect, but serotonergic neuron-specific rescue significantly restored the early egg-laying defect. Lastly, nhr-49 mutants displayed dramatically reduced on-food speed, but no single tissue rescued the phenotype.
Lifespan and brood size are complex phenotypes with many contributing factors. Instead of all neurons each contributing to the health of the animal, we found that NHR-49 in specific neuron types, cholinergic and serotonergic neurons, are sufficient to mediate restored lifespan, while other neuron types had no effect. This may indicate either that certain neuron types are more involved in influencing lifespan, or that NHR-49 serves an especially important role in those neurons to maintain their function. Further studies to narrow down the neurons responsible may help reveal the underlying mechanism.
Brood size in C. elegans is tightly linked to nutrient availability or past experience of dauer [47]. In general, brood size in unmated hermaphrodites is limited by the number of sperm, whose production occurs only during the L4 larval stage [48]. However, because both sperm and oocytes differentiate from the same pool of germline cells, general decrease in germline proliferation may be a better explanation for the reduced brood size seen in nhr-49 mutants, rather than a specific defect in sperm development. The fact that pan-neuronal NHR-49 can rescue this phenotype underscores the contribution of neuronal NHR-49 to the overall health of the animal.
The main neuron that governs vulval muscle contraction and egg-laying is the serotonergic HSN motor neuron. The HSN neuron cycles through a series of active and quiescent activity states, controlling the rate of egg-laying [37]. EGL-6 is an inhibitory GPCR, whose gain-of-function mutation results in suppression of HSN activity, leading to the characteristic accumulation of late-stage eggs in the mutants [20]. Mutations in the G-protein signaling pathway or inward rectifying potassium channel that works downstream of EGL-6 can suppress the egg retention phenotype of egl-6(gf) mutants [49]. The fact that nhr-49;egl-6(gf) double mutants can also relieve the egl-6(gf) egg retention phenotype strongly suggests that the absence of nhr-49 results in increased neuronal activity in HSN. Consistently, restoring NHR-49 expression selectively in the serotonergic neurons was sufficient to cause nhr-49 mutant worms to hold eggs until later embryonic stages. There are three serotonergic neurons in C. elegans: ADF, NSM, and HSN. Among these, only HSN is directly involved in egg-laying. Our data provide strong evidence that NHR-49 may directly impact HSN activity. Further investigation into the effect of NHR-49 and relevant downstream targets in HSN may lead to better understanding of how lipid metabolism affects neuronal activity and function.
C. elegans motor neurons control forward and backward locomotion to allow the worm to move sinusoidally to search for sources of bacterial food [50]. Movement of worms in bacterial lawns has been studied in C. elegans in relation to foraging behavior and food sensing, which are affected by external cues such as food concentration [43], as well as internal states such as hunger, and the nutritional quality of the food [41,51]. In addition to the general slowing down of speed on food, worms alternate between states of dwelling and roaming. Longer recording of worm movement is needed to determine whether their switch between dwelling and roaming is impacted, but the pattern of movement of nhr-49 on food may suggest increased dwelling behavior as well as reduced speed. Such reduced speed and increased dwelling are often observed in starved worms [27]. The fact that nhr-49 mutants display behavior similar to starved worms is consistent with having defective lipid metabolism. Surprisingly, none of the tissue-specific transgenic strains rescued the enhanced slowing response. This may indicate that the behavior requires NHR-49 in yet another tissue that we have not yet tested, such as the hypodermis, or that it requires NHR-49 in multiple tissues. Since sensing of starvation and nutrition is a whole-body response, it may require NHR-49 function in multiple tissues acting together, in order to assess organismal nutritional state and carry out the correct behavior.
Several studies using C. elegans have reported cell-non-autonomous effects from neurons that result in improvement in whole body proteostasis, stress resistance, and longevity [52,53,54,55]. In addition, Zullo et al. reported that inhibition of neural excitation and neurotransmission during aging increases longevity in both C. elegans and humans [56]. These studies demonstrate how brain function is intricately tied to the health and homeostasis of the whole body. What is still unclear, however, is how these cellular perturbations occurring in the neurons result in such whole-body effects. Although some specific signals are known, many are still unidentified [57,58,59]. Whether neuronal NHR-49 impacts lifespan and pathogenic infection via similar signals is yet to be determined. Recently, Savini et al. reported that neuronal NHR-49 is required for neuropeptide gene induction that mediates longevity in response to lysosomal lipolysis in the intestine [60], suggesting that NHR-49 may support neuropeptide signaling. Elucidating the nature of the cell-nonautonomous functions of the nervous system will enhance our understanding the intricate relationship between the brain and whole body.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/life13122346/s1, Figure S1: Neuron-type GFP expression in each transgenic strain (closed arrowhead). Open arrowheads indicate co-injection marker fluorescence; Figure S2: Trajectories and path angles of N2 and nhr-49 worms off-food; Table S1: Summary of all lifespan assays; Table S2: Strains used in this study; Table S3: Primers used for this study; Video S1: On-food movement of wild-type N2 worms; Video S2: On-food movement of nhr-49 mutants; Video S3: Off-food movement of wild-type N2 worms; Video S4: Off-food movement of nhr-49 mutants.
Author Contributions
Conceptualization, S.K., K.-S.P. and K.-h.Y.; investigation, S.K. and K.-h.Y.; writing—original draft, S.K. and K.-h.Y.; writing—review and editing, S.K., K.-S.P. and K.-h.Y.; funding acquisition, K.-S.P. and K.-h.Y.; supervision, K.-S.P. and K.-h.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was sponsored by the National Research Foundation of Korea 2017R1A5A2015369 (K.-S.P.) and 2019R1C1C1008708 (K.-h.Y.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request.
Acknowledgments
We thank the Caenorhabditis Genetics Center (CGC) (P40 OD010440) for several strains used in this study. We thank the Junho Lee lab for generously providing the myo-3p plasmid. We thank Jin Lee and the MRC members for providing helpful comments on the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cooper, O.; Hallett, P.; Isacson, O. Upstream lipid and metabolic systems are potential causes of Alzheimer’s disease, Parkinson’s disease and dementias. FEBS J. 2022; early view. [Google Scholar] [CrossRef]
- Antebi, A. Nuclear hormone recpetors in C. elegans. WormBook 2015, 1–49. [Google Scholar] [CrossRef] [PubMed]
- Van Gilst, M.R.; Hadjivassiliou, H.; Yamamoto, K.R. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc. Natl. Acad. Sci. USA 2005, 102, 13496–13501. [Google Scholar] [CrossRef] [PubMed]
- Pathare, P.P.; Lin, A.; Bornfeldt, K.E.; Taubert, S.; Van Gilst, M.R. Coordinate regulation of lipid metabolism by novel nuclear receptor partnerships. PLoS Genet. 2012, 8, e1002645. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; An, S.W.A.; Jung, Y.; Yamaoka, Y.; Ryu, Y.; Goh, G.Y.S.; Beigi, A.; Yang, J.S.; Jung, G.Y.; Ma, D.K.; et al. MDT-15/MED15 permits longevity at low temperature via enhancing lipidostasis and proteostasis. PLoS Biol. 2019, 17, e3000415. [Google Scholar] [CrossRef] [PubMed]
- Goh, G.Y.S.; Winter, J.J.; Bhanshali, F.; Doering, K.R.S.; Lai, R.; Lee, K.; Veal, E.A.; Taubert, S. NHR-49/HNF4 integrates regulation of fatty acid metabolism with a protective transcriptional response to oxidative stress and fasting. Aging Cell 2018, 17, e12743. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, M.; Shashikanth, M.; Gupta, A.; Sandhu, A.; De, A.; Javed, S.; Singh, V. NHR-49 Transcription Factor Regulates Immunometabolic Response and Survival of Caenorhabditis elegans during Enterococcus faecalis Infection. Infect. Immun. 2020, 88, e00130-20. [Google Scholar] [CrossRef]
- Naim, N.; Amrit, F.R.G.; Ratnappan, R.; DelBuono, N.; Loose, J.A.; Ghazi, A. Cell nonautonomous roles of NHR-49 in promoting longevity and innate immunity. Aging Cell 2021, 20, e13413. [Google Scholar] [CrossRef]
- Wani, K.A.; Goswamy, D.; Taubert, S.; Ratnappan, R.; Ghazi, A.; Irazoqui, J.E. NHR-49/PPAR-alpha and HLH-30/TFEB cooperate for C. elegans host defense via a flavin-containing monooxygenase. eLife 2021, 10, e62775. [Google Scholar] [CrossRef]
- Doering, K.R.S.; Cheng, X.; Milburn, L.; Ratnappan, R.; Ghazi, A.; Miller, D.L.; Taubert, S. Nuclear hormone receptor NHR-49 acts in parallel with HIF-1 to promote hypoxia adaptation in Caenorhabditis elegans. eLife 2022, 11, e67911. [Google Scholar] [CrossRef]
- Ralhan, I.; Chang, C.L.; Lippincott-Schwartz, J.; Ioannou, M.S. Lipid droplets in the nervous system. J. Cell Biol. 2021, 220, e202102136. [Google Scholar] [CrossRef]
- Liu, L.; MacKenzie, K.R.; Putluri, N.; Maletic-Savatic, M.; Bellen, H.J. The Glia-Neuron Lactate Shuttle and Elevated ROS Promote Lipid Synthesis in Neurons and Lipid Droplet Accumulation in Glia via APOE/D. Cell Metab. 2017, 26, 719–737.e6. [Google Scholar] [CrossRef]
- Burkewitz, K.; Morantte, I.; Weir, H.J.M.; Yeo, R.; Zhang, Y.; Huynh, F.K.; Ilkayeva, O.R.; Hirschey, M.D.; Grant, A.R.; Mair, W.B. Neuronal CRTC-1 governs systemic mitochondrial metabolism and lifespan via a catecholamine signal. Cell 2015, 160, 842–855. [Google Scholar] [CrossRef]
- Altun-Gultekin, Z.; Andachi, Y.; Tsalik, E.L.; Pilgrim, D.; Kohara, Y.; Hobert, O. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 2001, 128, 1951–1969. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.L.; Lutz, S.; Charlie, N.K.; Vettel, C.; Ailion, M.; Coco, C.; Tesmer, J.J.; Jorgensen, E.M.; Wieland, T.; Miller, K.G. Trio’s Rho-specific GEF domain is the missing Galpha q effector in C. elegans. Genes Dev. 2007, 21, 2731–2746. [Google Scholar] [CrossRef]
- Lee, R.Y.; Sawin, E.R.; Chalfie, M.; Horvitz, H.R.; Avery, L. EAT-4, a homolog of a mammalian sodium-dependent inorganic phosphate cotransporter, is necessary for glutamatergic neurotransmission in Caenorhabditis elegans. J. Neurosci. 1999, 19, 159–167. [Google Scholar] [CrossRef]
- Pocock, R.; Hobert, O. Hypoxia activates a latent circuit for processing gustatory information in C. elegans. Nat. Neurosci. 2010, 13, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Nass, R.; Hall, D.H.; Miller, D.M., 3rd; Blakely, R.D. Neurotoxin-induced degeneration of dopamine neurons in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2002, 99, 3264–3269. [Google Scholar] [CrossRef]
- Eastman, C.; Horvitz, H.R.; Jin, Y. Coordinated transcriptional regulation of the unc-25 glutamic acid decarboxylase and the unc-47 GABA vesicular transporter by the Caenorhabditis elegans UNC-30 homeodomain protein. J. Neurosci. 1999, 19, 6225–6234. [Google Scholar] [CrossRef]
- Ringstad, N.; Horvitz, H.R. FMRFamide neuropeptides and acetylcholine synergistically inhibit egg-laying by C. elegans. Nat. Neurosci. 2008, 11, 1168–1176. [Google Scholar] [CrossRef]
- de Bono, M.; Bargmann, C.I. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell 1998, 94, 679–689. [Google Scholar] [CrossRef]
- Nussbaum-Krammer, C.I.; Neto, M.F.; Brielmann, R.M.; Pedersen, J.S.; Morimoto, R.I. Investigating the spreading and toxicity of prion-like proteins using the metazoan model organism C. elegans. J. Vis. Exp. 2015, 95, 52321. [Google Scholar] [CrossRef]
- Han, S.K.; Lee, D.; Lee, H.; Kim, D.; Son, H.G.; Yang, J.S.; Lee, S.V.; Kim, S. OASIS 2: Online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget 2016, 7, 56147–56152. [Google Scholar] [CrossRef]
- Wang, H.; Liu, J.; Gharib, S.; Chai, C.M.; Schwarz, E.M.; Pokala, N.; Sternberg, P.W. cGAL, a temperature-robust GAL4-UAS system for Caenorhabditis elegans. Nat. Methods 2017, 14, 145–148. [Google Scholar] [CrossRef]
- Murata, D.; Nomura, K.H.; Dejima, K.; Mizuguchi, S.; Kawasaki, N.; Matsuishi-Nakajima, Y.; Ito, S.; Gengyo-Ando, K.; Kage-Nakadai, E.; Mitani, S.; et al. GPI-anchor synthesis is indispensable for the germline development of the nematode Caenorhabditis elegans. Mol. Biol. Cell 2012, 23, 982–995. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Jimenez, C.; Ayuso, C.; Dobrzynska, A.; Torres-Mendez, A.; Ruiz, P.C.; Askjaer, P. An Efficient FLP-Based Toolkit for Spatiotemporal Control of Gene Expression in Caenorhabditis elegans. Genetics 2017, 206, 1763–1778. [Google Scholar] [CrossRef] [PubMed]
- Sawin, E.R.; Ranganathan, R.; Horvitz, H.R. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 2000, 26, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Omura, D.T.; Clark, D.A.; Samuel, A.D.; Horvitz, H.R. Dopamine signaling is essential for precise rates of locomotion by C. elegans. PLoS ONE 2012, 7, e38649. [Google Scholar] [CrossRef]
- Hills, T.; Brockie, P.J.; Maricq, A.V. Dopamine and glutamate control area-restricted search behavior in Caenorhabditis elegans. J. Neurosci. 2004, 24, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, S.; Wintle, R.F.; Kindt, K.S.; Nuttley, W.M.; Arvan, R.; Fitzmaurice, P.; Bigras, E.; Merz, D.C.; Hebert, T.E.; van der Kooy, D.; et al. Dopamine modulates the plasticity of mechanosensory responses in Caenorhabditis elegans. EMBO J. 2004, 23, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Weinshenker, D.; Garriga, G.; Thomas, J.H. Genetic and pharmacological analysis of neurotransmitters controlling egg laying in C. elegans. J. Neurosci. 1995, 15, 6975–6985. [Google Scholar] [CrossRef]
- Segalat, L.; Elkes, D.A.; Kaplan, J.M. Modulation of serotonin-controlled behaviors by Go in Caenorhabditis elegans. Science 1995, 267, 1648–1651. [Google Scholar] [CrossRef]
- Jorgensen, E.M. GABA. WormBook 2005, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Zhang, X.; Liu, J.; He, P.; Zhang, S.; Zhang, Y.; Gao, J.; Yang, S.; Kang, N.; Afridi, M.I.; et al. GABAergic synapses suppress intestinal innate immunity via insulin signaling in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2021, 118, e2021063118. [Google Scholar] [CrossRef]
- Rand, J.B. Acetylcholine. WormBook 2006, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Horowitz, L.B.; Ringstad, N. Opponent vesicular transporters regulate the strength of glutamatergic neurotransmission in a C. elegans sensory circuit. Nat. Commun. 2021, 12, 6334. [Google Scholar] [CrossRef] [PubMed]
- Schafer, W.R. Egg-laying. WormBook 2005, 1–7. [Google Scholar] [CrossRef]
- Medrano, E.; Collins, K.M. Muscle-directed mechanosensory feedback activates egg-laying circuit activity and behavior in Caenorhabditis elegans. Curr. Biol. 2023, 33, 2330–2339.e8. [Google Scholar] [CrossRef]
- Gray, J.M.; Hill, J.J.; Bargmann, C.I. A circuit for navigation in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2005, 102, 3184–3191. [Google Scholar] [CrossRef]
- Pradhan, S.; Quilez, S.; Homer, K.; Hendricks, M. Environmental Programming of Adult Foraging Behavior in C. elegans. Curr. Biol. 2019, 29, 2867–2879.e4. [Google Scholar] [CrossRef]
- Shtonda, B.B.; Avery, L. Dietary choice behavior in Caenorhabditis elegans. J. Exp. Biol. 2006, 209, 89–102. [Google Scholar] [CrossRef]
- Fujiwara, M.; Sengupta, P.; McIntire, S.L. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron 2002, 36, 1091–1102. [Google Scholar] [CrossRef]
- Ben Arous, J.; Laffont, S.; Chatenay, D. Molecular and sensory basis of a food related two-state behavior in C. elegans. PLoS ONE 2009, 4, e7584. [Google Scholar] [CrossRef] [PubMed]
- Ratnappan, R.; Amrit, F.R.; Chen, S.W.; Gill, H.; Holden, K.; Ward, J.; Yamamoto, K.R.; Olsen, C.P.; Ghazi, A. Germline signals deploy NHR-49 to modulate fatty-acid beta-oxidation and desaturation in somatic tissues of C. elegans. PLoS Genet. 2014, 10, e1004829. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Cong, W.N.; Ji, S.; Rothman, S.; Maudsley, S.; Martin, B. Metabolic dysfunction in Alzheimer’s disease and related neurodegenerative disorders. Curr. Alzheimer Res. 2012, 9, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Podolsky, S.; Leopold, N.A.; Sax, D.S. Increased frequency of diabetes mellitus in patients with Huntington’s chorea. Lancet 1972, 1, 1356–1358. [Google Scholar] [CrossRef] [PubMed]
- Fielenbach, N.; Antebi, A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008, 22, 2149–2165. [Google Scholar] [CrossRef] [PubMed]
- Hodgkin, J.; Barnes, T.M. More is not better: Brood size and population growth in a self-fertilizing nematode. Proc. Biol. Sci. 1991, 246, 19–24. [Google Scholar] [CrossRef]
- Emtage, L.; Aziz-Zaman, S.; Padovan-Merhar, O.; Horvitz, H.R.; Fang-Yen, C.; Ringstad, N. IRK-1 potassium channels mediate peptidergic inhibition of Caenorhabditis elegans serotonin neurons via a G(o) signaling pathway. J. Neurosci. 2012, 32, 16285–16295. [Google Scholar] [CrossRef]
- Huang, K.M.; Cosman, P.; Schafer, W.R. Automated detection and analysis of foraging behavior in Caenorhabditis elegans. J. Neurosci. Methods 2008, 171, 153–164. [Google Scholar] [CrossRef]
- Avery, L.; You, Y.J. C. elegans feeding. WormBook 2012. [Google Scholar] [CrossRef]
- Prahlad, V.; Morimoto, R.I. Neuronal circuitry regulates the response of Caenorhabditis elegans to misfolded proteins. Proc. Natl. Acad. Sci. USA 2011, 108, 14204–14209. [Google Scholar] [CrossRef]
- van Oosten-Hawle, P.; Morimoto, R.I. Organismal proteostasis: Role of cell-nonautonomous regulation and transcellular chaperone signaling. Genes Dev. 2014, 28, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.C.; Dillin, A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell 2013, 153, 1435–1447. [Google Scholar] [CrossRef]
- Chauve, L.; Hodge, F.; Murdoch, S.; Masoudzadeh, F.; Mann, H.J.; Lopez-Clavijo, A.F.; Okkenhaug, H.; West, G.; Sousa, B.C.; Segonds-Pichon, A.; et al. Neuronal HSF-1 coordinates the propagation of fat desaturation across tissues to enable adaptation to high temperatures in C. elegans. PLoS Biol. 2021, 19, e3001431. [Google Scholar] [CrossRef]
- Zullo, J.M.; Drake, D.; Aron, L.; O’Hern, P.; Dhamne, S.C.; Davidsohn, N.; Mao, C.A.; Klein, W.H.; Rotenberg, A.; Bennett, D.A.; et al. Regulation of lifespan by neural excitation and REST. Nature 2019, 574, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Ozbey, N.P.; Imanikia, S.; Krueger, C.; Hardege, I.; Morud, J.; Sheng, M.; Schafer, W.R.; Casanueva, M.O.; Taylor, R.C. Tyramine Acts Downstream of Neuronal XBP-1s to Coordinate Inter-tissue UPR(ER) Activation and Behavior in C. elegans. Dev. Cell 2020, 55, 754–770.e6. [Google Scholar] [CrossRef] [PubMed]
- Berendzen, K.M.; Durieux, J.; Shao, L.W.; Tian, Y.; Kim, H.E.; Wolff, S.; Liu, Y.; Dillin, A. Neuroendocrine Coordination of Mitochondrial Stress Signaling and Proteostasis. Cell 2016, 166, 1553–1563.e10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wu, X.; Chen, P.; Liu, L.; Xin, N.; Tian, Y.; Dillin, A. The Mitochondrial Unfolded Protein Response Is Mediated Cell-Non-autonomously by Retromer-Dependent Wnt Signaling. Cell 2018, 174, 870–883.e17. [Google Scholar] [CrossRef]
- Savini, M.; Folick, A.; Lee, Y.T.; Jin, F.; Cuevas, A.; Tillman, M.C.; Duffy, J.D.; Zhao, Q.; Neve, I.A.; Hu, P.W.; et al. Lysosome lipid signalling from the periphery to neurons regulates longevity. Nat. Cell Biol. 2022, 24, 906–916. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).