Radiomics from Various Tumour Volume Sizes for Prognosis Prediction of Head and Neck Squamous Cell Carcinoma: A Voted Ensemble Machine Learning Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Data
2.2. Variation of Target Volumes
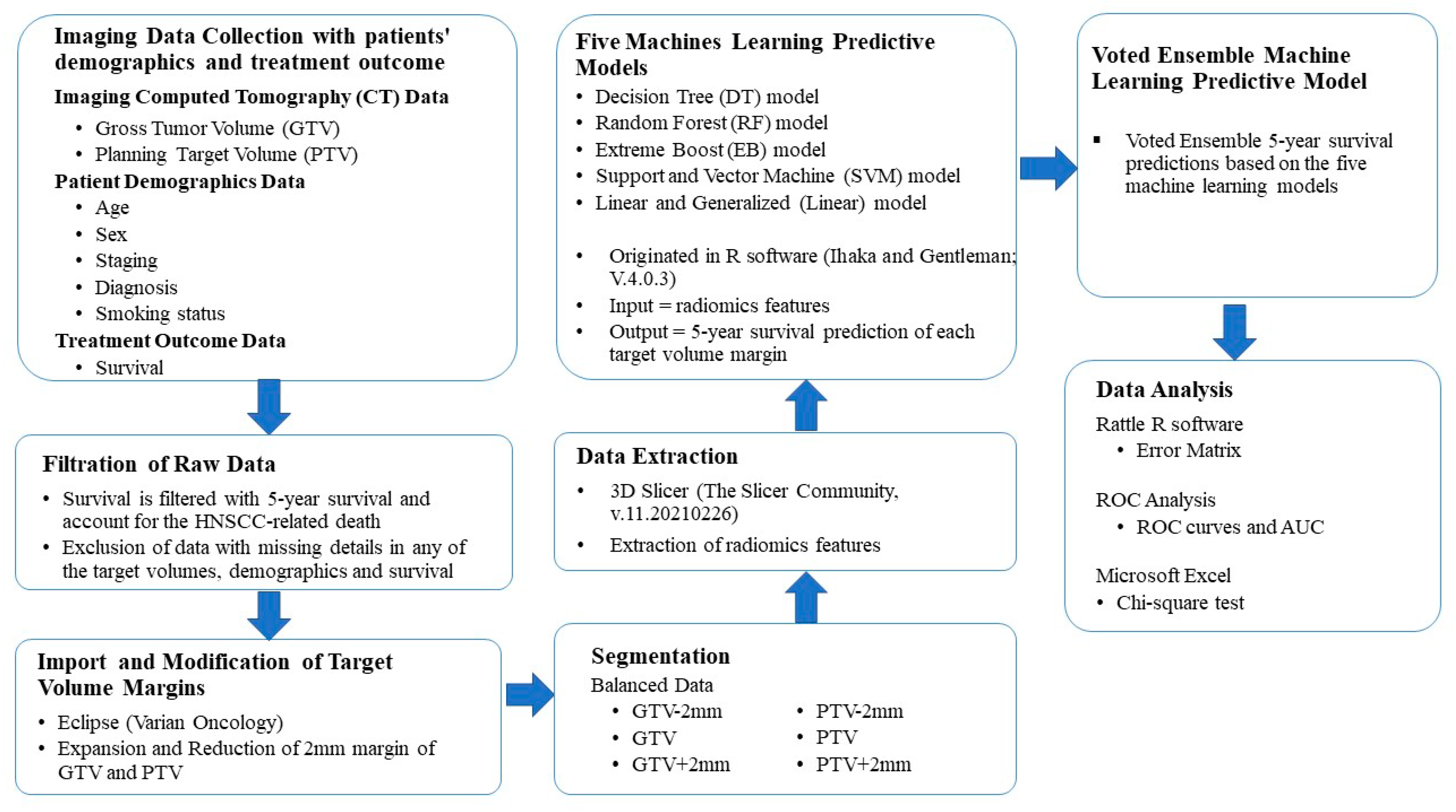
2.3. Study Workflow
2.4. Feature Extraction
2.5. Test to Avoid Overfitting
2.6. Machine Learning Algorithms
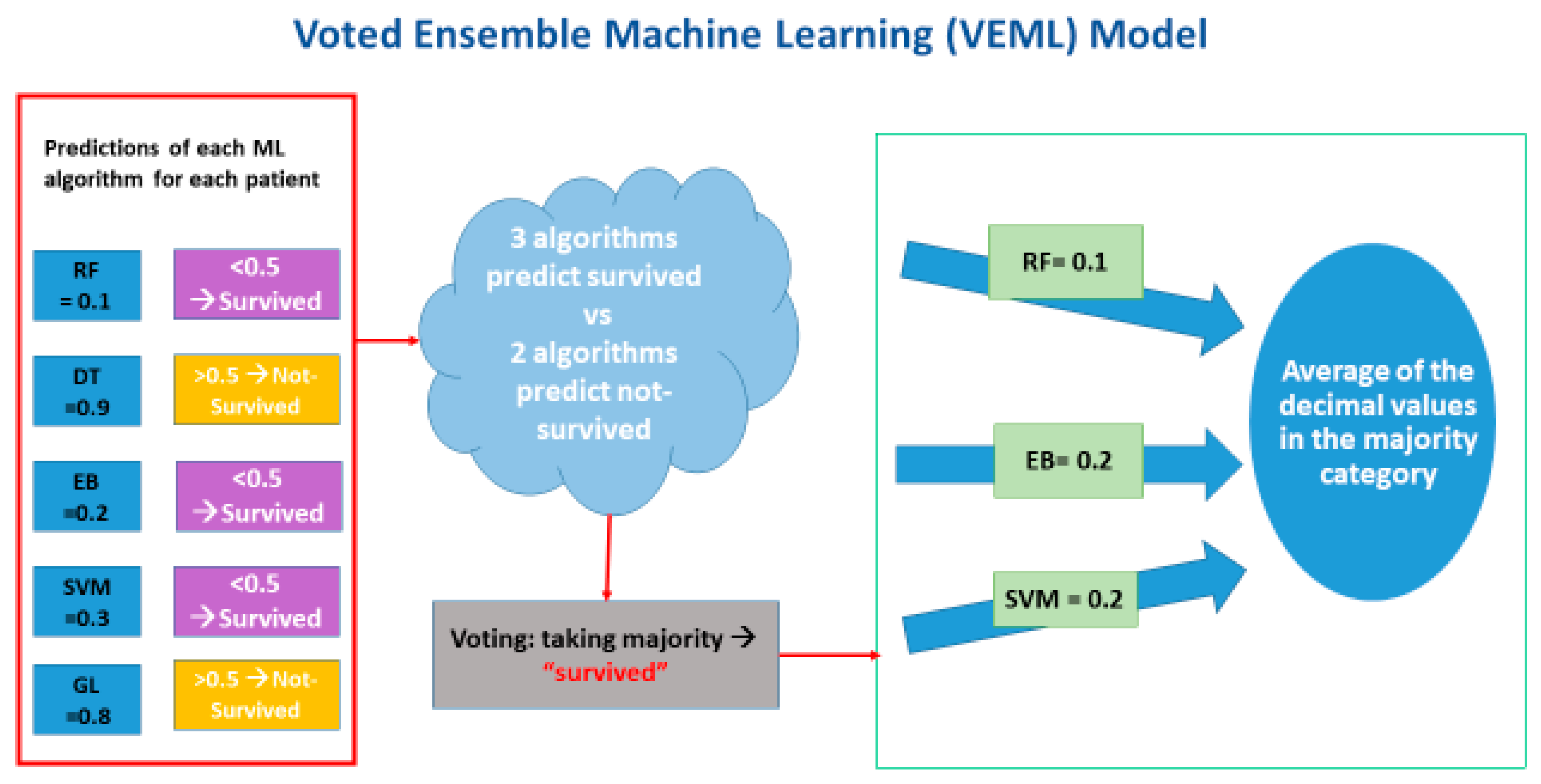
2.7. Voted Ensemble Machine Learning Model
2.8. Data Analysis
3. Results
3.1. Cohort Demographics
3.2. Prognosis Prediction of Five Machine Learning Algorithms and VEML Model
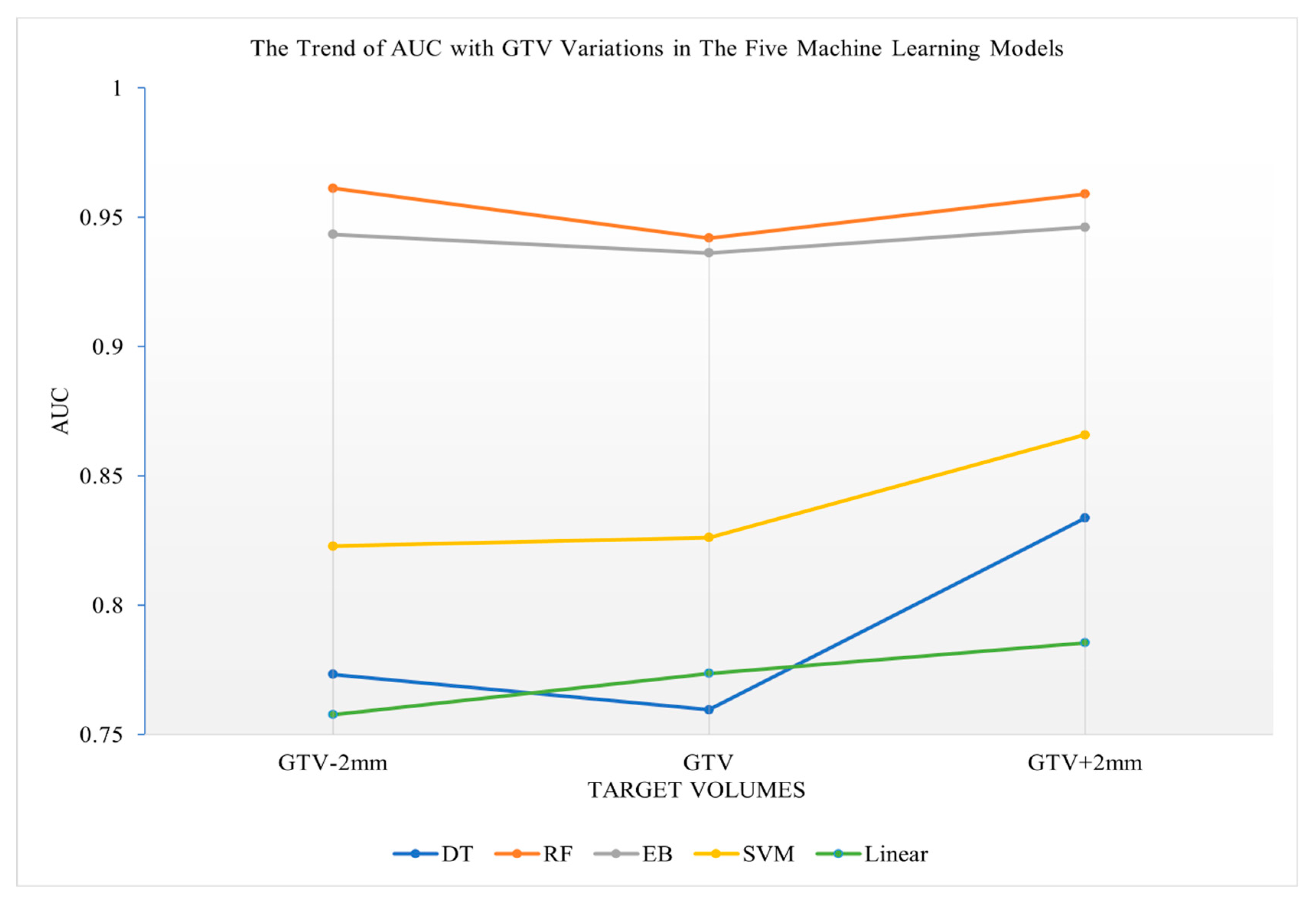
3.3. Prediction Performance of the Five Machine Learning Models Using Radiomics Features of Various Target Volumes
3.4. Performance of Voted Ensemble Machine Learning Model
4. Discussion
4.1. The Value of the VEML Model
4.2. Application of the VEML Model to Supplement TNM Staging for Prognosis Prediction
4.3. Potential Development of the VEML Model
4.4. Advantages of this Work Compared with Other Related Work
4.5. Study Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Definitions of Machine Learning Models [32]
References
- Diamant, A.; Chatterjee, A.; Vallières, M.; Shenouda, G.; Seuntjens, J. Deep Learning in Head & Neck Cancer Outcome Prediction. Sci. Rep. 2019, 9, 2764. [Google Scholar] [CrossRef]
- Cancer (IARC), T.I.A. for R. on Global Cancer Observatory. Available online: https://gco.iarc.fr/ (accessed on 4 July 2022).
- Hong Kong Cancer Registry, Hospital Authority. Available online: https://www3.ha.org.hk/cancereg/topten.html (accessed on 3 August 2021).
- Atun, R.; Jaffray, D.A.; Barton, M.B.; Bray, F.; Baumann, M.; Vikram, B.; Hanna, T.P.; Knaul, F.M.; Lievens, Y.; Lui, T.Y.M.; et al. Expanding Global Access to Radiotherapy. Lancet Oncol. 2015, 16, 1153–1186. [Google Scholar] [CrossRef] [PubMed]
- Cancer Facts & Figures 2021|American Cancer Society. Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2021.html (accessed on 4 July 2022).
- Shalev-Shwartz, S.; Ben-David, S. Understanding Machine Learning: From Theory to Algorithms, 1st ed.; Cambridge University Press: Cambridge, UK, 2014; ISBN 978-1-107-05713-5. [Google Scholar]
- Cheung, E.Y.W.; Chau, A.C.M.; Tang, F.H.; on behalf of the Alzheimer’s Disease Neuroimaging Initiative. Radiomics-Based Artificial Intelligence Differentiation of Neurodegenerative Diseases with Reference to the Volumetry. Life 2022, 12, 514. [Google Scholar] [CrossRef] [PubMed]
- Vicini, S.; Bortolotto, C.; Rengo, M.; Ballerini, D.; Bellini, D.; Carbone, I.; Preda, L.; Laghi, A.; Coppola, F.; Faggioni, L. A Narrative Review on Current Imaging Applications of Artificial Intelligence and Radiomics in Oncology: Focus on the Three Most Common Cancers. Radiol. Med. 2022, 127, 819–836. [Google Scholar] [CrossRef]
- Mahmood, H.; Shaban, M.; Indave, B.I.; Santos-Silva, A.R.; Rajpoot, N.; Khurram, S.A. Use of Artificial Intelligence in Diagnosis of Head and Neck Precancerous and Cancerous Lesions: A Systematic Review. Oral Oncol. 2020, 110, 104885. [Google Scholar] [CrossRef]
- Rahman, T.Y.; Mahanta, L.B.; Chakraborty, C.; Das, A.K.; Sarma, J.D. Textural Pattern Classification for Oral Squamous Cell Carcinoma. J. Microsc. 2018, 269, 85–93. [Google Scholar] [CrossRef]
- Lee, J.H.; Ha, E.J.; Kim, J.H. Application of Deep Learning to the Diagnosis of Cervical Lymph Node Metastasis from Thyroid Cancer with CT. Eur. Radiol. 2019, 29, 5452–5457. [Google Scholar] [CrossRef] [PubMed]
- Xiong, T.; Lv, X.-S.; Wu, G.-J.; Guo, Y.-X.; Liu, C.; Hou, F.-X.; Wang, J.-K.; Fu, Y.-F.; Liu, F.-Q. Single-Cell Sequencing Analysis and Multiple Machine Learning Methods Identified G0S2 and HPSE as Novel Biomarkers for Abdominal Aortic Aneurysm. Front. Immunol. 2022, 13, 907309. [Google Scholar] [CrossRef]
- Bogowicz, M.; Riesterer, O.; Ikenberg, K.; Stieb, S.; Moch, H.; Studer, G.; Guckenberger, M.; Tanadini-Lang, S. Computed Tomography Radiomics Predicts HPV Status and Local Tumor Control After Definitive Radiochemotherapy in Head and Neck Squamous Cell Carcinoma. Int. J. Radiat. Oncol. 2017, 99, 921–928. [Google Scholar] [CrossRef]
- Huang, C.; Cintra, M.; Brennan, K.; Zhou, M.; Colevas, A.D.; Fischbein, N.; Zhu, S.; Gevaert, O. Development and Validation of Radiomic Signatures of Head and Neck Squamous Cell Carcinoma Molecular Features and Subtypes. eBioMedicine 2019, 45, 70–80. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.-Y.; Lin, Y.-C.; Shen, W.-C.; Hsieh, T.-C.; Yen, K.-Y.; Chen, S.-W.; Kao, C.-H. Associations of Tumor PD-1 Ligands, Immunohistochemical Studies, and Textural Features in 18F-FDG PET in Squamous Cell Carcinoma of the Head and Neck. Sci. Rep. 2018, 8, 105. [Google Scholar] [CrossRef] [PubMed]
- Zwirner, K.; Hilke, F.J.; Demidov, G.; Socarras Fernandez, J.; Ossowski, S.; Gani, C.; Thorwarth, D.; Riess, O.; Zips, D.; Schroeder, C.; et al. Radiogenomics in Head and Neck Cancer: Correlation of Radiomic Heterogeneity and Somatic Mutations in TP53, FAT1 and KMT2D. Strahlenther. Onkol. 2019, 195, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Berthelsen, A.K. What’s New in Target Volume Definition for Radiologists in ICRU Report 71? How Can the ICRU Volume Definitions Be Integrated in Clinical Practice? Cancer Imaging 2007, 7, 104–116. [Google Scholar] [CrossRef]
- Tang, F.; Chu, Y.-W.; Cheung, E.Y.W. Radiomics AI Prediction for Head and Neck Squamous Cell Carcinoma (HNSCC) Prognosis and Recurrence with Target Volume Approach. BJR|Open 2021, 3, 20200073. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Grossberg, A.; Mohamed, A.; El Halawani, H.; Bennett, W.; Smith, K.; Nolan, T.; Chamchod, S.; Kantor, M.; Browne, T.; Hutcheson, K.; et al. Data from Head and Neck Cancer CT Atlas. 2017. Available online: https://wiki.cancerimagingarchive.net/display/Public/HNSCC (accessed on 4 July 2022).
- MICCAI/M.D. Anderson Cancer Center Head and Neck Quantitative Imaging Working Group Matched Computed Tomography Segmentation and Demographic Data for Oropharyngeal Cancer Radiomics Challenges. Sci. Data 2017, 4, 170077. [Google Scholar] [CrossRef]
- Grossberg, A.J.; Mohamed, A.S.R.; Elhalawani, H.; Bennett, W.C.; Smith, K.E.; Nolan, T.S.; Williams, B.; Chamchod, S.; Heukelom, J.; Kantor, M.E.; et al. Imaging and Clinical Data Archive for Head and Neck Squamous Cell Carcinoma Patients Treated with Radiotherapy. Sci. Data 2018, 5, 180173. [Google Scholar] [CrossRef]
- Edge, S.B.; American Joint Committee on Cancer; American Cancer Society (Eds.) AJCC Cancer Staging Handbook: From the AJCC Cancer Staging Manual, 7th ed.; Springer: New York, NY, USA, 2010; ISBN 978-0-387-88442-4. [Google Scholar]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Cavalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; et al. Decoding Tumour Phenotype by Noninvasive Imaging Using a Quantitative Radiomics Approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Shin, T. Ensemble Learning, Bagging, and Boosting Explained in 3 Minutes. Available online: https://towardsdatascience.com/ensemble-learning-bagging-and-boosting-explained-in-3-minutes-2e6d2240ae21 (accessed on 4 July 2022).
- Bryce, T.J.; Dewhirst, M.W.; Floyd, C.E.; Hars, V.; Brizel, D.M. Artificial Neural Network Model of Survival in Patients Treated with Irradiation with and without Concurrent Chemotherapy for Advanced Carcinoma of the Head and Neck. Int. J. Radiat. Oncol. Biol. Phys. 1998, 41, 339–345. [Google Scholar] [CrossRef]
- Wang, J.; Chen, X.; Tian, Y.; Zhu, G.; Qin, Y.; Chen, X.; Pi, L.; Wei, M.; Liu, G.; Li, Z.; et al. Six-Gene Signature for Predicting Survival in Patients with Head and Neck Squamous Cell Carcinoma. Aging 2020, 12, 767–783. [Google Scholar] [CrossRef]
- Yao, Y.; Yan, Z.; Lian, S.; Wei, L.; Zhou, C.; Feng, D.; Zhang, Y.; Yang, J.; Li, M.; Chen, Y. Prognostic Value of Novel Immune-Related Genomic Biomarkers Identified in Head and Neck Squamous Cell Carcinoma. J. Immunother. Cancer 2020, 8, e000444. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.-M.; Huang, B.-T.; Chen, C.-Z.; Wang, Y.; Su, C.-H.; Peng, G.-B.; Zeng, C.-B.; Wu, Y.-X.; Wang, R.-H.; Huang, K.; et al. A Combined Model to Improve the Prediction of Local Control for Lung Cancer Patients Undergoing Stereotactic Body Radiotherapy Based on Radiomic Signature Plus Clinical and Dosimetric Parameters. Front. Oncol. 2022, 11, 819047. [Google Scholar] [CrossRef] [PubMed]
- Bao, D.; Zhao, Y.; Liu, Z.; Zhong, H.; Geng, Y.; Lin, M.; Li, L.; Zhao, X.; Luo, D. Prognostic and Predictive Value of Radiomics Features at MRI in Nasopharyngeal Carcinoma. Discov. Onc. 2021, 12, 63. [Google Scholar] [CrossRef] [PubMed]
- Schnack, H.G.; Kahn, R.S. Detecting Neuroimaging Biomarkers for Psychiatric Disorders: Sample Size Matters. Front. Psychiatry 2016, 7, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amidi, A.; Amidi, S. Study Guide—Machine Learning with R. Available online: https://www.mit.edu/~amidi/teaching/modeling/study-guide/machine-learning-with-r/ (accessed on 4 July 2022).
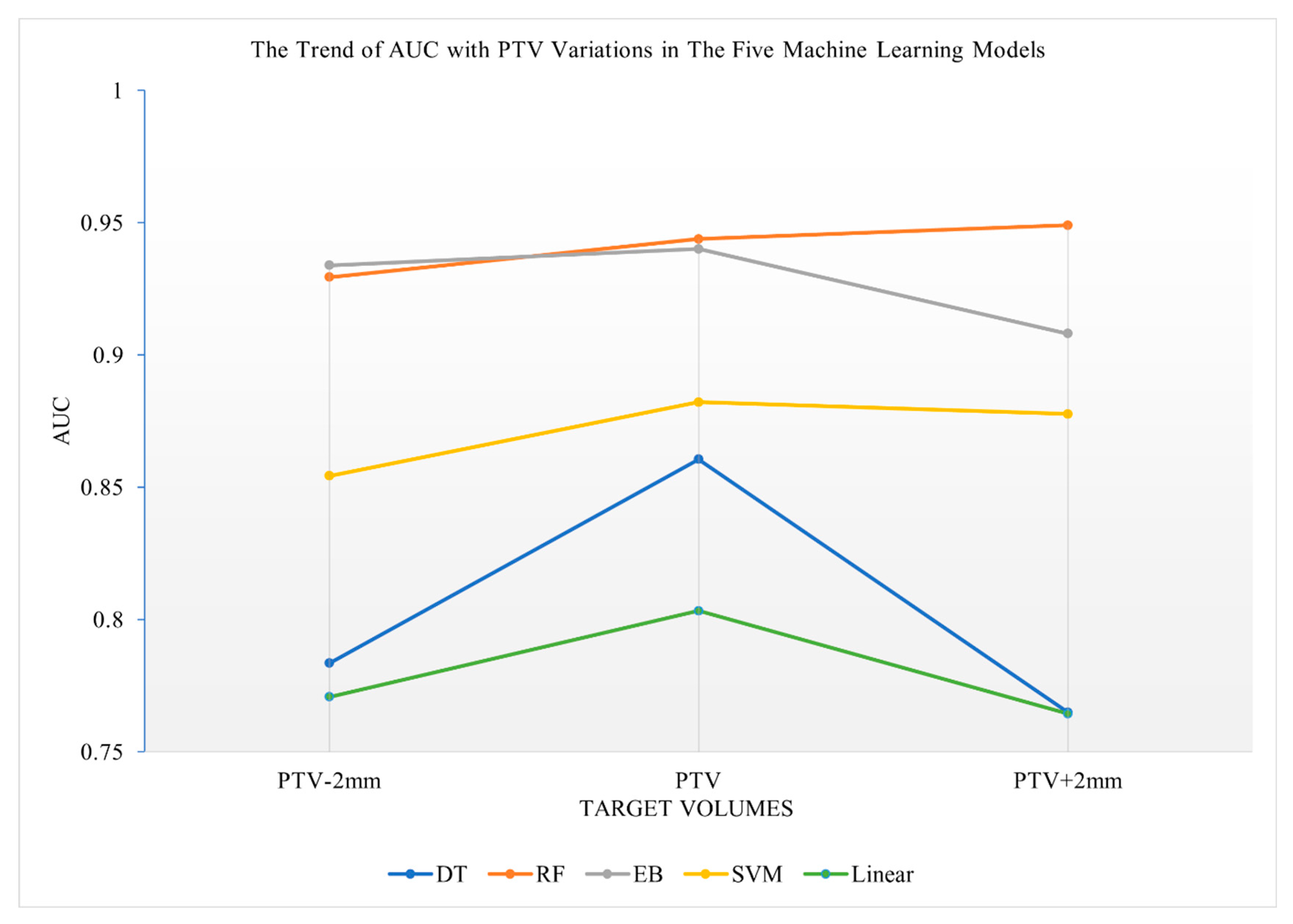
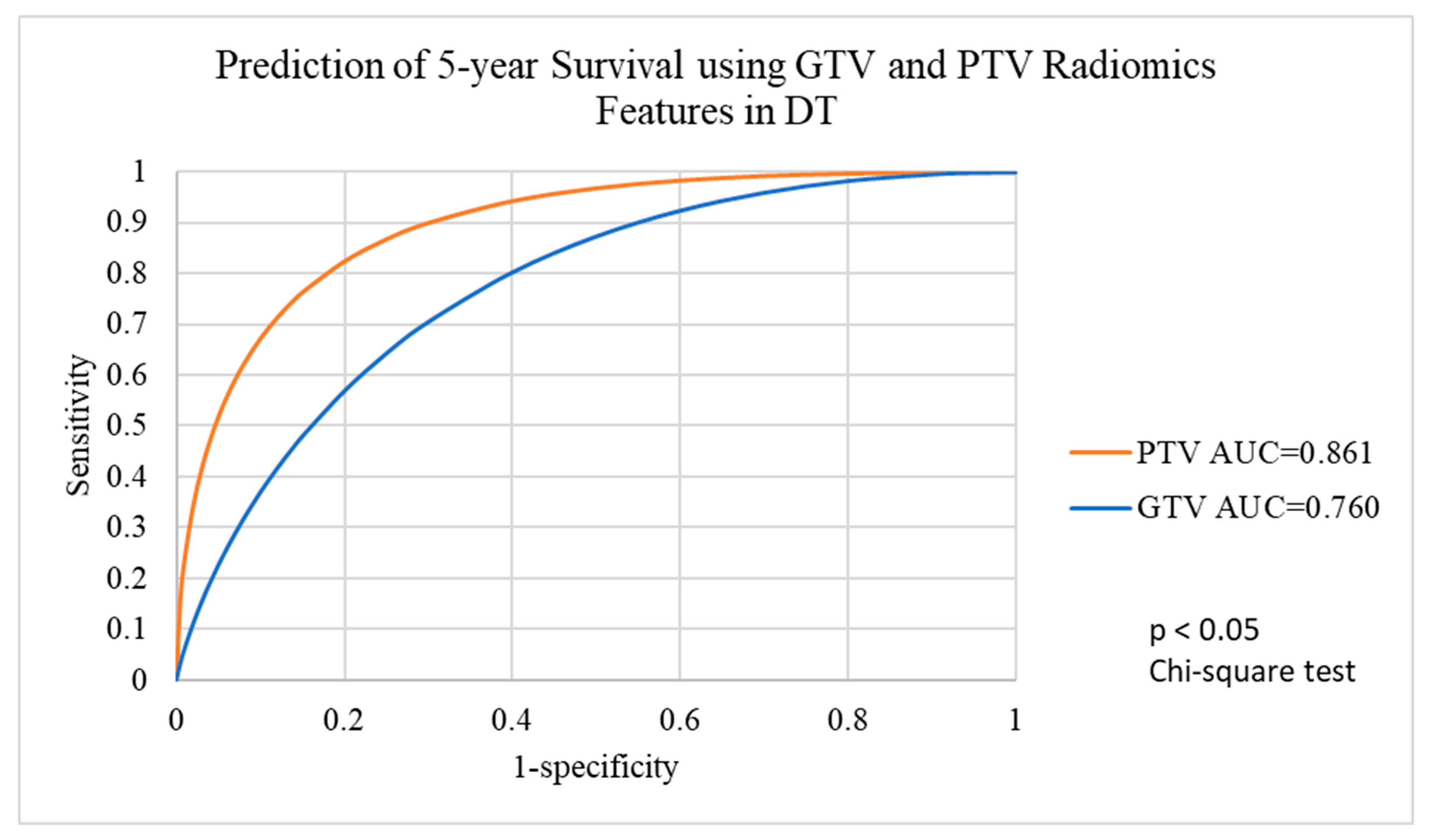
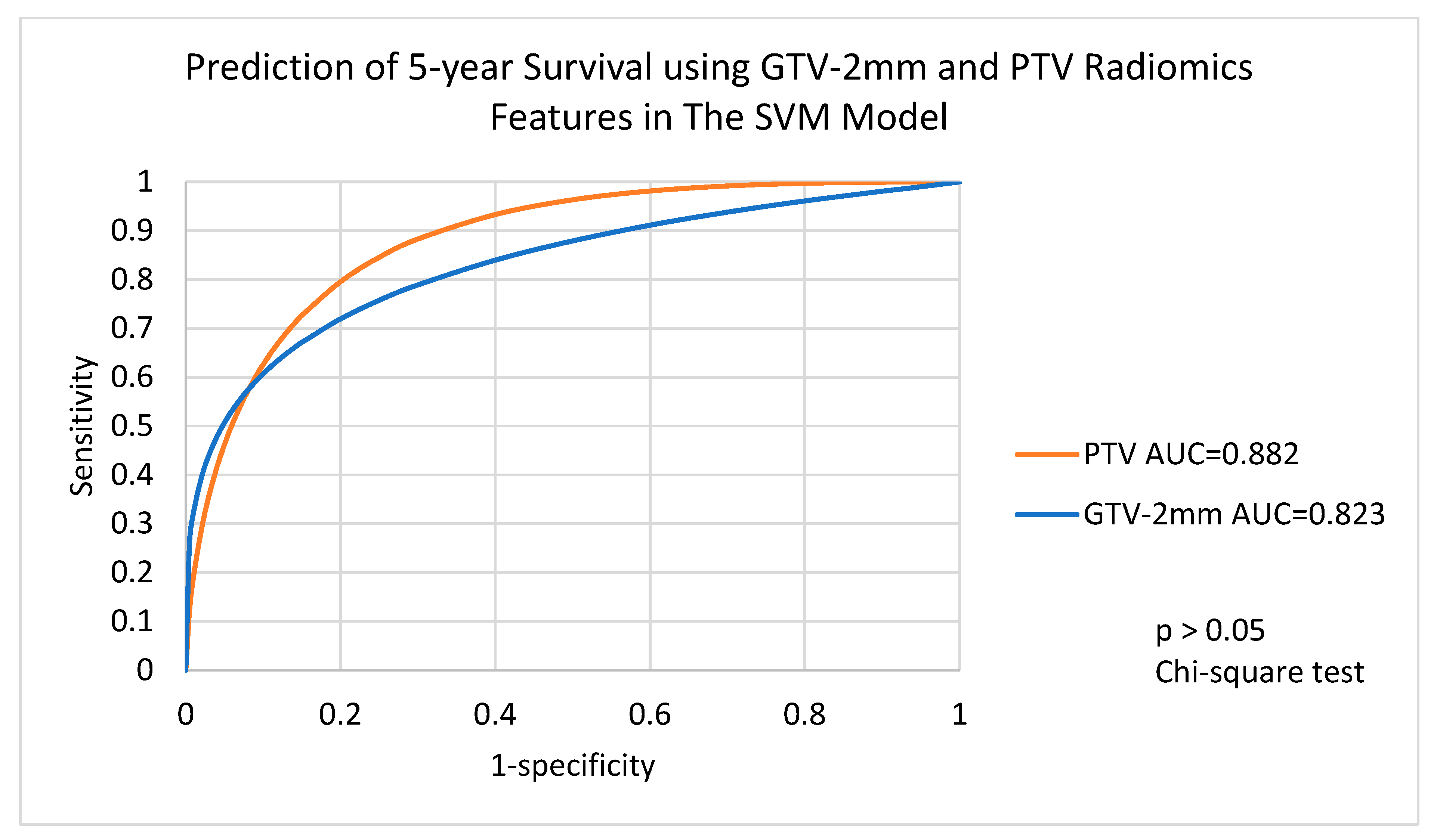

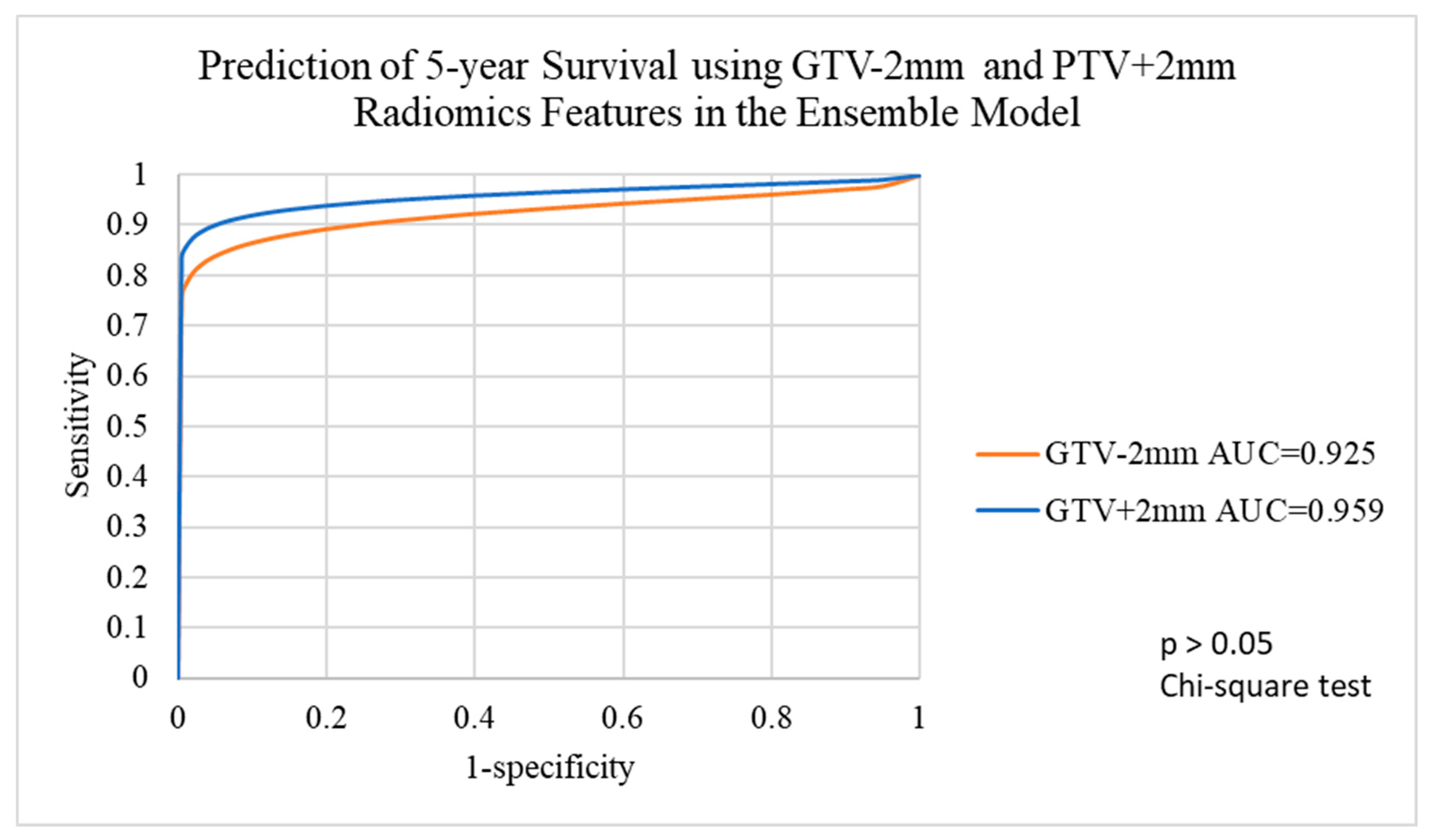
| Target Volumes | GTV-2mm | GTV | GTV+2mm | PTV-2mm | PTV | PTV+2mm | PTV+4mm |
|---|---|---|---|---|---|---|---|
| Sample Size | 92 | 92 | 92 | 98 | 98 | 98 | 98 |
| Balanced sample | 46 not-survived; 46 survived | 46 not-survived; 46 survived | 46 not-survived; 46 survived | 46 not-survived; 46 survived | 49 not-survived; 49 survived | 49 not-survived; 49 survived | 49 not-survived; 49 survived |
| Patient and Tumour Characteristics (All n = 135) | Data |
|---|---|
| Age range (years) | 34–91 |
| Sex | |
| Female | 20 |
| Male | 115 |
| Staging | |
| Stage I | 4 |
| Stage II | 2 |
| Stage III | 17 |
| Stage IVA | 102 |
| Stage IVB | 10 |
| Diagnosis | |
| Ca Base of Tongue | 48 |
| Ca Tonsil | 41 |
| Ca others | 46 |
| Smoking status | |
| Smoker | 86 |
| Non-smoker | 49 |
| Machine Learning Model | GTV -2mm | GTV | GTV +2mm | PTV-2mm | PTV | PTV +2mm | Average | |
|---|---|---|---|---|---|---|---|---|
| AUC | RF | 0.961 | 0.942 | 0.959 | 0.929 | 0.944 | 0.949 | - |
| EB | 0.943 | 0.936 | 0.946 | 0.934 | 0.940 | 0.908 | - | |
| DT | 0.773 | 0.760 | 0.834 | 0.783 | 0.861 | 0.765 | - | |
| SVM | 0.823 | 0.826 | 0.866 | 0.854 | 0.882 | 0.878 | - | |
| Linear | 0.757 | 0.774 | 0.785 | 0.771 | 0.803 | 0.764 | - | |
| VEMLM | 0.925 | 0.927 | 0.959 | 0.927 | 0.947 | 0.956 | - | |
| Accuracy | RF | 0.880 | 0.891 | 0.891 | 0.867 | 0.909 | 0.887 | 0.887 |
| EB | 0.870 | 0.880 | 0.880 | 0.867 | 0.867 | 0.888 | 0.876 | |
| DT | 0.750 | 0.739 | 0.782 | 0.755 | 0.806 | 0.786 | 0.770 | |
| SVM | 0.739 | 0.761 | 0.750 | 0.785 | 0.806 | 0.806 | 0.775 | |
| Linear | 0.859 | 0.848 | 0.859 | 0.847 | 0.867 | 0.857 | 0.856 | |
| VEMLM | 0.880 | 0.859 | 0.902 | 0.878 | 0.888 | 0.888 | 0.883 | |
| Sensitivity | RF | 0.761 | 0.783 | 0.804 | 0.816 | 0.857 | 0.837 | 0.810 |
| EB | 0.739 | 0.783 | 0.761 | 0.796 | 0.816 | 0.816 | 0.785 | |
| DT | 0.609 | 0.500 | 0.587 | 0.612 | 0.878 | 0.714 | 0.650 | |
| SVM | 0.587 | 0.652 | 0.717 | 0.735 | 0.776 | 0.816 | 0.714 | |
| Linear | 0.826 | 0.848 | 0.804 | 0.857 | 0.796 | 0.878 | 0.835 | |
| VEMLM | 0.761 | 0.739 | 0.804 | 0.816 | 0.837 | 0.837 | 0.799 | |
| Specificity | RF | 1.000 | 1.000 | 0.978 | 0.918 | 0.959 | 0.939 | 0.966 |
| EB | 1.000 | 0.978 | 1.000 | 0.938 | 0.918 | 0.959 | 0.966 | |
| DT | 0.581 | 0.978 | 0.978 | 0.898 | 0.735 | 0.857 | 0.838 | |
| SVM | 0.891 | 0.870 | 0.783 | 0.837 | 0.837 | 0.796 | 0.836 | |
| Linear | 0.891 | 0.848 | 0.913 | 0.837 | 0.939 | 0.837 | 0.877 | |
| VEMLM | 1.000 | 0.978 | 1.000 | 0.939 | 0.939 | 0.939 | 0.966 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, F.-H.; Cheung, E.-Y.-W.; Wong, H.-L.; Yuen, C.-M.; Yu, M.-H.; Ho, P.-C. Radiomics from Various Tumour Volume Sizes for Prognosis Prediction of Head and Neck Squamous Cell Carcinoma: A Voted Ensemble Machine Learning Approach. Life 2022, 12, 1380. https://doi.org/10.3390/life12091380
Tang F-H, Cheung E-Y-W, Wong H-L, Yuen C-M, Yu M-H, Ho P-C. Radiomics from Various Tumour Volume Sizes for Prognosis Prediction of Head and Neck Squamous Cell Carcinoma: A Voted Ensemble Machine Learning Approach. Life. 2022; 12(9):1380. https://doi.org/10.3390/life12091380
Chicago/Turabian StyleTang, Fuk-Hay, Eva-Yi-Wah Cheung, Hiu-Lam Wong, Chun-Ming Yuen, Man-Hei Yu, and Pui-Ching Ho. 2022. "Radiomics from Various Tumour Volume Sizes for Prognosis Prediction of Head and Neck Squamous Cell Carcinoma: A Voted Ensemble Machine Learning Approach" Life 12, no. 9: 1380. https://doi.org/10.3390/life12091380
APA StyleTang, F.-H., Cheung, E.-Y.-W., Wong, H.-L., Yuen, C.-M., Yu, M.-H., & Ho, P.-C. (2022). Radiomics from Various Tumour Volume Sizes for Prognosis Prediction of Head and Neck Squamous Cell Carcinoma: A Voted Ensemble Machine Learning Approach. Life, 12(9), 1380. https://doi.org/10.3390/life12091380