Primary and Metastatic Cutaneous Melanomas Discriminately Enrich Several Ligand-Receptor Interactions
Abstract
1. Introduction
2. Methods
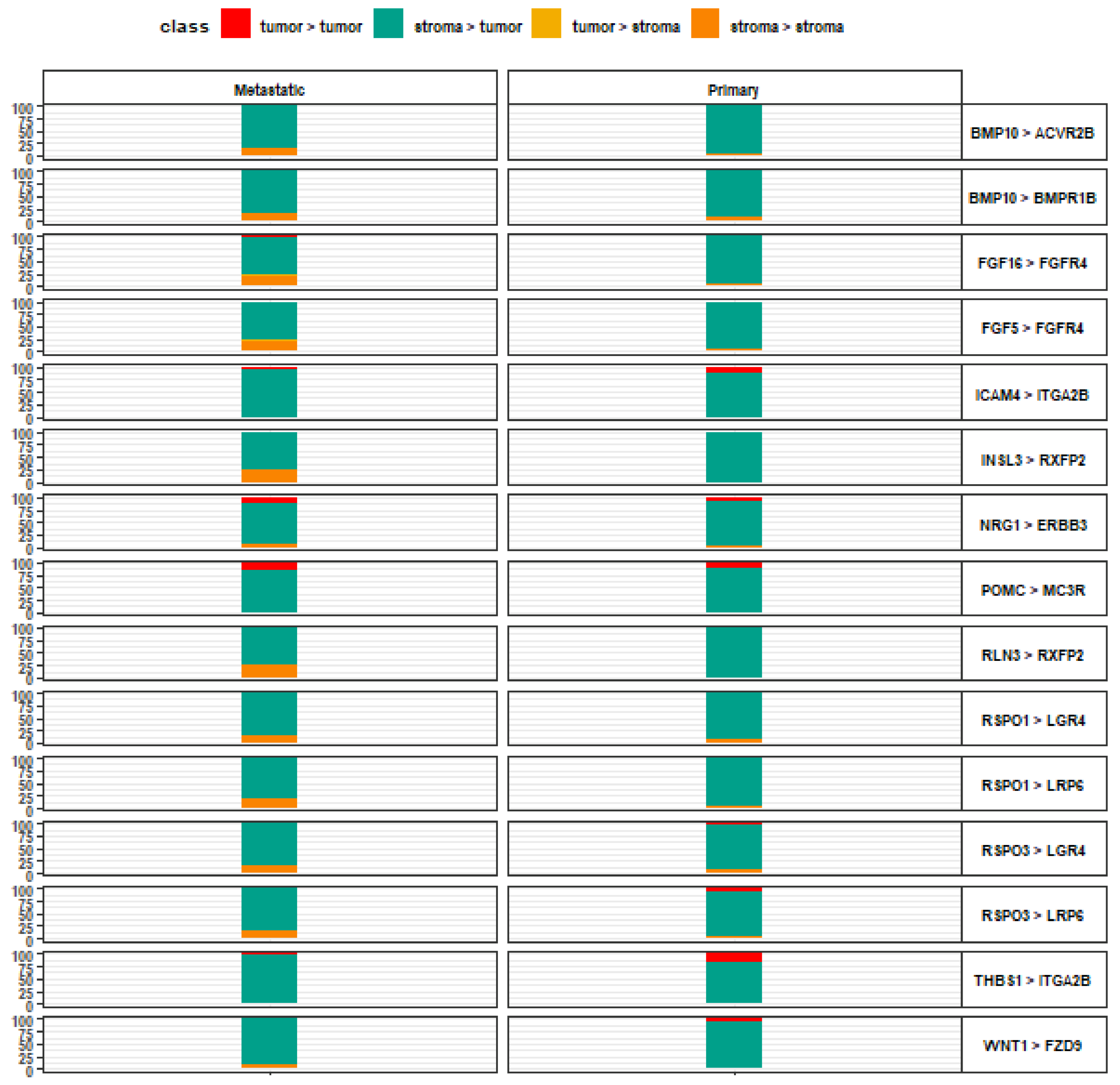
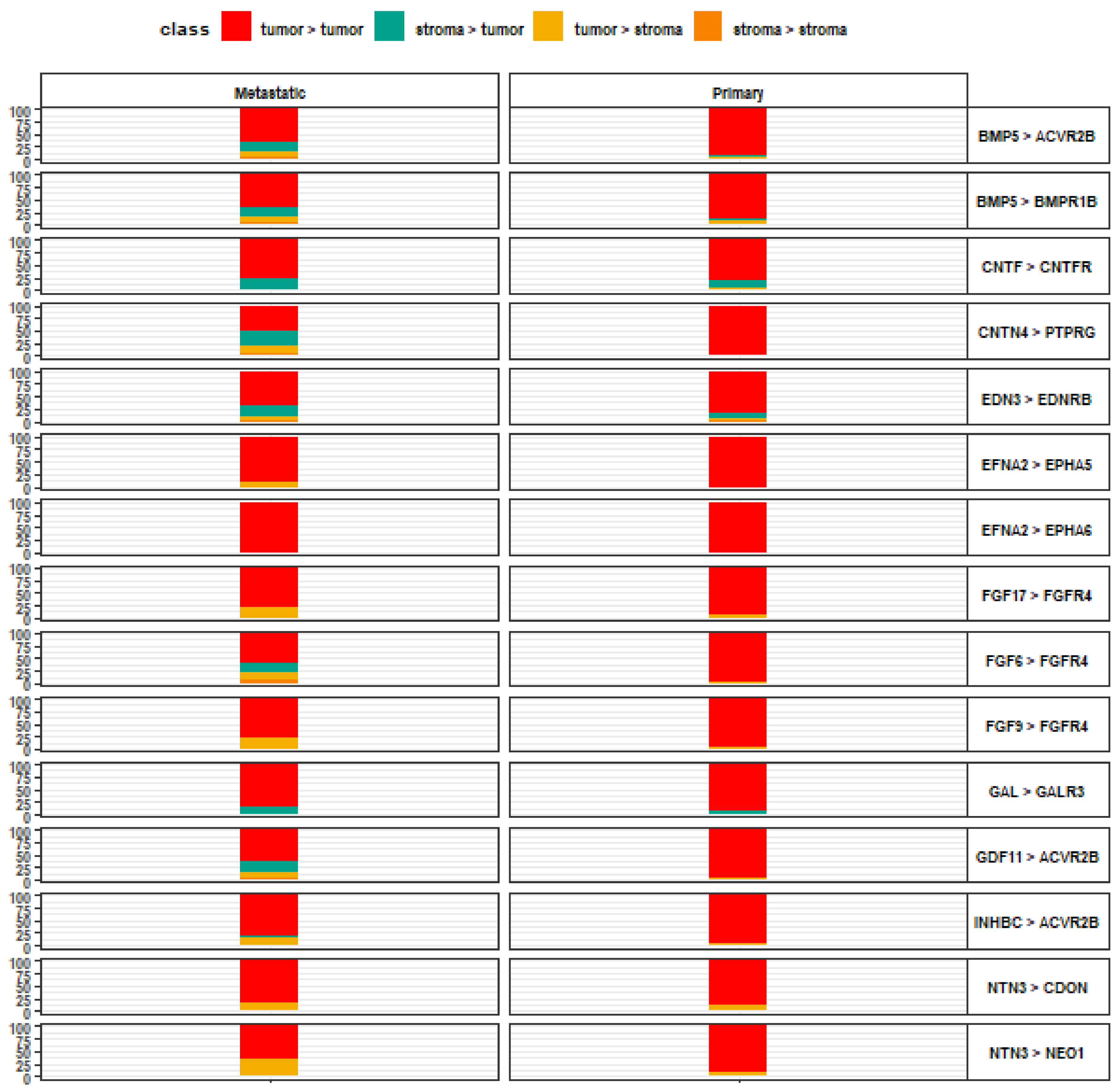
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Coricovac, D.; Dehelean, C.; Moaca, E.-A.; Pinzaru, I.; Bratu, T.; Navolan, D.; Boruga, O. Cutaneous Melanoma—A Long Road from Experimental Models to Clinical Outcome: A Review. Int. J. Mol. Sci. 2018, 19, 1566. [Google Scholar] [CrossRef]
- Zeng, N.; Ma, L.; Cheng, Y.; Xia, Q.; Li, Y.; Chen, Y.; Lu, Z.; Lu, Q.; Jiang, F.; Luo, D. Construction of a Ferroptosis-Related Gene Signature for Predicting Survival and Immune Microenvironment in Melanoma Patients. Int. J. Gen. Med. 2021, 14, 6423–6438. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Rong, R.; Xiong, S.; Song, W.; Ji, D.; Xia, X. Integrated analysis to reveal potential therapeutic targets and prognostic biomarkers of skin cutaneous melanoma. Front. Immunol. 2022, 13, 914108. [Google Scholar] [CrossRef]
- Yang, Y.; Qi, F.; Wei, C.; Liu, J.; Zhang, Y.; Luan, W.; Gu, J. PSMC2 knockdown suppressed tumor progression of skin cutaneous melanoma. Cell Death Discov. 2021, 7, 323. [Google Scholar] [CrossRef]
- Fei, H.; Chen, X. Establishment and validation of an autophagy-related prognostic signature for survival predicting in cutaneous melanoma. Am. J. Cancer Res. 2021, 11, 5979–5991. [Google Scholar] [PubMed]
- Cheng, S.; Li, Z.; Zhang, W.; Sun, Z.; Fan, Z.; Luo, J.; Liu, H. Identification of IL10RA by Weighted Correlation Network Analysis and in vitro Validation of Its Association With Prognosis of Metastatic Melanoma. Front. Cell Dev. Biol. 2021, 8, 630790. [Google Scholar] [CrossRef] [PubMed]
- Kiefel, H.; Bondong, S.; Hazin, J.; Ridinger, J.; Schirmer, U.; Riedle, S.; Altevogt, P. L1CAM: A major driver for tumor cell invasion and motility. Cell Adhes. Migr. 2012, 6, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Bencomo, T.; Das, I.; Lee, C.S. Unravelling the landscape of skin cancer through single-cell transcriptomics. Transl. Oncol. 2023, 27, 101557. [Google Scholar] [CrossRef]
- Wan, J.; Dai, H.; Zhang, X.; Liu, S.; Lin, Y.; Somani, A.-K.; Xie, J.; Han, J. Distinct transcriptomic landscapes of cutaneous basal cell carcinomas and squamous cell carcinomas. Genes Dis. 2019, 8, 181–192. [Google Scholar] [CrossRef]
- Chitsazzadeh, V.; Coarfa, C.; Drummond, J.A.; Nguyen, T.; Joseph, A.; Chilukuri, S.; Charpiot, E.; Adelmann, C.H.; Ching, G.; Nguyen, T.N.; et al. Cross-species identification of genomic drivers of squamous cell carcinoma development across preneoplastic intermediates. Nat. Commun. 2016, 7, 12601. [Google Scholar] [CrossRef]
- Kunz, M.; Löffler-Wirth, H.; Dannemann, M.; Willscher, E.; Doose, G.; Kelso, J.; Kottek, T.; Nickel, B.; Hopp, L.; Landsberg, J.; et al. RNA-seq analysis identifies different transcriptomic types and developmental trajectories of primary melanomas. Oncogene 2018, 37, 6136–6151. [Google Scholar] [CrossRef]
- Svedman, F.C.; Das, I.; Tuominen, R.; Ramqvist, E.D.; Höiom, V.; Brage, S.E. Proliferation and Immune Response Gene Signatures Associated with Clinical Outcome to Immunotherapy and Targeted Therapy in Metastatic Cutaneous Malignant Melanoma. Cancers 2022, 14, 3587. [Google Scholar] [CrossRef] [PubMed]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H., II; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef]
- Tran, M.; Yoon, S.; Teoh, M.; Andersen, S.; Lam, P.; Purdue, B.W.; Raghubar, A.; Hanson, S.; Devitt, K.; Jones, K.; et al. A robust experimental and computational analysis framework at multiple resolutions, modalities and coverages. Front. Immunol. 2022, 13, 911873. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Trevino, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Ghoshdastider, U.; Rohatgi, N.; Naeini, M.M.; Baruah, P.; Revkov, E.; Guo, Y.A.; Rizzetto, S.; Wong, A.M.; Solai, S.; Nguyen, T.T.; et al. Pan-Cancer Analysis of Ligand–Receptor Cross-talk in the Tumor Microenvironment. Cancer Res 2021, 81, 1802–1812. [Google Scholar] [CrossRef]
- Ramilowski, J.A.; Goldberg, T.; Harshbarger, J.; Kloppmann, E.; Lizio, M.; Satagopam, V.P.; Itoh, M.; Kawaji, H.; Carninci, P.; Rost, B.; et al. A draft network of ligand–receptor-mediated multicellular signalling in human. Nat. Commun. 2015, 6, 7866. [Google Scholar] [CrossRef]
- Xie, Y.; Su, N.; Yang, J.; Tan, Q.; Huang, S.; Jin, M.; Ni, Z.; Zhang, B.; Zhang, D.; Luo, F.; et al. FGF/FGFR signaling in health and disease. Signal Transduct. Target. Ther. 2020, 5, 181. [Google Scholar] [CrossRef]
- Seitz, T.; John, N.; Sommer, J.; Dietrich, P.; Thasler, W.E.; Hartmann, A.; Evert, K.; Lang, S.A.; Bosserhoff, A.; Hellerbrand, C. Role of Fibroblast Growth Factors in the Crosstalk of Hepatic Stellate Cells and Uveal Melanoma Cells in the Liver Metastatic Niche. Int. J. Mol. Sci. 2022, 23, 11524. [Google Scholar] [CrossRef]
- Czyz, M. Fibroblast Growth Factor Receptor Signaling in Skin Cancers. Cells 2019, 8, 540. [Google Scholar] [CrossRef]
- ter Steege, E.J.; Bakker, E.R.M. The role of R-spondin proteins in cancer biology. Oncogene 2021, 40, 6469–6478. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Shi, X.; Zhang, J.; Qin, J.; Zhang, N.; Ren, H.; Qian, M.; Siwko, S.; Carmon, K.; Liu, Q.; et al. Inhibition of Rspo-Lgr4 Facilitates Checkpoint Blockade Therapy by Switching Macrophage Polarization. Cancer Res 2018, 78, 4929–4942. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.-X.; Xie, Y.; Zhang, Y.; Charlat, O.; Oster, E.; Avello, M.; Lei, H.; Mickanin, C.; Liu, D.; Ruffner, H.; et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 2012, 485, 195–200. [Google Scholar] [CrossRef]
- O'Connell, M.; Weeraratna, A.T. Hear the Wnt Ror: How melanoma cells adjust to changes in Wnt. Pigment. Cell Melanoma Res. 2009, 22, 724–739. [Google Scholar] [CrossRef] [PubMed]
- Gajos-Michniewicz, A.; Czyz, M. WNT Signaling in Melanoma. Int. J. Mol. Sci. 2020, 21, 4852. [Google Scholar] [CrossRef]
- Dodelet, V.C.; Pasquale, E.B. Eph receptors and ephrin ligands: Embryogenesis to tumorigenesis. Oncogene 2000, 19, 5614–5619. [Google Scholar] [CrossRef] [PubMed]
- Walsh, R.; Blumenberg, M. Specific and Shared Targets of Ephrin A Signaling in Epidermal Keratinocytes. J. Biol. Chem. 2011, 286, 9419–9428. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, R.; Kataoka, H.; Sato, N.; Kanamori, M.; Ihara, M.; Igarashi, H.; Ravshanov, S.; Wang, Y.-J.; Li, Z.-Y.; Shimamura, T.; et al. EPHA2/EFNA1 expression in human gastric cancer. Cancer Sci. 2005, 96, 42–47. [Google Scholar] [CrossRef]
- Zhao, Y.; Cai, C.; Zhang, M.; Shi, L.; Wang, J.; Zhang, H.; Ma, P.; Li, S. Ephrin-A2 promotes prostate cancer metastasis by enhancing angiogenesis and promoting EMT. J. Cancer Res. Clin. Oncol. 2021, 147, 2013–2023. [Google Scholar] [CrossRef]
- Aykul, S.; Martinez-Hackert, E. Transforming Growth Factor-β Family Ligands Can Function as Antagonists by Competing for Type II Receptor Binding. J. Biol. Chem. 2016, 291, 10792–10804. [Google Scholar] [CrossRef] [PubMed]
- Barreto, R.; Kitase, Y.; Matsumoto, T.; Pin, F.; Colston, K.C.; Couch, K.E.; O’Connell, T.M.; Couch, M.E.; Bonewald, L.F.; Bonetto, A. ACVR2B/Fc counteracts chemotherapy-induced loss of muscle and bone mass. Sci. Rep. 2017, 7, 14470. [Google Scholar] [CrossRef] [PubMed]
- Huot, J.R.; Pin, F.; Narasimhan, A.; Novinger, L.J.; Keith, A.S.; Zimmers, T.A.; Willis, M.S.; Bonetto, A. ACVR2B antagonism as a countermeasure to multi-organ perturbations in metastatic colorectal cancer cachexia. J. Cachex-Sarcopenia Muscle 2020, 11, 1779–1798. [Google Scholar] [CrossRef]
- Tschernia, N.P.; Gulley, J.L. Tumor in the Crossfire: Inhibiting TGF-β to Enhance Cancer Immunotherapy. Biodrugs 2022, 36, 153–180. [Google Scholar] [CrossRef] [PubMed]
- Conlon, K.C.; Miljkovic, M.D.; Waldmann, T.A. Cytokines in the Treatment of Cancer. J. Interferon Cytokine Res. 2019, 39, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Petrella, T.M.; Tozer, R.; Belanger, K.; Savage, K.J.; Wong, R.; Smylie, M.; Kamel-Reid, S.; Tron, V.; Chen, B.E.; Hunder, N.N.; et al. Interleukin-21 Has Activity in Patients With Metastatic Melanoma: A Phase II Study. J. Clin. Oncol. 2012, 30, 3396–3401. [Google Scholar] [CrossRef] [PubMed]
- Jacquelot, N.; Duong, C.P.M.; Belz, G.T.; Zitvogel, L. Targeting Chemokines and Chemokine Receptors in Melanoma and Other Cancers. Front. Immunol. 2018, 9, 2480. [Google Scholar] [CrossRef]
- Ugurel, S.; Röhmel, J.; Ascierto, P.A.; Flaherty, K.T.; Grob, J.J.; Hauschild, A.; Larkin, J.; Long, G.V.; Lorigan, P.; McArthur, G.A.; et al. Survival of patients with advanced metastatic melanoma: The impact of novel therapies–update 2017. Eur. J. Cancer 2017, 83, 247–257. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diaz, M.J.; Fadil, A.; Tran, J.T.; Batchu, S.; Root, K.T.; Tran, A.X.; Lucke-Wold, B. Primary and Metastatic Cutaneous Melanomas Discriminately Enrich Several Ligand-Receptor Interactions. Life 2023, 13, 180. https://doi.org/10.3390/life13010180
Diaz MJ, Fadil A, Tran JT, Batchu S, Root KT, Tran AX, Lucke-Wold B. Primary and Metastatic Cutaneous Melanomas Discriminately Enrich Several Ligand-Receptor Interactions. Life. 2023; 13(1):180. https://doi.org/10.3390/life13010180
Chicago/Turabian StyleDiaz, Michael J., Angela Fadil, Jasmine T. Tran, Sai Batchu, Kevin T. Root, Andrew X. Tran, and Brandon Lucke-Wold. 2023. "Primary and Metastatic Cutaneous Melanomas Discriminately Enrich Several Ligand-Receptor Interactions" Life 13, no. 1: 180. https://doi.org/10.3390/life13010180
APA StyleDiaz, M. J., Fadil, A., Tran, J. T., Batchu, S., Root, K. T., Tran, A. X., & Lucke-Wold, B. (2023). Primary and Metastatic Cutaneous Melanomas Discriminately Enrich Several Ligand-Receptor Interactions. Life, 13(1), 180. https://doi.org/10.3390/life13010180







