Incidental Detection of a Chromosomal Aberration by Array-CGH in an Early Prenatal Diagnosis for Monogenic Disease on Coelomic Fluid
Abstract
1. Introduction
2. Materials and Methods
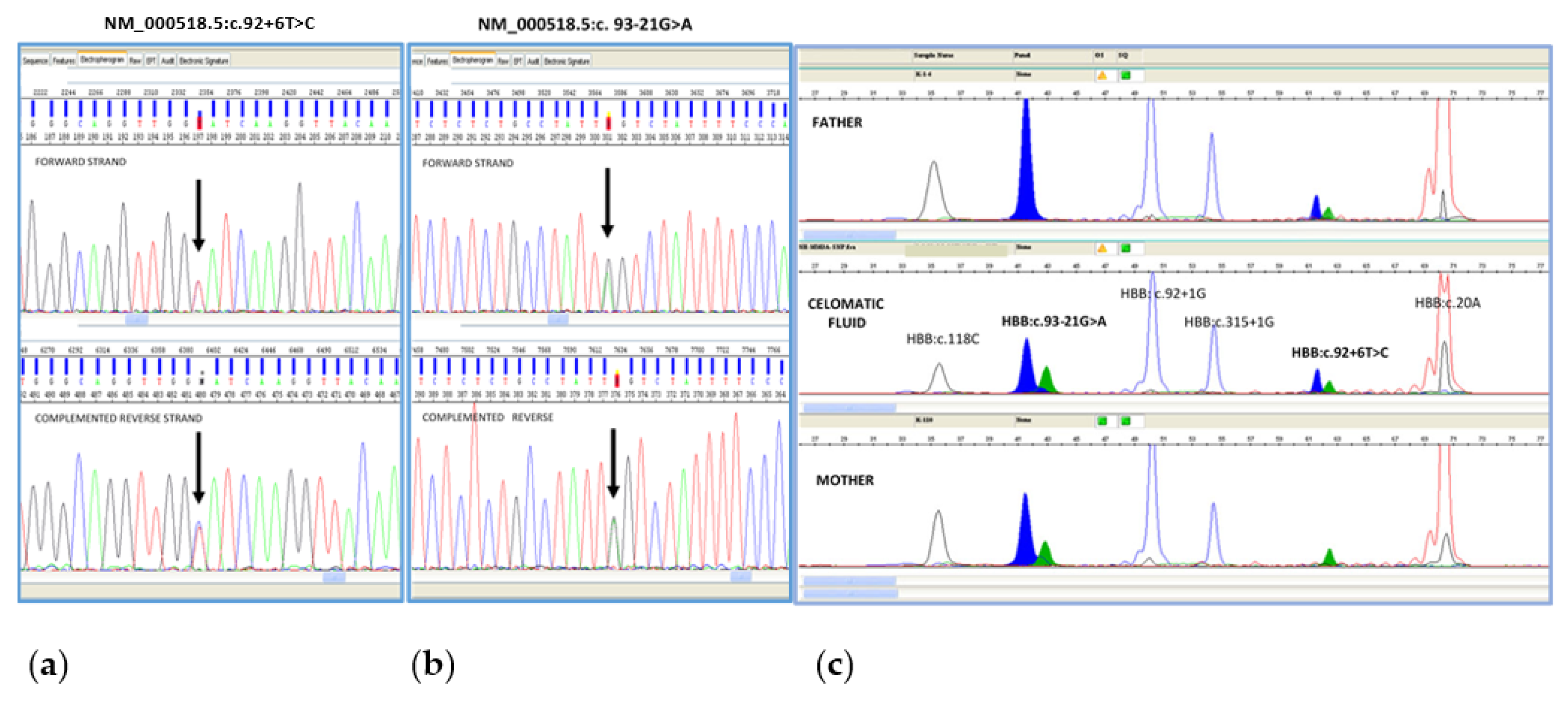
2.1. Molecular Analysis for Prenatal Diagnosis of Beta Thalassemia on CF
2.2. Molecular Analysis for Prenatal Diagnosis of Chromosomal Aberrations on CF
2.2.1. Whole Genome Amplification
2.2.2. Quantitative Fluorescent PCR (QF-PCR) of WGA Products
2.2.3. Array Comparative Genomic Hybridization
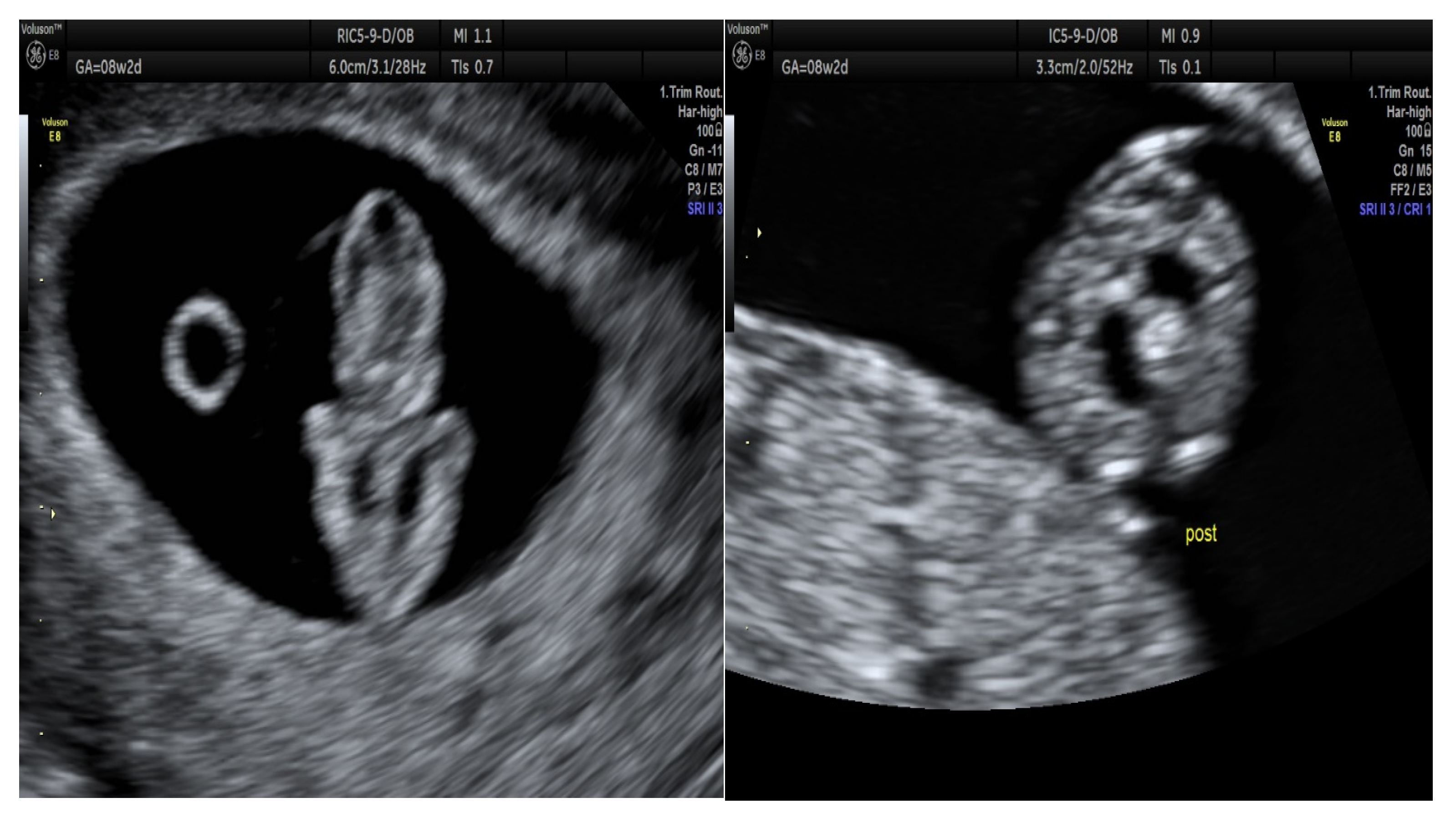
3. Results
3.1. Molecular Analysis for Prenatal Diagnosis of Beta Thalassemia on CF
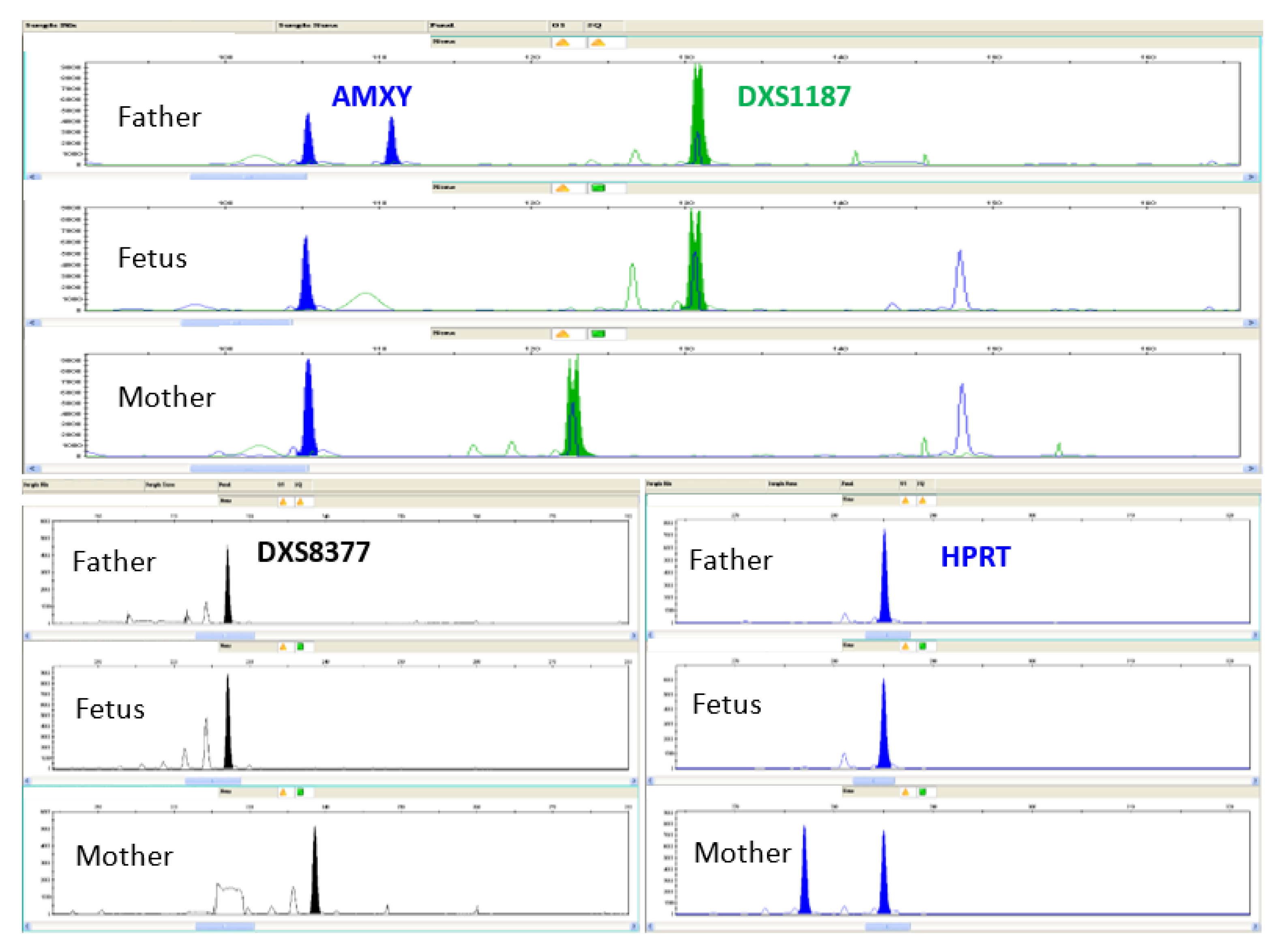
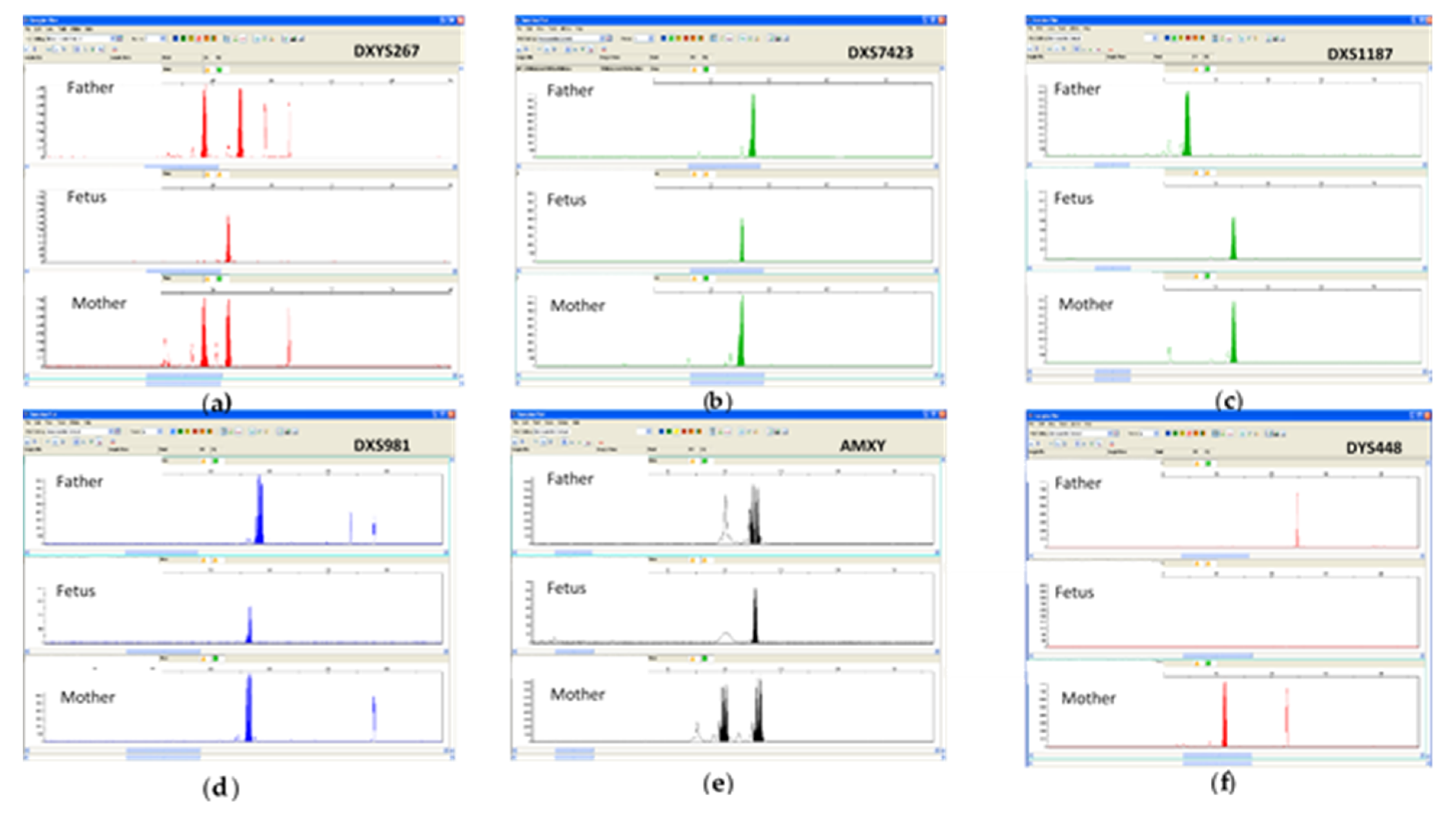
3.2. Molecular Analysis for Prenatal Diagnosis of Chromosomal Aberrations on CF
3.2.1. Whole Genome Amplification
3.2.2. Quantitative Fluorescent PCR (QF-PCR) of WGA Products
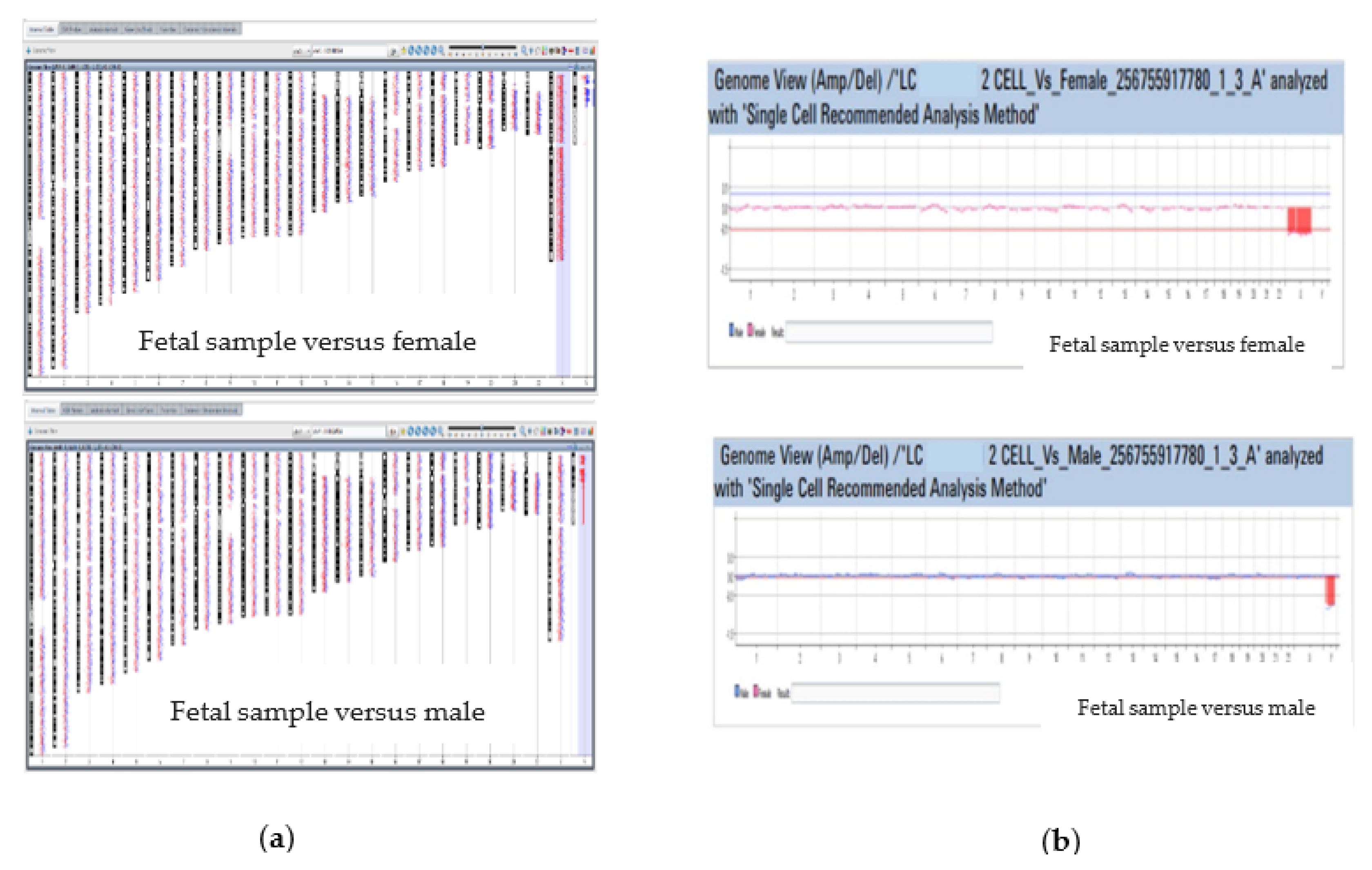
3.2.3. Array Comparative Genomic Hybridization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morgan, T. Turner syndrome: Diagnosis and management. Am. Fam. Phys. 2007, 76, 405–410. [Google Scholar]
- Slagel, D.D.; Bromley, C.M.; Benda, J.A. Detection of chromosomal abnormalities in the dysmorphic fetus using fluorescence in situ hybridization: Evaluation for monosomy X genotype. Hum. Pathol. 1995, 26, 1241–1244. [Google Scholar] [CrossRef] [PubMed]
- Makrydimas, G.; Damiani, G.; Jakil, C.; Cigna, V.; Orlandi, M.; Picciotto, F.; Schillaci, G.; Cassarà, F.; Vinciguerra, M.; Leto, F.; et al. Celocentesis for early prenatal diagnosis of hemoglobinopathy. Ultrasound Obstet. Gynecol. 2020, 56, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Giambona, A.; Makrydimas, G.; Leto, F.; Damiani, G.; Jakil, M.C.; Picciotto, F.; Renda, D.; Fiorino, R.; Renda, M.C.; Schillaci, G.; et al. Feasibility of DNA diagnosis of haemoglobinopathies on coelocentesis. Br. J. Haematol. 2011, 153, 268–272. [Google Scholar] [CrossRef]
- Giambona, A.; Leto, F.; Passarello, C.; Vinciguerra, M.; Cigna, V.; Schillaci, G.; Picciotto, F.; Lauricella, S.; Nicolaides, K.H.; Makrydimas, G.; et al. Fetal aneuploidy diagnosed at celocentesis for early prenatal diagnosis of congenital hemoglobinopathies. Acta Obstet. Gynecol. Scand. 2018, 97, 312–321. [Google Scholar] [CrossRef]
- Jurkovic, D.; Jauniaux, E.; Campbell, S.; Pandya, P.; Cardy, D.L.; Nicolaides, K.H. Coelocentesis: A new technique for early prenatal diagnosis. Lancet 1993, 341, 1623–1624. [Google Scholar] [CrossRef]
- Findlay, I.; Atkinson, G.; Chambers, M.; Quirke, P.; Campbell, J.; Rutherford, A. Rapid genetic diagnosis at 7-9weeks gestation: Diagnosis of sex, single gene defects and DNA fingerprint from coelomic samples. Hum. Reprod. 1996, 11, 2548–2553. [Google Scholar] [CrossRef]
- Jurkovic, D.; Jauniaux, E.; Campbell, S.; Mitchell, M.; Lees, C.; Layton, M. Detection of sickle gene by coelocentesis in early pregnancy: A new approach to prenatal diagnosis of single gene disorders. Hum. Reprod. 1995, 10, 1287–1289. [Google Scholar] [CrossRef]
- Makrydimas, G.; Georgiou, I.; Kranas, V.; Zikopoulos, K.; Lolis, D. Prenatal diagnosis of beta-thalassaemia by coelocentesis. Mol. Hum. Reprod. 1997, 3, 729–731. [Google Scholar] [CrossRef]
- Makrydimas, G.; Georgiou, I.; Kranas, V.; Kaponis, A.; Lolis, D. Prenatal paternity testing using DNA extracted from coelomic cells. Fetal Diagn. Ther. 2004, 19, 75–77. [Google Scholar] [CrossRef]
- Makrydimas, G.; Georgiou, I.; Bouba, I.; Lolis, D.; Nicolaides, K.H. Early prenatal diagnosis by celocentesis. Ultrasound Obstet. Gynecol. 2004, 23, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Giambona, A.; Vinciguerra, M.; Leto, F.; Cassarà, F.; Tartaglia, V.; Cigna, V.; Orlandi, E.; Picciotto, F.; Al Qahtani, N.H.; Alsulmi, E.S.; et al. Celomic fluid: Laboratory workflow for prenatal diagnosis of monogenic diseases. Mol. Diagn. Ther. 2022, 26, 239–252. [Google Scholar] [CrossRef]
- Tchirikov, M.; Arnold, C.; Oshovskyy, V.; Heinrich, U.R.; Thäle, V. Three years’ experience of using a 29-gauge atraumatic needle for amniocentesis. J. Perinat. Med. 2012, 40, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Tchirikov, M.; Scheler, C.; Gericke, M.; Wienke, A.; Entezami, M. Genetic amniocentesis using atraumatic 29 gauge needle in patients having a chorioamniotic separation. J. Perinat. Med. 2022. Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Renda, M.C.; Giambona, A.; Fecarotta, E.; Leto, F.; Makrydimas, G.; Renda, D.; Damiani, G.; Jakil, M.C.; Picciotto, F.; Piazza, A.; et al. Embryo-fetal erythroid megaloblasts in the human coelomic cavity. J. Cell Physiol. 2010, 225, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Giambona, A.; Damiani, G.; Leto, F.; Jakil, C.; Renda, D.; Cigna, V.; Schillaci, G.; Picciotto, F.; Nicolaides, K.H.; Passarello, C.; et al. Embryo-fetal erythroid cell selection from celomic fluid allows earlier prenatal diagnosis of hemoglobinopathies. Prenat. Diagn. 2016, 36, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Giambona, A.; Leto, F.; Damiani, G.; Jakil, C.; Cigna, V.; Schillaci, G.; Stampone, G.; Volpes, A.; Allegra, A.; Nicolaides, K.H.; et al. Identification of embryo-fetal cells in celomic fluid using morphological and short-tandem repeats analysis. Prenat. Diagn. 2016, 36, 973–978. [Google Scholar] [CrossRef]
- Pugh, T.J.; Delaney, A.D.; Farnoud, N.; Flibotte, S.; Griffith, M.; Li, H.I.; Qian, H.; Farinha, P.; Gascoyne, R.D.; Marra, M.A. Impact of whole genome amplification on analysis of copy number variants. Nucleic Acids Res. 2008, 36, e80. [Google Scholar] [CrossRef]
- Jasmine, F.; Ahsan, H.; Andrulis, I.L.; John, E.M.; Chang-Claude, J.; Kibriya, M.G. Whole Genome Amplification enables accurate genotyping for microarray-based high density SNP array. Cancer Epidemiol. Biomark. Prev. 2008, 17, 3499–3508. [Google Scholar] [CrossRef]
- Lovrečić, L.; Pereza, N.; Jaklič, H.; Ostojić, S.; Peterlin, B. Combination of QF-PCR and aCGH is an efficient diagnostic strategy for the detection of chromosome aberrations in recurrent miscarriage. Mol. Genet. Genomic Med. 2019, 7, e980. [Google Scholar] [CrossRef]
- Kashork, C.D.; Theisen, A.; Shaffer, L.G. Prenatal diagnosis using array CGH. Methods Mol. Biol. 2008, 444, 59–69. [Google Scholar] [PubMed]
- Lichtenbelt, K.D.; Knoers, N.V.; Schuring-Blom, G.H. From karyotyping to array-CGH in prenatal diagnosis. Cytogenet. Genome Res. 2011, 135, 241–250. [Google Scholar] [CrossRef] [PubMed]
| Scanner C Scan Settings | |
|---|---|
| Dye channel | R + G (red + green) |
| Scan region | Agilent HD (61 × 21.6 mm) |
| Scan resolution | 3µm |
| Tiff file dynamic range | 16 bit |
| Red PMT gain | 100% |
| Green PMT gain | 100% |
| XDR | <noXDR> |
| Aberration Filter Settings for Single Cell Recommended Analysis Method |
|---|
| Minimum Size: 5 Mb |
| Minimum log2 ratio: 0.35 for amplification/gain −0.45 for deletion/loss |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vinciguerra, M.; Leto, F.; Cassarà, F.; Tartaglia, V.; Malacarne, M.; Coviello, D.; Cigna, V.; Orlandi, E.; Picciotto, F.; Cucinella, G.; et al. Incidental Detection of a Chromosomal Aberration by Array-CGH in an Early Prenatal Diagnosis for Monogenic Disease on Coelomic Fluid. Life 2023, 13, 20. https://doi.org/10.3390/life13010020
Vinciguerra M, Leto F, Cassarà F, Tartaglia V, Malacarne M, Coviello D, Cigna V, Orlandi E, Picciotto F, Cucinella G, et al. Incidental Detection of a Chromosomal Aberration by Array-CGH in an Early Prenatal Diagnosis for Monogenic Disease on Coelomic Fluid. Life. 2023; 13(1):20. https://doi.org/10.3390/life13010020
Chicago/Turabian StyleVinciguerra, Margherita, Filippo Leto, Filippo Cassarà, Viviana Tartaglia, Michela Malacarne, Domenico Coviello, Valentina Cigna, Emanuela Orlandi, Francesco Picciotto, Gaspare Cucinella, and et al. 2023. "Incidental Detection of a Chromosomal Aberration by Array-CGH in an Early Prenatal Diagnosis for Monogenic Disease on Coelomic Fluid" Life 13, no. 1: 20. https://doi.org/10.3390/life13010020
APA StyleVinciguerra, M., Leto, F., Cassarà, F., Tartaglia, V., Malacarne, M., Coviello, D., Cigna, V., Orlandi, E., Picciotto, F., Cucinella, G., Salzano, E., Piccione, M., Maggio, A., & Giambona, A. (2023). Incidental Detection of a Chromosomal Aberration by Array-CGH in an Early Prenatal Diagnosis for Monogenic Disease on Coelomic Fluid. Life, 13(1), 20. https://doi.org/10.3390/life13010020









