Microbial Ecosystems in Movile Cave: An Environment of Extreme Life
Abstract
1. Introduction
2. Materials and Methods
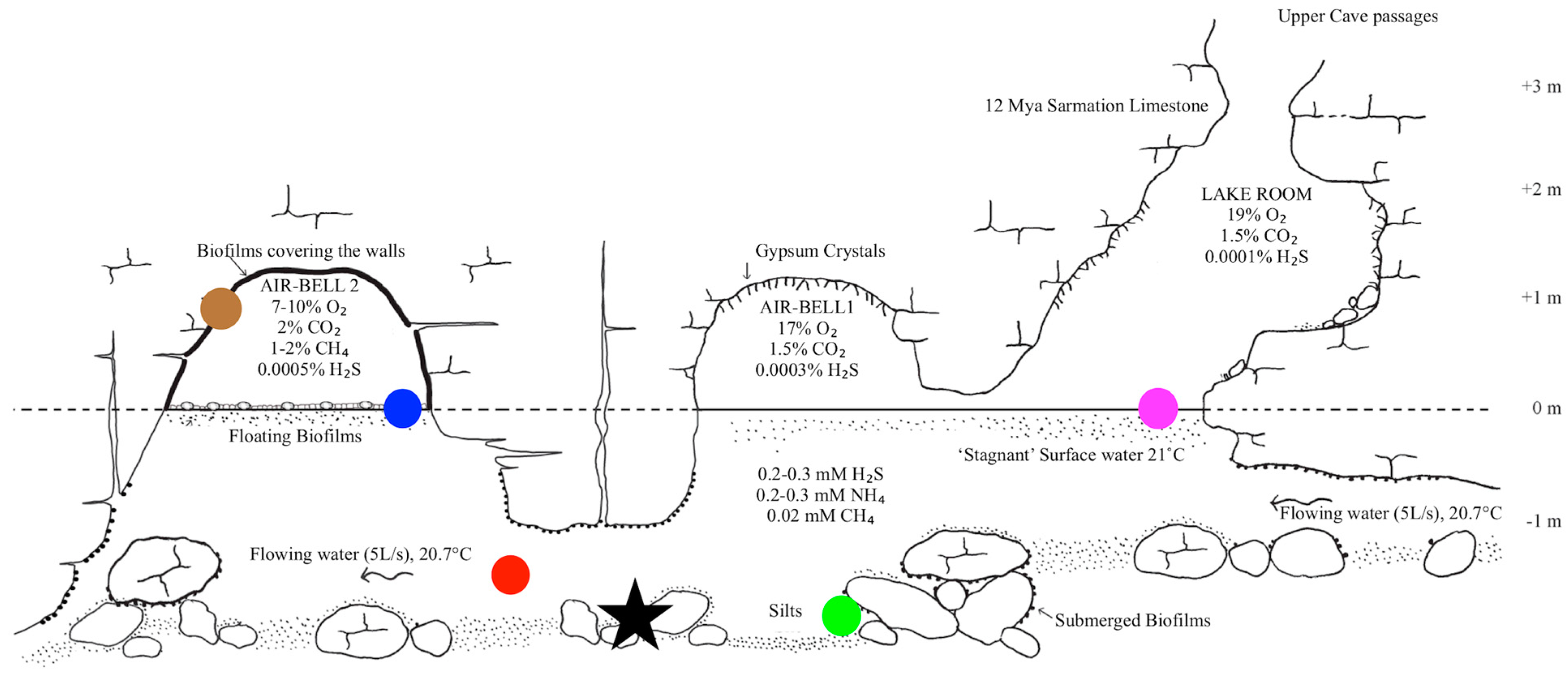
2.1. Visits to Movile Cave
2.2. Sample Collection
2.3. Site Measurements
2.4. Microcosm Experiment
2.5. DNA Extraction
2.6. Illumina 16S Amplicon Sequencing
2.7. Data Analysis and Processing
3. Results
3.1. Environmental Measurements and Observations in the Cave
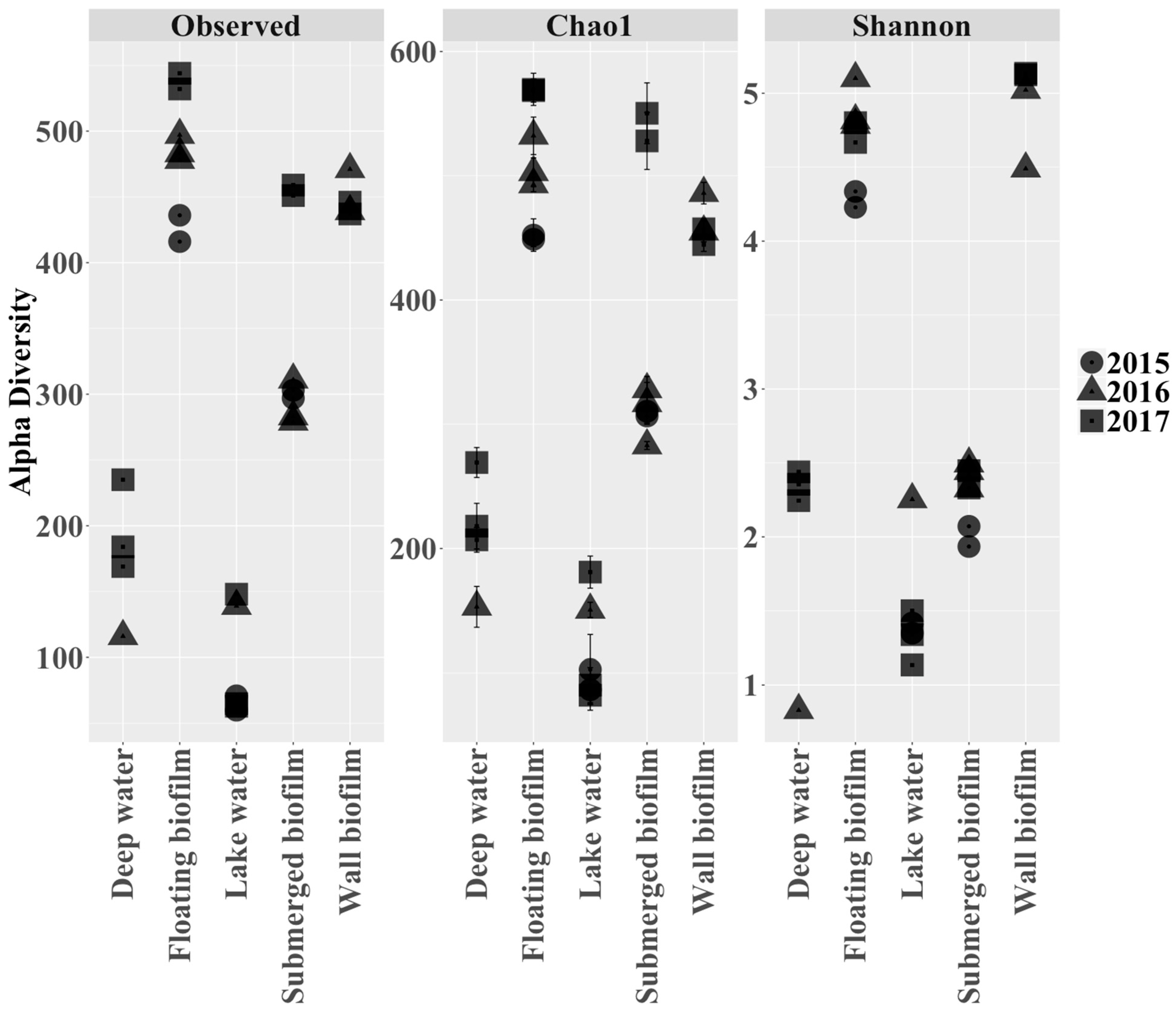
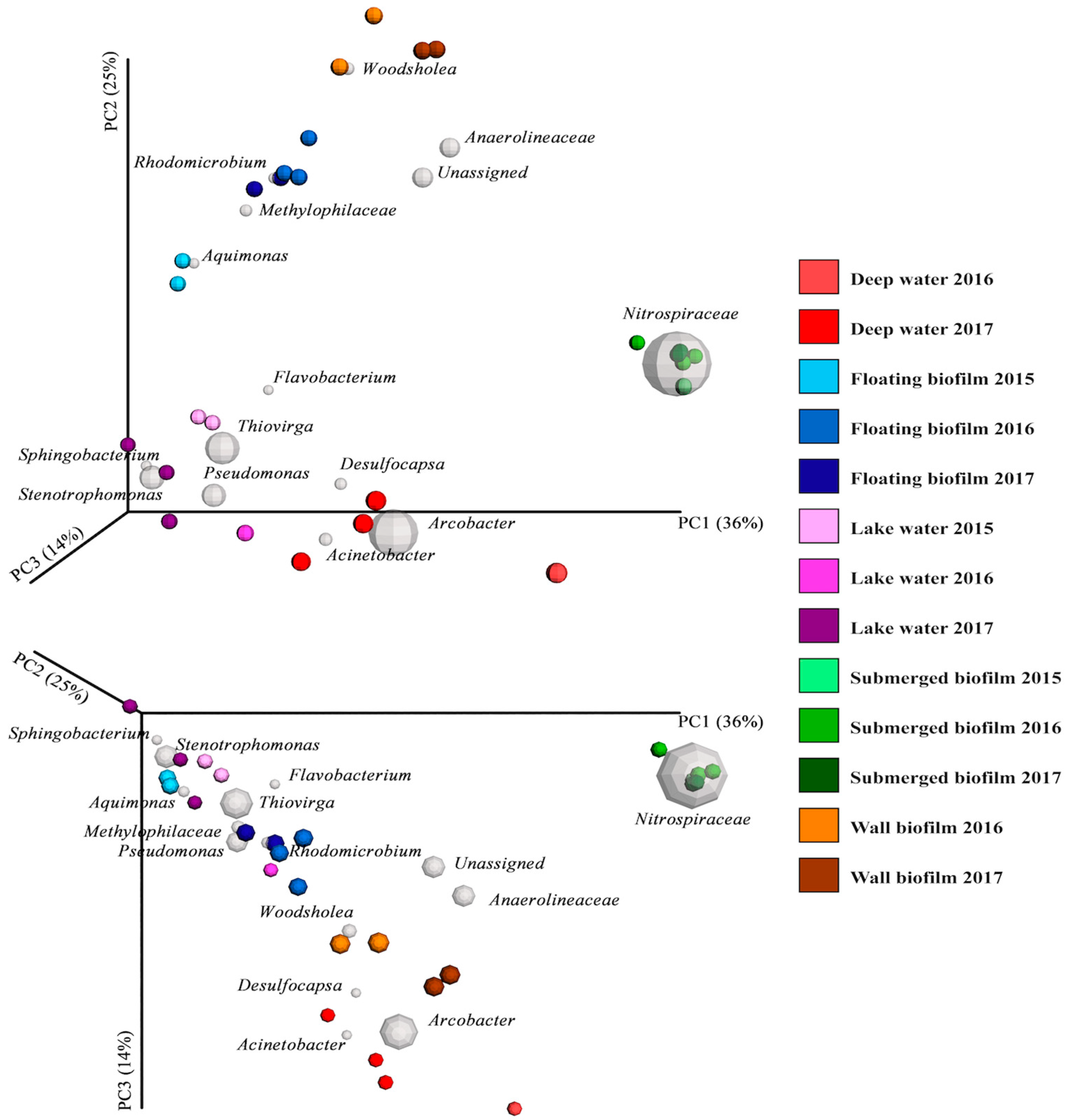
3.2. Endemic Microbial Ecology in Movile Cave: Diversity within and between the Sublocations
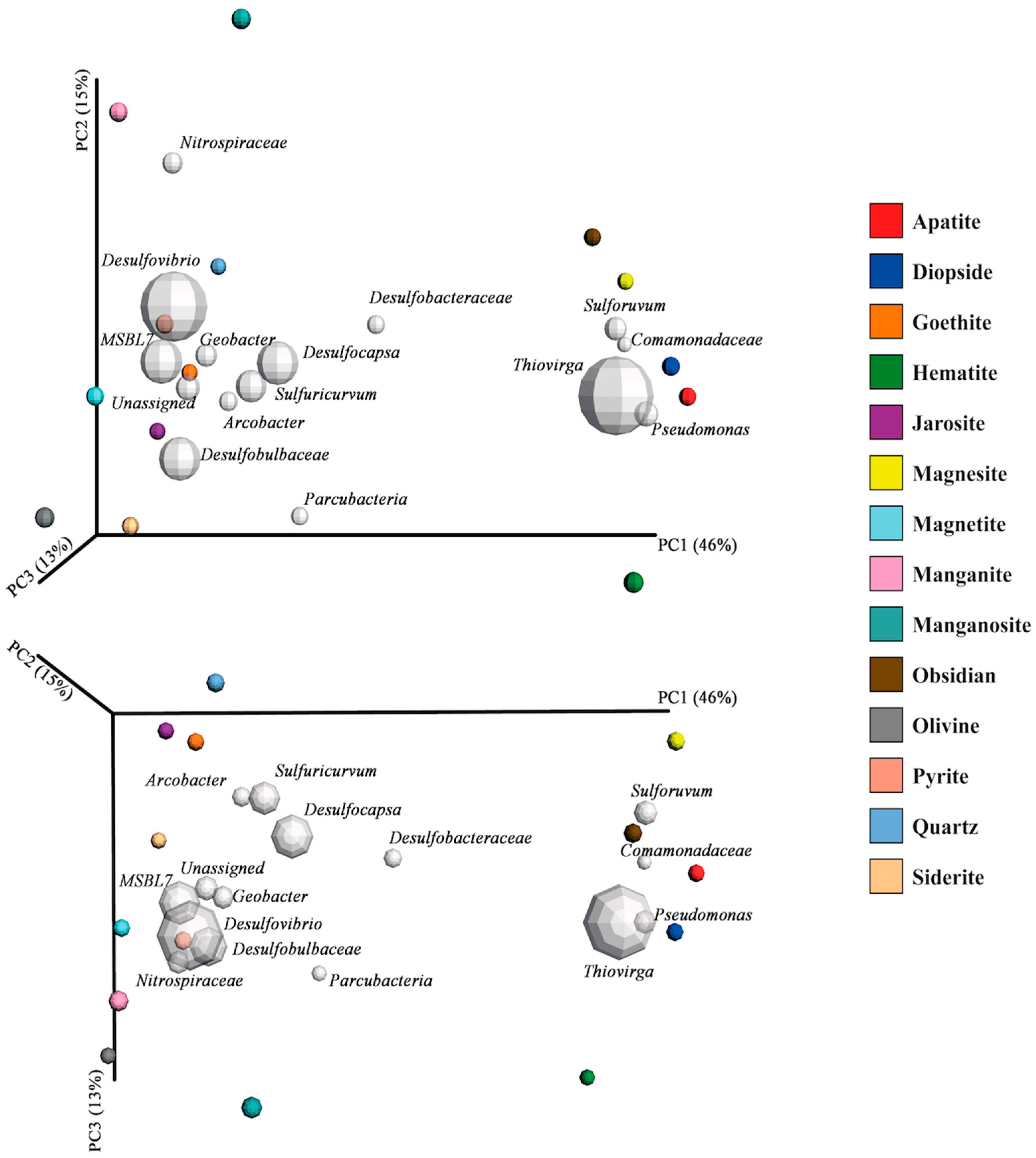
3.3. Mineral Microcosms
4. Discussion
4.1. General Findings and Limitations of Our Methodology
4.2. Potential of the Endemic Microbial Communities
| FeII + CO2 → [Leptospirillum] → FeIII + organic carbon (or S + CO2 → [Thiobacillus] → SO42− + organic carbon) | (1) |
| Organic carbon + SO42− → [Thermodesulfofibrio]→ CO2 + S2−, | |
| with as feeding reaction CH4 + O2 → [Methanotrophs (e.g., Methylococcales, Methylococcaceae)] → organic carbon + CO2 |
| NO → [Pseudomonas] →N2O → [Pseudomonas] → N2 → [Azospirillum] → NH4+ → | (2) |
| [COMAMMOX; Nitrospira, Beijerinckiaceae] → NO2− → [COMAMMOX; Nitrospira, | |
| Beijerinckiaceae] → NO3− → [ANME] → NO2− → [ANME] →NO |
| S→ [Thiobacillus; Sulfuricurvumkujinese]→ SO42−→ [Desulfobulbaceae] → SO32− → | (3) |
| [Desulfobulbaceae] →S2− (H2S) → [Arcobacter sulfidicus, Thiovirga] → S |
| H2 + S → [Desulfurellaceae] → CO2 + H2S | (4) |
4.3. Explanations of the Beta Diversity in the Endemic Microbial Communities
4.4. Comparison to Previous Findings on Microbial Communities in Movile Cave
4.5. The Effects of Minerals
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sarbu, S.M.; Kane, T.C.; Kinkle, B.K. A Chemoautotrophically Based Cave Ecosystem. Science 1996, 272, 1953–1955. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, L.J.; Mancinelli, R.L. Life in extreme environments. Nature 2001, 409, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Aerts, J.; Röling, W.; Elsaesser, A.; Ehrenfreund, P. Biota and Biomolecules in Extreme Environments on Earth: Implications for Life Detection on Mars. Life 2014, 4, 535. [Google Scholar] [CrossRef] [PubMed]
- Sarbu, S.M.; Vlasceanu, L.; Popa, R.; Sheridan, P.; Kinkle, B.K.; Kane, T.C. Microbial mats in a thermomineral sulfurous cave. In Microbial Mats; Stal, L.J., Caumette, P., Eds.; Springer: Berlin, Germany, 1994; Volume 35, pp. 45–50. [Google Scholar]
- Sarbu, S.M.; Lascu, C. Condensation corrosion in Movile Cave, Romania. J. Cave Karst Stud. 1997, 59, 99–102. [Google Scholar]
- Chen, Y.; Wu, L.; Boden, R.; Hillebrand, A.; Kumaresan, D.; Moussard, H.; Baciu, M.; Lu, Y.; Colin Murrell, J. Life without light: Microbial diversity and evidence of sulfur- and ammonium-based chemolithotrophy in Movile Cave. ISME J. 2009, 3, 1093–1104. [Google Scholar] [CrossRef]
- Hutchens, E.; Radajewski, S.; Dumont, M.G.; McDonald, I.R.; Murrell, J.C. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ. Microbiol. 2004, 6, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Riess, W.; Giere, O.; Kohls, K.; Sarbu, S. Anoxic thermomineral cave waters and bacterial mats as habitat for freshwater nematodes. Aquat. Microb. Ecol. 1999, 18, 157–164. [Google Scholar] [CrossRef]
- Röling, W.F.M.; Aerts, J.W.; Patty, C.H.L.; ten Kate, I.L.; Ehrenfreund, P.; Direito, S.O.L. The Significance of Microbe-Mineral-Biomarker Interactions in the Detection of Life on Mars and Beyond. Astrobiology 2015, 15, 492–507. [Google Scholar] [CrossRef]
- Carson, J.K.; Campbell, L.; Rooney, D.; Clipson, N.; Gleeson, D.B. Minerals in soil select distinct bacterial communities in their microhabitats. FEMS Microbiol. Ecol. 2009, 67, 381–388. [Google Scholar] [CrossRef]
- Jones, A.A.; Bennett, P.C. Mineral Microniches Control the Diversity of Subsurface Microbial Populations. Geomicrobiol. J. 2014, 31, 246–261. [Google Scholar] [CrossRef]
- Smith, A.R.; Fisk, M.R.; Thurber, A.R.; Flores, G.E.; Mason, O.U.; Popa, R.; Colwell, F.S. Deep Crustal Communities of the Juan de Fuca Ridge Are Governed by Mineralogy. Geomicrobiol. J. 2017, 34, 147–156. [Google Scholar] [CrossRef]
- Sarbu, S.M.; Popa, R. A unique chemoautotrophically based cave ecosystem. In The Natural History of Biospeleology; Camacho, A., Ed.; Luis Arguero: Madrid, Spain, 1992; pp. 637–666. [Google Scholar]
- Herlemann, D.P.R.; Labrenz, M.; Jurgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef]
- Muyzer, G.; Dewaal, E.C.; Uitterlinden, A.G. Profiling of Complex Microbial-Populations by Denaturing Gradient Gel-Electrophoresis Analysis of Polymerase Chain Reaction-Amplified Genes-Coding for 16s Ribosomal-RNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Pylro, V.S.; Roesch, L.F.W.; Morais, D.K.; Clark, I.M.; Hirsch, P.R.; Totola, M.R. Data analysis for 16S microbial profiling from different benchtop sequencing platforms. J. Microbiol. Methods 2014, 107, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2009, 26, 266–267. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Clemente, J.C.; Kuczynski, J.; Rideout, J.R.; Stombaugh, J.; Wendel, D.; Wilke, A.; Huse, S.; Hufnagle, J.; Meyer, F.; et al. The Biological Observation Matrix (BIOM) format or: How I learned to stop worrying and love the ome-ome. GigaScience 2012, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Kita, K.; Konishi, K.; Anraku, Y. Terminal oxidases of Escherichia coli aerobic respiratory chain. II. Purification and properties of cytochrome b558-d complex from cells grown with limited oxygen and evidence of branched electron-carrying systems. J. Biol. Chem. 1984, 259, 3375–3381. [Google Scholar] [CrossRef]
- Baas Becking, L. Geobiologie of Inleiding tot de Milieukunde; Willem van Stockum & Zoon: The Hague, The Netherlands, 1934. (In Dutch) [Google Scholar]
- Macalady, J.L.; Dattagupta, S.; Schaperdoth, I.; Jones, D.S.; Druschel, G.K.; Eastman, D. Niche differentiation among sulfur-oxidizing bacterial populations in cave waters. ISME J. 2008, 2, 590–601. [Google Scholar] [CrossRef]
- Engel, A.S.; Porter, M.L.; Stern, L.A.; Quinlan, S.; Bennett, P.C. Bacterial diversity and ecosystem function of filamentous microbial mats from aphotic (cave) sulfidic springs dominated by chemolithoautotrophic “Epsilonproteobacteria”. FEMS Microbiol. Ecol. 2004, 51, 31–53. [Google Scholar] [CrossRef]
- Edwards, K.J.; Bach, W.; McCollom, T.M. Geomicrobiology in oceanography: Microbe–mineral interactions at and below the seafloor. Trends Microbiol. 2005, 13, 449–456. [Google Scholar] [CrossRef]
- Hutchens, E.; Gleeson, D.; McDermott, F.; Miranda-CasoLuengo, R.; Clipson, N. Meter-Scale Diversity of Microbial Communities on a Weathered Pegmatite Granite Outcrop in the Wicklow Mountains, Ireland; Evidence for Mineral Induced Selection? Geomicrobiol. J. 2010, 27, 1–14. [Google Scholar] [CrossRef]
- Lennon, J.T.; Locey, K.J. The Underestimation of Global Microbial Diversity. mBio 2016, 7, e01298-16. [Google Scholar] [CrossRef] [PubMed]
- Porter, M.L.; Engel, A.S.; Kane, T.C.; Kinkle, B.K. Productivity-Diversity Relationships from Chemolithoautotrophically Based Sulfidic Karst Systems. Int. J. Speleol. 2009, 38, 27–40. [Google Scholar] [CrossRef]
- Rodríguez-Ramos, T.; Dornelas, M.; Marañón, E.; Cermeño, P. Conventional sampling methods severely underestimate phytoplankton species richness. J. Plankton Res. 2013, 36, 334–343. [Google Scholar] [CrossRef]
- Friedrich, C.G.; Mitrenga, G. Oxidation of thiosulfate by Paracoccus denitrificans and other hydrogen bacteria. FEMS Microbiol. Lett. 1981, 10, 209–212. [Google Scholar] [CrossRef][Green Version]
- Mahmood, Q.; Zheng, P.; Hu, B.; Jilani, G.; Azim, M.R.; Wu, D.; Liu, D. Isolation and characterization of Pseudomonas stutzeri QZ1 from an anoxic sulfide-oxidizing bioreactor. Anaerobe 2009, 15, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Lü, C.; Hou, N.; Xin, Y.; Liu, J.; Liu, H.; Xun, L. Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J. 2017, 11, 2754–2766. [Google Scholar] [CrossRef] [PubMed]
- Sievert, S.M.; Wieringa, E.B.A.; Wirsen, C.O.; Taylor, C.D. Growth and mechanism of filamentous-sulfur formation by Candidatus Arcobacter sulfidicus in opposing oxygen-sulfide gradients. Environ. Microbiol. 2007, 9, 271–276. [Google Scholar] [CrossRef]
- Wirsen, C.O.; Sievert, S.M.; Cavanaugh, C.M.; Molyneaux, S.J.; Ahmad, A.; Taylor, L.T.; DeLong, E.F.; Taylor, C.D. Characterization of an Autotrophic Sulfide-Oxidizing Marine Arcobacter sp. That Produces Filamentous Sulfur. Appl. Environ. Microbiol. 2022, 68, 316–325. [Google Scholar] [CrossRef]
- Tamas, I.; Smirnova, A.V.; He, Z.; Dunfield, P.F. The (d)evolution of methanotrophy in the Beijerinckiaceae—A comparative genomics analysis. ISME J. 2014, 8, 369–382. [Google Scholar] [CrossRef]
- Pohlman, J.W.; Iliffe, T.M.; Cifuentes, L.A. A stable isotope study of organic cycling and the ecology of an anchialine cave ecosystem. Mar. Ecol. Prog. Ser. 1997, 155, 17–27. [Google Scholar] [CrossRef]
- Costa, E.; Pérez, J.; Kreft, J.U. Why is metabolic labour divided in nitrification? Trends Microbiol. 2006, 14, 213–219. [Google Scholar] [CrossRef] [PubMed]
- van Kessel, M.A.H.J.; Speth, D.R.; Albertsen, M.; Nielsen, P.H.; Op den Camp, H.J.M.; Kartal, B.; Jetten, M.S.M.; Lücker, S. Complete nitrification by a single microorganism. Nature 2015, 528, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Schrenk, M.O.; Edwards, K.J.; Goodman, R.M.; Hamers, R.J.; Banfield, J.F. Distribution of Thiobacillus ferrooxidans and Leptospirillum ferrooxidans: Implications for Generation of Acid Mine Drainage. Science 1998, 279, 1519–1522. [Google Scholar] [CrossRef] [PubMed]
- Frank, Y.A.; Kadnikov, V.V.; Lukina, A.P.; Banks, D.; Beletsky, A.V.; Mardanov, A.V.; Sen’kina, E.I.; Avakyan, M.R.; Karnachuk, O.V.; Ravin, N.V. Characterization and Genome Analysis of the First Facultatively Alkaliphilic Thermodesulfovibrio Isolated from the Deep Terrestrial Subsurface. Front. Microbiol. 2016, 7, 2000. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Liu, J.; Fang, Y.; Hedlund, B.P.; Lian, Z.-H.; Huang, L.-Y.; Li, J.-T.; Li, W.-J.; Jiang, H.-C.; Dong, H.-L.; et al. Insights into ecological role of a new deltaproteobacterial order Candidatus Acidulodesulfobacterales by metagenomics and metatranscriptomics. ISME J. 2019, 13, 2044–2057. [Google Scholar] [CrossRef] [PubMed]
- Holler, T.; Widdel, F.; Knittel, K.; Amann, R.; Kellermann, M.Y.; Hinrichs, K.-U.; Teske, A.; Boetius, A.; Wegener, G. Thermophilic anaerobic oxidation of methane by marine microbial consortia. ISME J. 2011, 5, 1946–1956. [Google Scholar] [CrossRef]
- Knittel, K.; Boetius, A. Anaerobic Oxidation of Methane: Progress with an Unknown Process. Annu. Rev. Microbiol. 2009, 63, 311–334. [Google Scholar] [CrossRef]
- Kodama, Y.; Watanabe, K. Sulfuricurvum kujiense gen. nov., sp. nov., a facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium isolated from an underground crude-oil storage cavity. Int. J. Syst. Evol. Microbiol. 2004, 54, 2297–2300. [Google Scholar] [CrossRef]
- Engel, A.S.; Meisinger, D.B.; Porter, M.L.; Payn, R.A.; Schmid, M.; Stern, L.A.; Schleifer, K.H.; Lee, N.M. Linking phylogenetic and functional diversity to nutrient spiraling in microbial mats from Lower Kane Cave (USA). ISME J. 2010, 4, 98–110. [Google Scholar] [CrossRef]
- Popkova, A.; Mazina, S. Microbiota of Otap Head Cave. Environ. Res. Eng. Manag. 2019, 75, 71–82. [Google Scholar] [CrossRef][Green Version]
- Marschall, E.; Jogler, M.; Henßge, U.; Overmann, J. Large-scale distribution and activity patterns of an extremely low-light-adapted population of green sulfur bacteria in the Black Sea. Environ. Microbiol. 2010, 12, 1348–1362. [Google Scholar] [CrossRef] [PubMed]
- Engel, A.S. Observations on the biodiversity of sulfidic Karst habitats. J. Cave Karst Stud. 2007, 69, 187–206. [Google Scholar]
- Berg, J.S.; Pjevac, P.; Sommer, T.; Buckner, C.R.; Philippi, M.; Hach, P.F.; Liebeke, M.; Holtappels, M.; Danza, F.; Tonolla, M.; et al. Dark aerobic sulfide oxidation by anoxygenic phototrophs in anoxic waters. Environ. Microbiol. 2019, 21, 1611–1626. [Google Scholar] [CrossRef] [PubMed]
- Rohwerder, T.; Sand, W.; Lascu, C. Preliminary Evidence for a Sulphur Cycle in Movile Cave, Romania. Acta Biotechnol. 2003, 23, 101–107. [Google Scholar] [CrossRef]
- Sarbu, S.M. Movile Cave: A chemoautotrophically based ground water system. In Ecosystems of the World: Subterranean Ecosystems; Wilkens, H., Culver, D.C., Humphreys, W.F., Eds.; Elsevier: Amsterdam, The Netherlands, 2000; pp. 319–343. [Google Scholar]
- Wischer, D.; Kumaresan, D.; Johnston, A.; El Khawand, M.; Stephenson, J.; Hillebrand-Voiculescu, A.M.; Chen, Y.; Murrell, J.C. Bacterial metabolism of methylated amines and identification of novel methylotrophs in Movile Cave. ISME J. 2015, 9, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Steenhoudt, O.; Vanderleyden, J. Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: Genetic, biochemical and ecological aspects. FEMS Microbiol. Rev. 2000, 24, 487–506. [Google Scholar] [CrossRef]
- Mello, R.; Hill, S.; Poole, R.K. Determination of the oxygen affinities of terminal oxidases in Azotobacter vinelandii using the deoxygenation of oxyleghaemoglobin and oxymyoglobin: Cytochrome bd is a low-affinity oxidase. Microbiology 1994, 140, 1395–1402. [Google Scholar] [CrossRef][Green Version]
- Krab, K.; Kempe, H.; Wikström, M. Explaining the enigmatic KM for oxygen in cytochrome c oxidase: A kinetic model. Biochim. Biophys. Acta (BBA)-Bioenerg. 2011, 1807, 348–358. [Google Scholar] [CrossRef]
- Kumaresan, D.; Stephenson, J.; Doxey, A.C.; Bandukwala, H.; Brooks, E.; Hillebrand-Voiculescu, A.; Whiteley, A.S.; Murrell, J.C. Aerobic proteobacterial methylotrophs in Movile Cave: Genomic and metagenomic analyses. Microbiome 2018, 6, 1. [Google Scholar] [CrossRef]
- Bizic, M.; Brad, T.; Barbu-Tudoran, L.; Aerts, J.; Ionescu, D.; Popa, R.; Ody, J.; Flot, J.-F.; Tighe, S.; Vellone, D.; et al. Genomic and morphologic characterization of a planktonic Thiovulum (Campylobacterota) dominating the surface waters of the sulfidic Movile Cave, Romania. bioRxiv 2020. [Google Scholar] [CrossRef]
- Fletcher, M. Bacterial attachment in aquatic environments: A diversity of surfaces and adhesion strategies. In Bacterial Adhesion: Molecular and Ecological Diversity; Fletcher, M., Ed.; John Wiley & Sons, Inc.: New York, NY, USA, 1996; pp. 1–24. [Google Scholar]
- Gounot, A.-M. Microbial oxidation and reduction of manganese: Consequences in groundwater and applications. FEMS Microbiol. Rev. 1994, 14, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Osorio, H.; Mangold, S.; Denis, Y.; Ñancucheo, I.; Esparza, M.; Johnson, D.B.; Bonnefoy, V.; Dopson, M.; Holmes, D.S. Anaerobic Sulfur Metabolism Coupled to Dissimilatory Iron Reduction in the Extremophile Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 2013, 79, 2172. [Google Scholar] [CrossRef]
- Thiel, J.; Byrne, J.M.; Kappler, A.; Schink, B.; Pester, M. Pyrite formation from FeS and H2S is mediated through microbial redox activity. Proc. Natl. Acad. Sci. USA 2019, 116, 6897–6902. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Vannela, R.; Hayes, K.F.; Rittmann, B.E. Effect of growth conditions on microbial activity and iron-sulfide production by Desulfovibrio vulgaris. J. Hazard. Mater. 2014, 272, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Weber, K.A.; Urrutia, M.M.; Churchill, P.F.; Kukkadapu, R.K.; Roden, E.E. Anaerobic redox cycling of iron by freshwater sediment microorganisms. Environ. Microbiol. 2006, 8, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, A.; Weinberg, R.F.; Wright, H.M.N. Magma fracturing and degassing associated with obsidian formation: The explosive–effusive transition. J. Volcanol. Geotherm. Res. 2015, 298, 71–84. [Google Scholar] [CrossRef]
- Ericson, J.E.; Makishima, A.; Mackenzie, J.D.; Berger, R. Chemical and physical properties of obsidian: A naturally occurring glass. J. Non-Cryst. Solids 1975, 17, 129–142. [Google Scholar] [CrossRef]
- McSween, H.Y.; Arvidson, R.E.; Bell, J.F.; Blaney, D.; Cabrol, N.A.; Christensen, P.R.; Clark, B.C.; Crisp, J.A.; Crumpler, L.S.; Marais, D.J.D.; et al. Basaltic Rocks Analyzed by the Spirit Rover in Gusev Crater. Science 2004, 305, 842–845. [Google Scholar] [CrossRef]
- Rubie, D.C.; Gessmann, C.K.; Frost, D.J. Partitioning of oxygen during core formation on the Earth and Mars. Nature 2004, 429, 58–61. [Google Scholar] [CrossRef]
- Klingelhöfer, G.; Morris, R.V.; De Souza, P.A.; Rodionov, D.; Schröder, C. Two earth years of Mössbauer studies of the surface of Mars with MIMOS II. Hyperfine Interact. 2006, 170, 169–177. [Google Scholar] [CrossRef]
- Klingelhöfer, G.; Morris, R.V.; Bernhardt, B.; Schröder, C.; Rodionov, D.S.; de Souza, P.A.; Yen, A.; Gellert, R.; Evlanov, E.N.; Zubkov, B.; et al. Jarosite and Hematite at Meridiani Planum from Opportunity’s Mössbauer Spectrometer. Science 2004, 306, 1740–1745. [Google Scholar] [CrossRef]
- Zolotov, M.Y.; Shock, E.L. Formation of jarosite-bearing deposits through aqueous oxidation of pyrite at Meridiani Planum, Mars. Geophys. Res. Lett. 2005, 32. [Google Scholar] [CrossRef]
- Vaniman, D.T.; Bish, D.L.; Ming, D.W.; Bristow, T.F.; Morris, R.V.; Blake, D.F.; Chipera, S.J.; Morrison, S.M.; Treiman, A.H.; Rampe, E.B.; et al. Mineralogy of a Mudstone at Yellowknife Bay, Gale Crater, Mars. Science 2014, 343, 1243480. [Google Scholar] [CrossRef] [PubMed]
- Ehlmann, B.L.; Mustard, J.F.; Murchie, S.L.; Poulet, F.; Bishop, J.L.; Brown, A.J.; Calvin, W.M.; Clark, R.N.; Marais, D.J.D.; Milliken, R.E.; et al. Orbital Identification of Carbonate-Bearing Rocks on Mars. Science 2008, 322, 1828–1832. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.V.; Ruff, S.W.; Gellert, R.; Ming, D.W.; Arvidson, R.E.; Clark, B.C.; Golden, D.C.; Siebach, K.; Klingelhöfer, G.; Schröder, C.; et al. Identification of Carbonate-Rich Outcrops on Mars by the Spirit Rover. Science 2010, 329, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Ody, A.; Poulet, F.; Bibring, J.-P.; Loizeau, D.; Carter, J.; Gondet, B.; Langevin, Y. Global investigation of olivine on Mars: Insights into crust and mantle compositions. J. Geophys. Res. Planets 2013, 118, 234–262. [Google Scholar] [CrossRef]
- Cannon, K.M.; Mustard, J.F. Preserved glass-rich impactites on Mars. Geology 2015, 43, 635–638. [Google Scholar] [CrossRef]
- Herrera, A.; Cockell, C.S.; Self, S.; Blaxter, M.; Reitner, J.; Arp, G.; Dröse, W.; Thorsteinsson, T.; Tindle, A.G. Bacterial Colonization and Weathering of Terrestrial Obsidian in Iceland. Geomicrobiol. J. 2008, 25, 25–37. [Google Scholar] [CrossRef]

| Location | Total | Unique | % |
|---|---|---|---|
| Lake water | 359 | 2 | 0.5 |
| Deep water | 424 | 0 | 0 |
| Floating biofilm | 734 | 317 | 43 |
| Submerged biofilm | 752 | 122 | 16 |
| Wall biofilm | 547 | 120 | 21 |
| Mineral | Chemical Formula |
|---|---|
| Apatite | Ca5(PO4)3(OH, F, Cl) |
| Diopside | MgCaSi2O6 |
| Goethite | FeIIIO(OH) |
| Hematite | Fe(III)2O3 |
| Jarosite | Kfe(III)3(OH)6(SO4)2 |
| Magnesite | MgCO3 |
| Magnetite | FeIIFeIII2O4 (Fe3O4) |
| Manganite | Mn(III)O(OH) |
| Manganosite | Mn(II)O |
| Obsidian | SiO2 (65–85%) * |
| Olivine | (Mg(II), Fe(II))2 SiO4 |
| Pyrite | Fe(II)S2 |
| Quartz beads | SiO2 |
| Siderite | Fe(II)CO3 |
| Component | p | R2 |
|---|---|---|
| Mineral type | 0.001 | 0.79 * |
| Fe (Fe2+, Fe3+) | 0.001 | 0.16 * |
| Ca | 0.001 | 0.16 |
| Fe2+ | 0.001 | 0.14 |
| Redox-active | 0.002 | 0.13 * |
| Mn (Mn2+, Mn3+) | 0.004 | 0.10 |
| Si | 0.005 | 0.10 |
| F | 0.003 | 0.08 |
| Mn2+ | 0.015 | 0.07 |
| P | 0.005 | 0.07 |
| Cl | 0.005 | 0.07 |
| Fe3+ | 0.031 | 0.064 * |
| Mg | 0.03 | 0.056 * |
| Mn3+ | 0.03 | 0.06 |
| S | 0.10 | 0.04 |
| CO32− | 0.5 | 0.02 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aerts, J.W.; Sarbu, S.M.; Brad, T.; Ehrenfreund, P.; Westerhoff, H.V. Microbial Ecosystems in Movile Cave: An Environment of Extreme Life. Life 2023, 13, 2120. https://doi.org/10.3390/life13112120
Aerts JW, Sarbu SM, Brad T, Ehrenfreund P, Westerhoff HV. Microbial Ecosystems in Movile Cave: An Environment of Extreme Life. Life. 2023; 13(11):2120. https://doi.org/10.3390/life13112120
Chicago/Turabian StyleAerts, Joost W., Serban M. Sarbu, Traian Brad, Pascale Ehrenfreund, and Hans V. Westerhoff. 2023. "Microbial Ecosystems in Movile Cave: An Environment of Extreme Life" Life 13, no. 11: 2120. https://doi.org/10.3390/life13112120
APA StyleAerts, J. W., Sarbu, S. M., Brad, T., Ehrenfreund, P., & Westerhoff, H. V. (2023). Microbial Ecosystems in Movile Cave: An Environment of Extreme Life. Life, 13(11), 2120. https://doi.org/10.3390/life13112120







