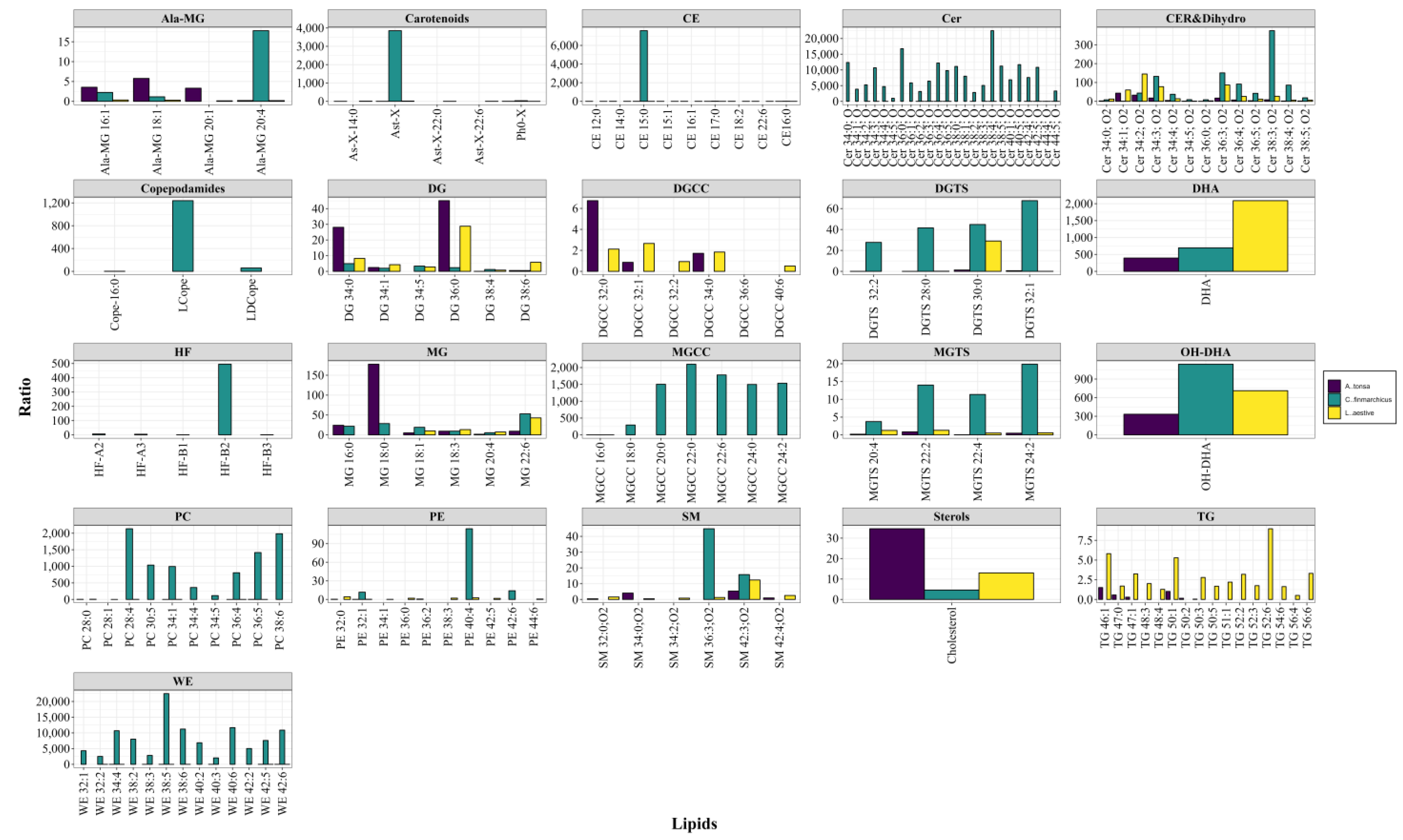
Figure 1.
Relative levels (R) of lipids in copepods. Levels are presented (mean values) as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA) corrected for the sample dry weight. (n = 5). Ala, alanine; ASt-X, astaxanthin; CE, cholesterol ester; Cer, ceramide; Cope, copepodamide; DG, diacylglycerol; DGCC, diacylglyceryl carboxyhydroxymethylcholine; DGTS, diacylglyceryl trimethylhomoserine; DHA, FA 22:6; HF, heterofibrin; L, lyso; MG, monoacylglycerol; OH-DHA, hydroxy FA 22:6; PC, phosphatidylcholine; PE, phosphatidylethanolamine; SM, sphingomyelin; TG, triacylglycerol; WE, wax ester.
Figure 1.
Relative levels (R) of lipids in copepods. Levels are presented (mean values) as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA) corrected for the sample dry weight. (n = 5). Ala, alanine; ASt-X, astaxanthin; CE, cholesterol ester; Cer, ceramide; Cope, copepodamide; DG, diacylglycerol; DGCC, diacylglyceryl carboxyhydroxymethylcholine; DGTS, diacylglyceryl trimethylhomoserine; DHA, FA 22:6; HF, heterofibrin; L, lyso; MG, monoacylglycerol; OH-DHA, hydroxy FA 22:6; PC, phosphatidylcholine; PE, phosphatidylethanolamine; SM, sphingomyelin; TG, triacylglycerol; WE, wax ester.
Table 1.
Relative levels of carotenoids and carotenoid fatty esters in copepods and microalga. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [13C3]triacylglycerol 48:0), corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5). -, not detected.
Table 1.
Relative levels of carotenoids and carotenoid fatty esters in copepods and microalga. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [13C3]triacylglycerol 48:0), corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5). -, not detected.
| Carotenoids | [M+H]+ | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
|---|
| Phoenicoxanthin | 581.3989 | 566.9 ± 124.8 | 3.34 ± 1.23 | 32.8 ± 15.3 | 1.04 ± 0.72 |
| Astaxanthin | 597.3938 | 18.56 ± 4.32 | 2.46 ± 1.07 | 3854.0 ± 200.8 | 16.5 ± 7.0 |
| Astaxanthin-14:0 | 807.5922 | - | 0.99 ± 0.12 | - | 10.2 ± 3.6 |
| Astaxanthin-22:0 | 919.7174 | - | - | - | 2.34 ± 1.34 |
| Astaxanthin-22:6 | 907.6235 | - | - | - | 3.63 ± 0.66 |
Table 2.
Relative levels of copepodamides in C. finmarchicus. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA), corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5).
Table 2.
Relative levels of copepodamides in C. finmarchicus. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA), corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5).
| Copepodamides | [M-H]− | Calanus finmarchicus |
|---|
| Lysocopepodamide | 448.2739 | 1240 ± 329 |
| Lysodihydrocopepodamide | 450.2895 | 59 ± 13 |
| Copepodamide-16:0 | 686.5035 | 2.11 ± 1.20 |
Table 3.
Relative levels of wax esters (WE) and triacylglycerols (TG) in copepods and microalga. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [13C3]triacylglycerol 48:0), corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5). -, not detected.
Table 3.
Relative levels of wax esters (WE) and triacylglycerols (TG) in copepods and microalga. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [13C3]triacylglycerol 48:0), corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5). -, not detected.
| Wax Esters | [M+NH4]+ | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
|---|
| WE 32:1 | 496.5088 | - | - | 4318 ± 2414 | - |
| WE 32:2 | 494.4932 | - | - | 2516 ± 1392 | 0.058 ± 0.017 |
| WE 34:4 | 518.4932 | - | 0.54 ± 0.25 | 10,686 ± 5960 | 1.50 ± 0.28 |
| WE 38:2 | 578.5871 | 18.35 ± 4.84 | - | 8009 ± 4530 | - |
| WE 38:3 | 576.5714 | 2.67 ± 0.77 | 0.091 ± 0.008 | 2835 ± 1618 | - |
| WE 38:5 | 572.5401 | 2.41 ± 1.23 | 0.13 ± 0.019 | 22,497 ± 12,673 | 0.16 ± 0.017 |
| WE 38:6 | 570.5245 | - | - | 11,219 ± 6374 | 0.018 ± 0.006 |
| WE 40:2 | 606.6184 | 0.20 ± 0.026 | - | 6865 ± 3916 | - |
| WE 40:3 | 604.6027 | 0.16 ± 0.024 | 0.71 ± 0.062 | 2042 ± 1423 | - |
| WE 40:6 | 598.5558 | 1.21 ± 0.034 | - | 11,687 ± 6611 | - |
| WE 42:2 | 634.6497 | 0.31 ± 0.051 | - | 5028 ± 2764 | - |
| WE 42:5 | 628.6027 | 0.038 ± 0.008 | 4.63 ± 2.25 | 7584 ± 4321 | 2.64 ± 0.065 |
| WE 42:6 | 626.5871 | 1.20 ± 0.23 | 1.08 ± 0.23 | 10,882 ± 6056 | 0.66 ± 0.18 |
| Triacylglycerols | [M+NH4]+ | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
| TG 46:1 | 780.7076 | - | 1.51 ± 0.75 | - | 5.81 ± 3.00 |
| TG 47:0 | 810.7545 | - | 0.57 ± 0.26 | - | 1.70 ± 0.88 |
| TG 47:1 | 808.7389 | - | 0.29 ± 0.15 | - | 3.24 ± 1.57 |
| TG 48:3 | 818.7232 | - | - | - | 2.01 ± 1.00 |
| TG 48:4 | 816.7076 | - | - | - | 1.28 ± 0.65 |
| TG 50:1 | 850.7858 | - | 1.02 ± 0.17 | - | 5.29 ± 2.89 |
| TG 50:2 | 848.7702 | - | 0.15 ± 0.05 | - | - |
| TG 50:3 | 846.7545 | - | 0.042 ± 0.021 | - | 2.79 ± 1.45 |
| TG 50:5 | 842.7232 | - | - | - | 1.68 ± 1.02 |
| TG 51:1 | 864.8015 | - | - | - | 2.20 ± 1.01 |
| TG 52:2 | 876.8015 | - | - | - | 3.19 ± 1.66 |
| TG 52:3 | 874.7858 | - | - | - | 1.75 ± 0.090 |
| TG 52:6 | 868.7389 | - | - | - | 8.97 ± 3.12 |
| TG 54:6 | 896.7702 | - | - | - | 1.63 ± 0.83 |
| TG 56:4 | 928.8328 | - | - | - | 0.51 ± 0.15 |
| TG 56:6 | 924.8015 | - | - | - | 3.31 ± 1.53 |
Table 4.
Relative levels of monoacylglycerols (MG) and modified MGs in copepods and microalga. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA for NESI and 2 nanomoles [13C3]triacylglycerol 48:0 for PESI), corrected for the sample dry weight (Mean ± SD; n = 5). -, not detected.
Table 4.
Relative levels of monoacylglycerols (MG) and modified MGs in copepods and microalga. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA for NESI and 2 nanomoles [13C3]triacylglycerol 48:0 for PESI), corrected for the sample dry weight (Mean ± SD; n = 5). -, not detected.
| Monoacylglycerols | [M+Cl]− | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
|---|
| MG 16:0 | 365.2469 | 1.76 ± 0.094 | 24.09 ± 14.64 | 21.70 ± 3.08 | - |
| MG 18:0 | 393.2782 | 0.73 ± 0.079 | 177.7 ± 88.2 | 28.43 ± 6.91 | - |
| MG 18:1 | 391.2625 | 3.31 ± 0.061 | 5.07 ± 0.79 | 19.07 ± 8.00 | 10.11 ± 4.78 |
| MG 18:3 | 387.2312 | 0.29 ± 0.011 | 9.35 ± 4.64 | 9.42 ± 1.95 | 13.26 ± 4.20 |
| MG 20:4 | 413.2469 | 1.20 ± 0.13 | 1.59 ± 0.58 | 5.45 ± 1.49 | 7.28 ± 3.73 |
| MG 22:6 | 437.2469 | 1.32 ± 0.10 | 9.26 ± 0.26 | 52.88 ± 12.05 | 42.74 ± 20.38 |
| Alanyl-monoacyl-glycerols | [M+H]+ | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
| Ala-MG 16:1 | 400.3057 | 0.20 ± 0.063 | 3.55 ± 0.57 | 2.24 ± 0.81 | 0.34 ± 0.012 |
| Ala-MG 18:1 | 428.3370 | 0.27 ± 0.084 | 5.77 ± 0.35 | 1.14 ± 0.26 | 0.30 ± 0.11 |
| Ala-MG 20:1 | 456.3683 | 0.029 ± 0.013 | 3.31 ± 1.72 | - | 0.13 ± 0.070 |
| Ala-MG 20:4 | 450.3214 | - | 0.20 ± 0.11 | 17.8 ± 7.10 | 0.18 ± 0.051 |
| MGTS | [M+H]+ | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
| MGTS 20:4 | 522.3789 | - | 0.19 ± 0.028 | 3.74 ± 2.56 | 1.26 ± 0.58 |
| MGTS 22:2 | 554.4415 | 1.77 ± 0.71 | 0.86 ± 0.51 | 14.01 ± 4.07 | 1.29 ± 0.85 |
| MGTS 22:4 | 550.4102 | - | 0.044 ± 0.021 | 11.37 ± 3.32 | 0.52 ± 0.20 |
| MGTS 24:2 | 582.4728 | 0.83 ± 0.38 | 0.48 ± 0.21 | 19.88 ± 5.12 | 0.59 ± 0.20 |
| MGCC | [M+H]+ | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
| MGCC 16:0 | 490.3738 | - | 0.95 ± 0.03 | 0.22 ± 0.23 | 0.83 ± 0.40 |
| MGCC 18:0 | 518.4051 | - | - | 293.0 ± 69.6 | - |
| MGCC 20:0 | 546.4364 | - | - | 1504 ± 1002 | - |
| MGCC 22:0 | 574.4677 | - | - | 2100 ± 66 | - |
| MGCC 22:6 | 562.3738 | 0.18 ± 0.12 | - | 1781 ± 798 | - |
| MGCC 24:0 | 602.4990 | - | - | 1498 ± 516 | - |
| MGCC 24:2 | 598.4677 | - | - | 1534 ± 424 | - |
Table 5.
Relative levels of diacylglycerols (DG) and modified DGs in copepods and microalga. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA) for NESI and 2 nanomoles [13C3]triacylglycerol 48:0 for PESI), corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5). -, not detected.
Table 5.
Relative levels of diacylglycerols (DG) and modified DGs in copepods and microalga. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA) for NESI and 2 nanomoles [13C3]triacylglycerol 48:0 for PESI), corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5). -, not detected.
| Diacylglycerols | [M+Cl]− | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
|---|
| DG 34:0 | 631.5078 | 0.12 ± 0.022 | 28.1 ± 14.6 | 4.94 ± 1.35 | 8.26 ± 4.40 |
| DG 34:1 | 629.4922 | 1.14 ± 0.14 | 2.45 ± 1.24 | 1.99 ± 0.20 | 4.25 ± 1.29 |
| DG 34:5 | 621.4296 | 0.18 ± 0.039 | - | 3.38 ± 0.92 | 2.81 ± 0.64 |
| DG 36:0 | 659.5391 | - | 45.1 ± 5.24 | 2.37 ± 1.01 | 28.8 ± 10.1 |
| DG 38:4 | 679.5078 | 0.012 ± 0.005 | 0.022 ± 004 | 1.19 ± 0.46 | 0.78 ± 0.44 |
| DG 38:6 | 675.4765 | 0.37 ± 0.04 | 0.54 ± 0.10 | 0.52 ± 0.11 | 5.81 ± 2.12 |
| DGTS | [M+H]+ | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
| DGTS 28:0 | 656.5460 | 1.99 ± 0.64 | 0.019 ± 0.009 | 41.6 ± 22.5 | 0.16 ± 0.025 |
| DGTS 30:0 | 684.5773 | 0.67 ± 0.23 | 1.29 ± 0.67 | 44.8 ± 21.9 | 029 ± 0.18 |
| DGTS 32:1 | 710.5929 | 39.7 ± 5.8 | 0.63 ± 0.075 | 67.6 ± 23.7 | 0.069 ± 0.033 |
| DGTS 32:2 | 708.5773 | 1.97 ± 0.37 | 0.024 ± 0.004 | 27.7 ± 6.2 | - |
| DGCC | [M+H]+ | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
| DGCC 32:0 | 728.6035 | 0.81 ± 0.045 | 6.74 ± 2.12 | - | 2.12 ± 1.2 |
| DGCC 32:1 | 726.5878 | 4.23 ± 0.94 | 0.86 ± 0.51 | - | 2.66 ± 1.37 |
| DGCC 32:2 | 724.5722 | 2.18 ± 1.0 | - | - | 0.93 ± 0.13 |
| DGCC 34:0 | 756.6348 | - | 1.72 ± 0.91 | - | 1.84 ± 1.00 |
| DGCC 36:6 | 772.5722 | 5.41 ± 1.05 | - | - | - |
| DGCC 40:6 | 828.6348 | 0.18 ± 0.051 | - | - | 0.51 ± 0.11 |
| Monogalactosyl DG | [M+Cl]− | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
| MGDG 32:1 | 763.5137 | 12.31 ± 3.91 | - | - | - |
| MGDG 32:2 | 761.4981 | 4.07 ± 1.12 | - | - | - |
| MGDG 32:3 | 759.4824 | 2.04 ± 0.65 | - | - | - |
| MGDG 32:4 | 757.4668 | 3.90 ± 1.26 | - | - | - |
| MGDG 32:5 | 755.4511 | 1.22 ± 0.42 | - | - | - |
| MGDG 34:1 | 791.5450 | 4.25 ± 1.29 | - | - | - |
| MGDG 34:2 | 789.5294 | 3.96 ± 1.16 | - | - | - |
| MGDG 34:6 | 781.4668 | 1.57 ± 0.48 | - | - | - |
| MGDG 36:2 | 817.5607 | 11.13 ± 3.40 | - | - | - |
| MGDG 36:3 | 815.5450 | 1.92 ± 0.51 | - | - | - |
| MGDG 36:4 | 813.5294 | 1.39 ± 0.39 | - | - | - |
| MGDG 36:5 | 811.5137 | 2.55 ± 0.86 | - | - | - |
| MGDG 36:6 | 809.4981 | 2.64 ± 0.76 | - | - | - |
| Digalactosyl DG | [M+Cl]− | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
| DGDG 30:1 | 897.5352 | 1.11 ± 0.60 | - | - | - |
| DGDG 30:2 | 895.5196 | 0.49 ± 0.16 | - | - | - |
| DGDG 32:1 | 925.5665 | 0.51 ± 0.04 | - | - | - |
| DGDG 32:4 | 919.5196 | 0.33 ± 0.01 | - | - | - |
| DGDG 34:3 | 949.5665 | 0.22 ± 0.11 | - | - | - |
| DGDG 34:5 | 945.5396 | 2.79 ± 1.25 | - | - | - |
| DGDG 36:3 | 977.5978 | 3.96 ± 1.16 | - | - | - |
| DGDG 36:4 | 975.5822 | 0.64 ± 0.17 | - | - | - |
| DGDG 36:5 | 973.5665 | 0.42 ± 0.19 | - | - | - |
| DGDG 36:6 | 971.5509 | 0.22 ± 0.05 | - | - | - |
Table 6.
Relative levels of docosahexaenoic acid (DHA; PUFA 22:6), cholesterol, and cholesterol esters (CE) in copepods and microalga. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA for NESI and 2 nanomoles [13C3]triacylglycerol 48:0 for PESI), corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5). -, not detected.
Table 6.
Relative levels of docosahexaenoic acid (DHA; PUFA 22:6), cholesterol, and cholesterol esters (CE) in copepods and microalga. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA for NESI and 2 nanomoles [13C3]triacylglycerol 48:0 for PESI), corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5). -, not detected.
| PUFA | [M-H]− | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
|---|
| DHA | 327.2330 | 0.57 ± 0.055 | 394 ± 153 | 696 ± 149 | 2090 ± 769 |
| Hydroxy-DHA | 343.2279 | 0.36 ± 0.10 | 333 ± 67 | 1139 ± 278 | 712 ± 310 |
| Cholesterol | [MH-H2O]+ | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
| Cholesterol | 369.3516 | - | 34.58 ± 15.34 | 4.59 ± 2.12 | 12.98 ± 2.43 |
| Cholesterol Esters | [M+NH4]+ | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
| Hexosyl-Cholesterol * | 566.4415 | 591 ± 252 | - | - | - |
| CE 12:0 | 586.5558 | - | 0.66 ± 0.13 | - | 0.11 ± 0.04 |
| CE 14:0 | 614.5871 | - | 4.11 ± 1.83 | - | 1.51 ± 0.74 |
| CE 15:0 | 628.6027 | - | 4.63 ± 2.24 | 7584 ± 4521 | 2.64 ± 0.66 |
| CE 15:1 | 626.5871 | - | 1.08 ± 0.40 | - | 0.66 ± 0.18 |
| CE16:0 | 642.6184 | - | 1.76 ± 0.89 | 12.44 ± 4.75 | 0.90 ± 0.22 |
| CE 16:1 | 640.6027 | - | 4.31 ± 0.23 | - | 3.63 ± 0.69 |
| CE 17:0 | 656.6340 | - | 0.30± 0.99 | 20.70 ± 1.78 | 1.03 ± 0.26 |
| CE 18:2 | 668.6340 | - | 4.94 ± 1.26 | - | 2.90 ± 0.10 |
| CE 22:6 | 714.6184 | - | 2.28 ± 0.93 | - | 1.71 ± 0.84 |
Table 7.
Relative levels of glycerophospholipids in copepods and microalga. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA) for NESI and 2 nanomoles [13C3]triacylglycerol 48:0 for PESI), corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5). -, not detected.
Table 7.
Relative levels of glycerophospholipids in copepods and microalga. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA) for NESI and 2 nanomoles [13C3]triacylglycerol 48:0 for PESI), corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5). -, not detected.
| Phosphatidylcholine (PC) | [M+H]+ | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
|---|
| PC 28:0 (14:0/14:0) | 678.5068 | - | 3.44 ± 1.23 | - | 5.91 ± 2.34 |
| PC 28:1 | 676.4912 | - | - | - | 0.39 ± 0.21 |
| PC 28:4 | 670.4442 | - | - | 2134 ± 658 | 3.03 ± 0.75 |
| PC 30:5 | 696.4599 | - | - | 1037 ± 324 | - |
| PC 34:1 (16:0/18:1) | 760.5851 | - | 0.92 ± 0.21 | 995 ± 473 | 2.24 ± 0.90 |
| PC 34:4 | 754.5381 | - | - | 360 ± 114 | - |
| PC 34:5 | 752.5225 | - | - | 116 ± 56 | - |
| PC 36:4 | 782.5694 | - | 0.73 ± 0.11 | 806 ± 52 | 1.60 ± 0.87 |
| PC 36:5 | 780.5538 | - | 2.70 ± 0.34 | 1416 ± 745 | 1.37 ± 0.29 |
| PC 38:6 (18:1/20:5) | 806.5694 | - | - | 1984 ± 868 | - |
| Phosphatidylethanolamine (PE) | [M-H]− | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
| PE 32:0 (16:0/16:0; 18:0/14:0) | 690.5079 | - | 0.35 ± 0.05 | - | 4.27 ± 2.1 |
| PE 32:1 | 688.4923 | 0.14 ± 0.04 | 0.19 ± 0.06 | 11.70 ± 1.25 | 0.29 ± 0.12 |
| PE 34:1 | 716.5236 | 0.025 ± 0.011 | - | - | 0.17 ± 0.08 |
| PE 36:0 | 746.5705 | - | - | - | 1.94 ± 0.93 |
| PE 36:2 | 742.5392 | 0.051 ± 0.020 | 0.68 ± 0.21 | - | - |
| PE 38:3 | 768.5549 | - | - | - | 2.07 ± 0.88 |
| PE 40:4 | 794.5702 | - | - | 114 ± 51 | 2.59 ± 0.11 |
| PE 42:5 | 820.5862 | - | - | - | 1.69 ± 0.79 |
| PE 42:6 (20:0/22:6) | 818.5705 | - | - | 13.99 ± 3.81 | - |
| PE 44:6 | 846.6018 | - | - | - | 0.67 ± 0.33 |
| Phosphatidylglycerol (PG) | [M-H]− | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
| PG 30:0 (15:0/15:0) | 693.4712 | 1.23 ± 0.62 | - | - | - |
| PG 32:1 | 719.4869 | 0.63 ± 0.23 | - | - | - |
| PG 34:1 (16:0/18:1) | 747.5182 | 3.51 ± 1.35 | - | - | - |
| PG 34:2 | 745.5025 | 2.51 ± 0.99 | - | - | - |
| PG 36:2 (18:1/18:1) | 773.5338 | 7.58 ± 3.06 | - | - | - |
| PG 36:3 | 771.5182 | 1.27 ± 0.51 | - | - | - |
Table 8.
Relative levels of sphingolipids (ceramides and sphingomyelins) in copepods and microalga. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA for NESI and 2 nanomoles [13C3]triacylglycerol 48:0 for PESI), corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5). -, not detected.
Table 8.
Relative levels of sphingolipids (ceramides and sphingomyelins) in copepods and microalga. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA for NESI and 2 nanomoles [13C3]triacylglycerol 48:0 for PESI), corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5). -, not detected.
| Deoxyceramides | [M+H]+ | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
|---|
| Cer 34:0; O | 524.5401 | - | - | 12,363 ± 6942 | - |
| Cer 34:1; O | 522.5245 | - | - | 3871 ± 2188 | - |
| Cer 34:2; O | 520.5088 | - | - | 5258 ± 2970 | 0.42 ± 0.064 |
| Cer 34:3; O | 518.4932 | - | - | 10,686 ± 5960 | 1.50 ± 0.28 |
| Cer 34:4; O | 516.4775 | - | - | 4711 ± 1458 | 0.70 ± 0.17 |
| Cer 34:5; O | 514.4619 | - | - | 978 ± 562 | - |
| Cer 36:0; O | 552.5714 | - | - | 16,760 ± 9450 | - |
| Cer 36:1; O | 550.5558 | - | - | 5883 ± 3285 | - |
| Cer 36:2; O | 548.5401 | - | 0.14 ± 0.04 | 3146 ± 1805 | 0.11 ± 0.05 |
| Cer 36:3; O | 546.5245 | - | 0.11 ± 0.06 | 6435 ± 3759 | 0.25 ± 0.13 |
| Cer 36:4; O | 544.5088 | - | 0.61 ± 0.31 | 12,199 ± 6893 | 0.76 ± 0.02 |
| Cer 36:5; O | 542.4932 | - | 0.058 ± 0.023 | 9800 ± 5450 | 0.11 ± 0.02 |
| Cer 38:0; O | 580.6027 | - | - | 11,112 ± 6221 | - |
| Cer 38:1; O | 578.5871 | - | - | 8009 ± 4500 | - |
| Cer 38:2; O | 576.5714 | - | 0.091 ± 0.008 | 2835 ± 1618 | - |
| Cer 38:3; O | 574.5558 | - | - | 5059 ± 2750 | 0.079 ± 0.039 |
| Cer 38:4; O | 572.5401 | - | 0.13 ± 0.01 | 22,497 ± 12,676 | 0.16 ± 0.01 |
| Cer 38:5; O | 570.5245 | - | - | 11,220 ± 6341 | 0.018 ± 0.006 |
| Cer 40:1; O | 606.6184 | - | - | 6865 ± 3716 | - |
| Cer 40:5; O | 598.5558 | - | - | 11,687 ± 6211 | - |
| Cer 42:4; O | 628.6027 | - | 4.63 ± 2.25 | 7584 ± 4012 | 2.64 ± 0.66 |
| Cer 42:5; O | 626.5871 | - | 1.08 ± 0.22 | 10,822 ± 6156 | 0.66 ± 0.18 |
| Cer 44:4; O | 656.6340 | - | 0.21 ± 0.11 | 20.7 ± 1.7 | 1.03 ± 0.26 |
| Cer 44:5; O | 654.6184 | - | 0.094 ± 0.01 | 3289 ± 1642 | 0.18 ± 0.04 |
| Ceramides | [M+Cl]− | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
| Cer 34:0; O2 | 574.49758 | 0.69 ± 0.09 | 2.33 ± 0.10 | 8.17 ± 3.2 | 12.16 ± 6.31 |
| Cer 34:1; O2 | 572.48193 | 0.25 ± 0.09 | 43.45 ± 5.56 | 0.86 ± 0.23 | 60.3 ± 23.0 |
| Cer 34:2; O2 | 570.46628 | 8.93 ± 0.28 | 32.91 ± 15.17 | 43.8 ± 10.4 | 145 ± 43 |
| Cer 34:3; O2 | 568.45063 | - | 17.50 ± 6.56 | 132.4 ± 29.0 | 77.9 ± 19.7 |
| Cer 34:4; O2 | 566.43498 | - | 4.75 ± 1.22 | 37.5 ± 7.6 | 13.7 ± 6.63 |
| Cer 34:5; O2 | 564.41933 | - | 1.22 ± 0.34 | 9.00 ± 1.31 | 1.38 ± 0.52 |
| Cer 36:0; O2 | 602.52888 | 1.93 ± 0.37 | 0.63 ± 0.24 | 8.38 ± 2.56 | 2.67 ± 1.89 |
| Cer 36:3; O2 | 596.48193 | - | 17.25 ± 6.59 | 151 ± 30 | 86.8 ± 28.0 |
| Cer 36:4; O2 | 594.46628 | - | 7.18 ± 3.77 | 91.5 ± 20.5 | 26.03 ± 14.63 |
| Cer 36:5; O2 | 592.45063 | - | 3.44 ± 1.56 | 42.5 ± 8.4 | 11.69 ± 4.96 |
| Cer 38:3; O2 | 568.45063 | - | 8.28 ± 1.86 | 375 ± 86 | 27.03 ± 8.33 |
| Cer 38:4; O2 | 566.43498 | - | 1.80 ± 0.34 | 85.9 ± 22.5 | 6.63 ± 2.21 |
| Cer 38:5; O2 | 564.41933 | - | 2.12 ± 0.45 | 18.6 ± 3.0 | 6.72 ± 3.23 |
| Cer 40:4; O2 | 622.49758 | - | 3.98 ± 1.45 | 68.5 ± 16.4 | 6.62 ± 3.78 |
| Cer 42:3; O2 | 652.54453 | - | 4.00 ± 1.10 | 421 ± 104 | 326 ± 54 |
| Cer 42:4; O2 | 650.52888 | - | 18.81 ± 5.84 | 771 ± 210 | 66.3 ± 4.1 |
| Cer 42:5; O2 | 648.51323 | - | 0.92 ± 0.26 | 24.2 ± 6.1 | 2.26 ± 1.0 |
| Cer 44:3; O2 | 680.57583 | - | 16.76 ± 8.49 | 12.9 ± 3.7 | 130 ± 2 |
| Cer 44:4; O2 | 678.56018 | - | 6.32 ± 2.30 | 274 ± 72 | 44.30 ± 1.05 |
| Cer 44:5; O2 | 676.54453 | - | - | 9.77 ± 2.04 | 0.45 ± 0.17 |
| Cer 46:4; O2 | 706.59148 | - | - | 19.5 ± 5.7 | 0.16 ± 0.09 |
| Sphingomyelins (SM) | [M+H]+ | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
| SM 32:0; O2 | 677.5592 | - | 0.42 ± 0.17 | - | 1.55 ± 0.38 |
| SM 34:0; O2 | 705.5905 | - | 4.04 ± 2.03 | - | 0.44 ± 0.06 |
| SM 34:2; O2 | 701.5592 | - | - | - | 0.81 ± 0.37 |
| SM 36:3; O2 | 727.5749 | - | - | 44.83 ± 21.90 | 1.23 ± 0.60 |
| SM 42:3; O2 | 811.6688 | - | 5.31 ± 1.89 | 15.76 ± 5.39 | 12.35 ± 5.62 |
| SM 42:4; O2 | 809.6531 | - | 0.96 ± 0.48 | - | 2.51 ± 0.79 |
| SM d18:1/25:3 | 823.6688 | - | 2.87 ± 1.21 | - | 20.21 ± 10.39 |
| SM d18:1/25:4 | 821.6531 | - | 0.47 ± 0.14 | - | 1.45 ± 0.72 |
| SM d18:1/26:2 | 839.7001 | - | - | - | 5.72 ± 2.39 |
| SM d18:1/26:3 | 837.6844 | - | 0.40 ± 0.15 | 19.04 ± 8.32 | 1.46 ± 0.47 |
Table 9.
Relative levels of heterofibrins in C. finmarchicus and chlorophylls in I. galabana. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA for NESI and 2 nanomoles [13C3]triacylglycerol 48:0 for PESI), corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5).
Table 9.
Relative levels of heterofibrins in C. finmarchicus and chlorophylls in I. galabana. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA for NESI and 2 nanomoles [13C3]triacylglycerol 48:0 for PESI), corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5).
| Heterofibrins (HF) | [M-H]− | Calanus finmarchicus |
|---|
| HF-A2 | 345.2071 | 6.40 ± 1.33 |
| HF-A3 | 417.2283 | 4.50 ± 1.54 |
| HF-B1 | 287.2017 | 0.82 ± 0.15 |
| HF-B2 | 359.2228 | 495 ± 105 |
| HF-B3 | 431.2439 | 1.10 ± 0.58 |
| Chlorophylls | [M+H]+ | Isochrysis galbana |
| Chlorophyll a | 911.5532 | 0.40 ± 0.12 |
| Pheophytin a | 872.5765 | 12.6 ± 3.7 |
Table 10.
Relative levels of bacillariolides in copepods. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA) corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5). -, not detected.
Table 10.
Relative levels of bacillariolides in copepods. Levels are presented as a ratio of the peak intensity of each lipid to the peak intensity of the internal standard (2 nanomoles [2H5]DHA) corrected for the sample dry weight. Results are presented as Mean ± SD (n = 5). -, not detected.
| Bacillariolides | [M-H]− | Isochrysis galbana | Acartia tonsa | Calanus finmarchicus | Labidocerca aestiva |
|---|
| Bacillariolide II | 315.1966 | - | 10.7 ± 4.8 | 27.6 ± 6.2 | 10.7 ± 3.1 |
| Methoxymethyl-Bacillariolide II | 359.2228 | - | 70.4 ± 38.4 | 494.8 ± 105.1 | 120.8 ± 60.2 |