Some Factors from Theory, Simulation, Experiment and Proteomes in the Current Biosphere Supporting Deep Oceans as the Location of the Origin of Terrestrial Life
Abstract
:1. Introduction
2. Results
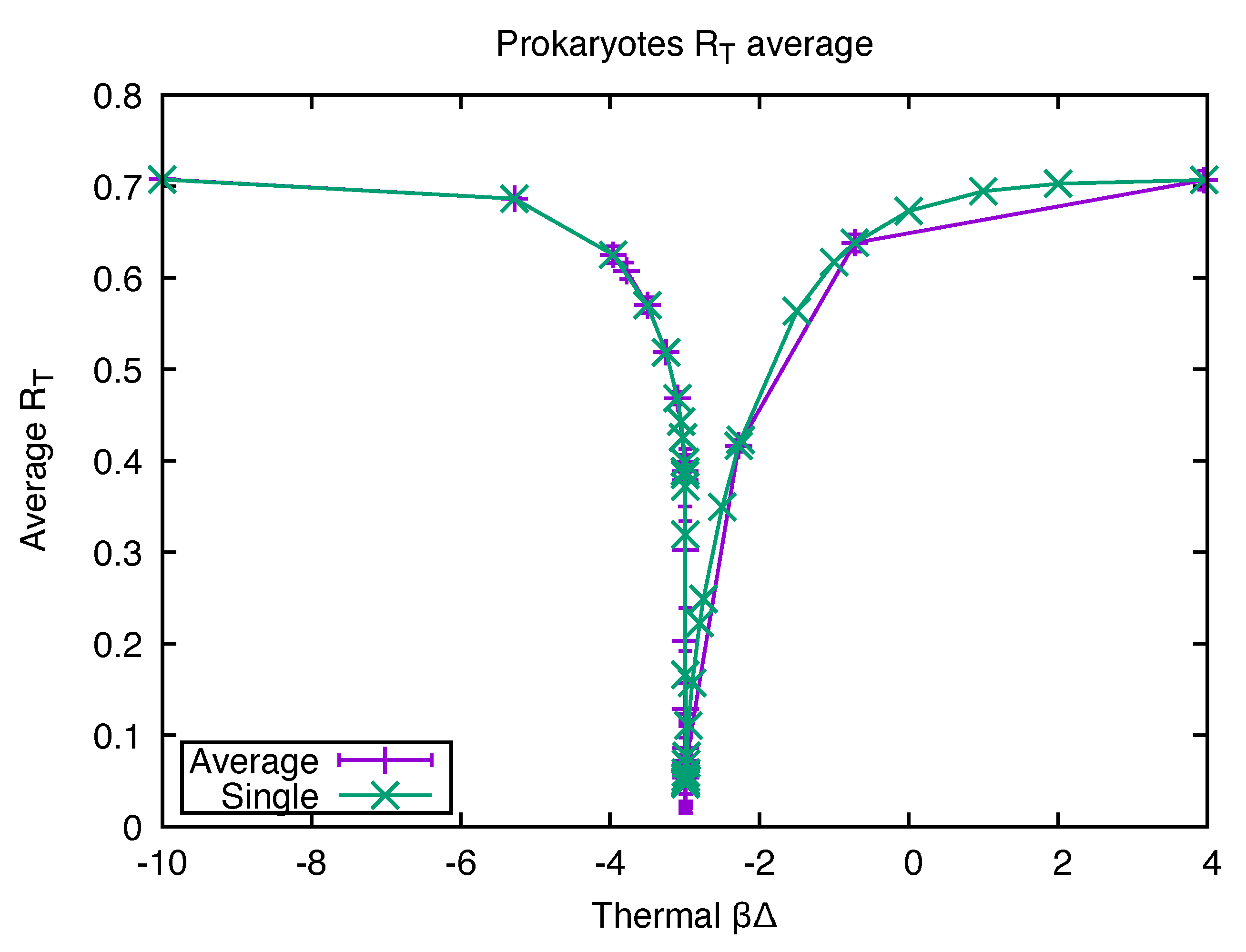
2.1. Evidence from Biodata for an Origin in a Quench
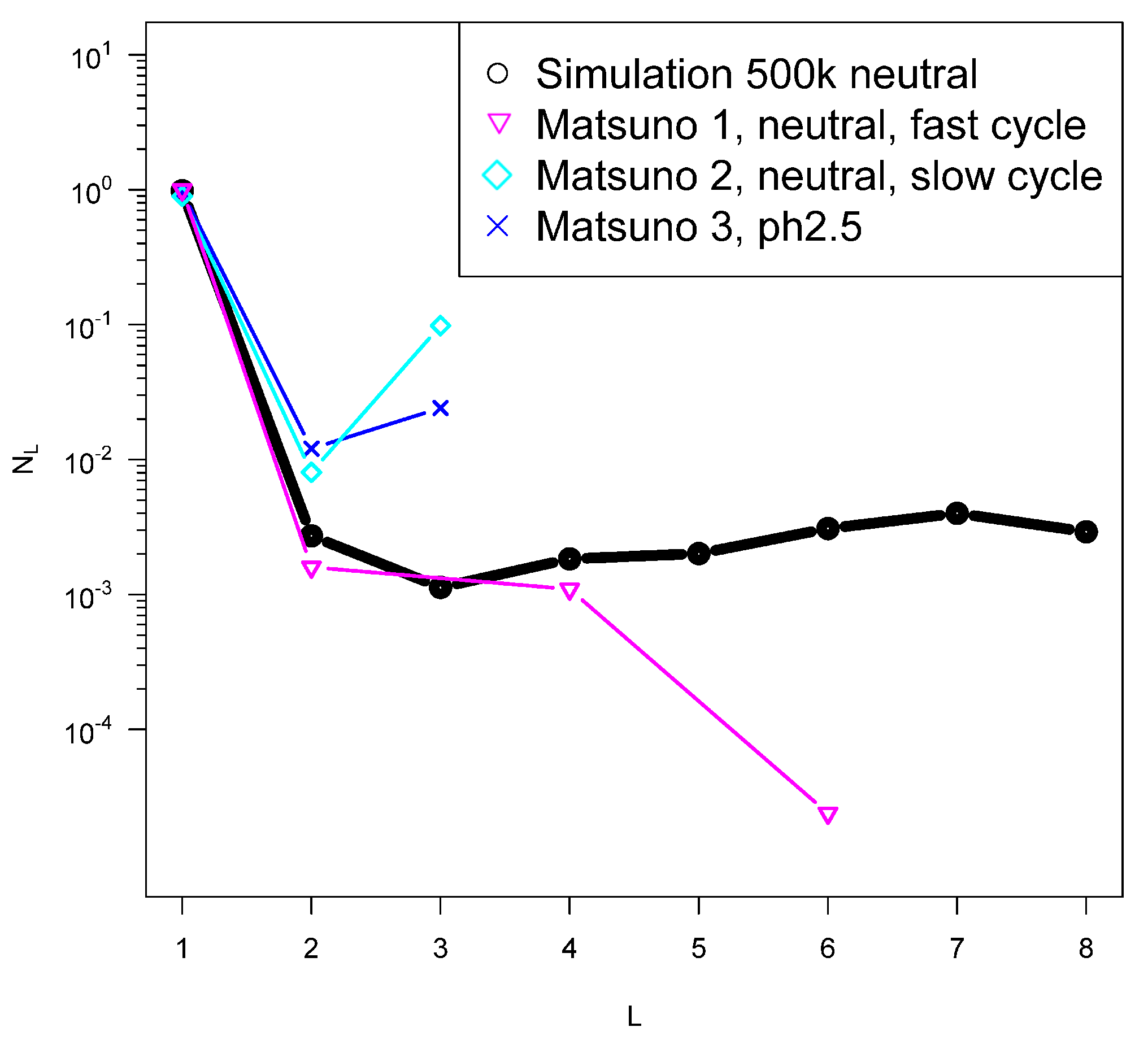
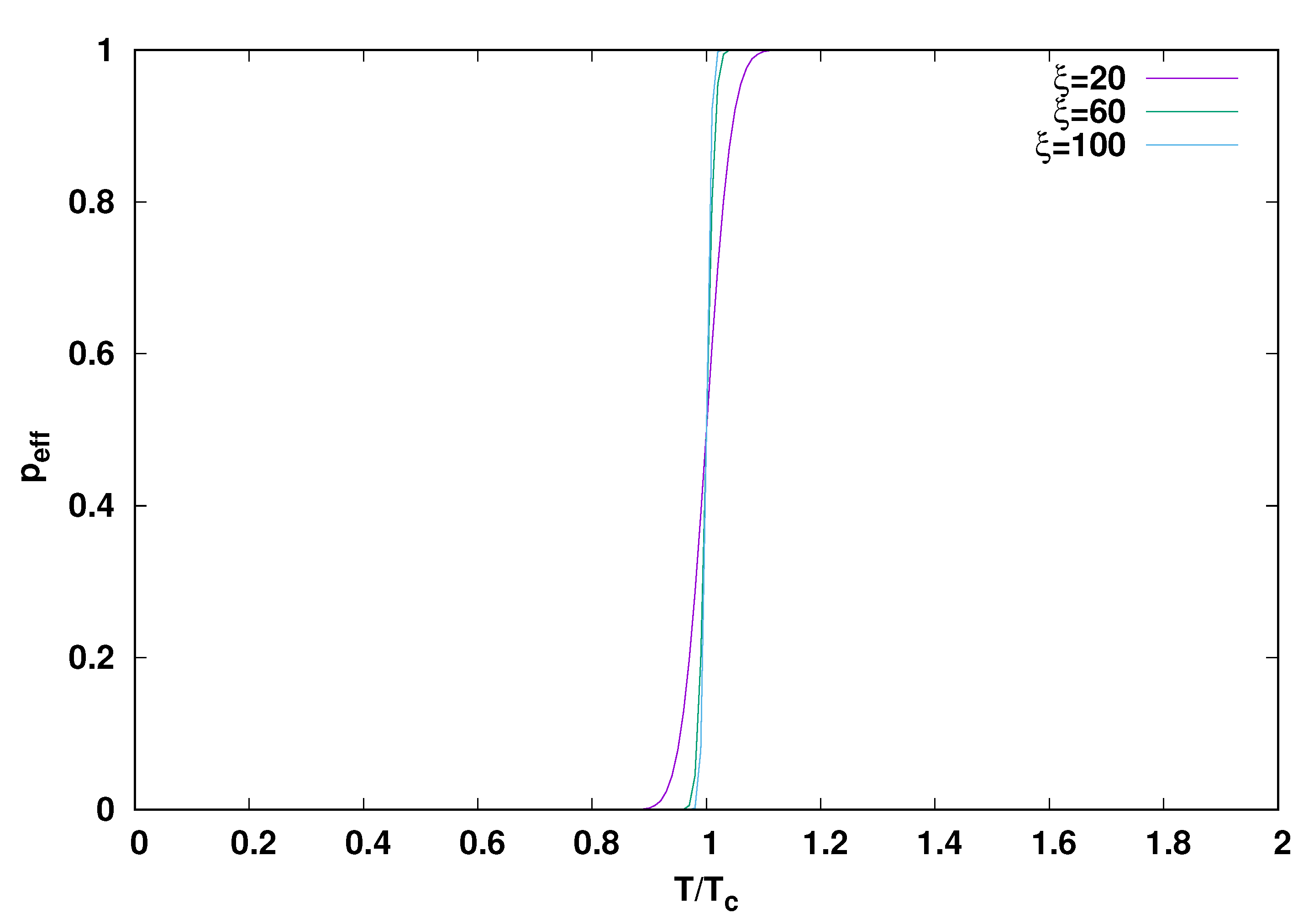
2.2. Evidence from Theory and Simulation for an Origin in a Quench
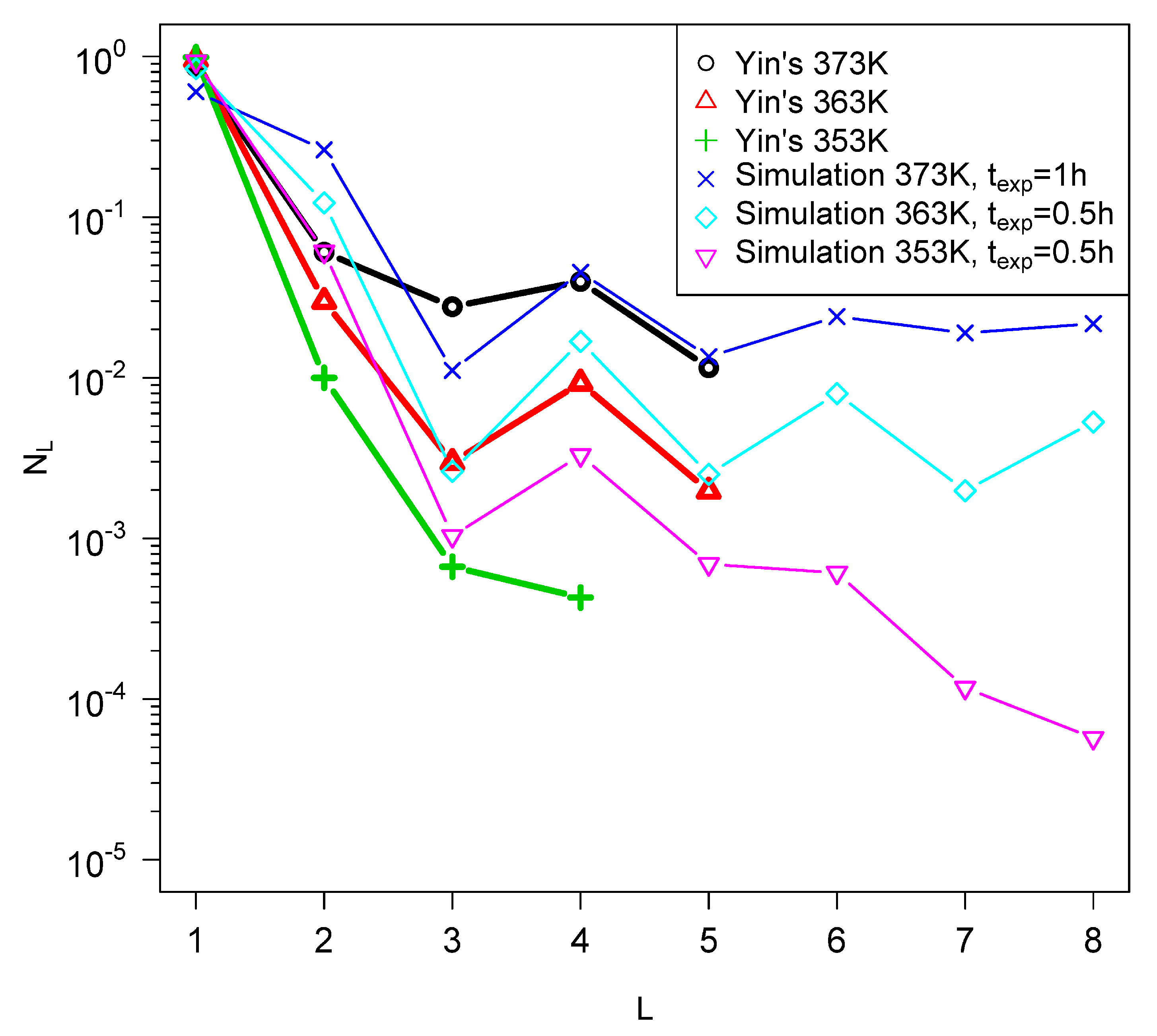
2.3. Comparison of Model Predictions with Laboratory Results of Quenches
3. Discussion and Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eigen, M. Selforganization of Matter and the Evolution of Biological Macromolecules. Naturwissenschaften 1971, 58, 465–523. [Google Scholar] [CrossRef] [PubMed]
- Hart, M. Atmosphereic Evolution, the Drake Equation and DNA: Sparse Life in an Infinite Universe, Chapter 22 in Extraterrestrials, Where Are They? 2nd ed.; Zuckerman, B., Hart, M.H., Eds.; Cambridge University Press: Cambridge, UK, 1995. [Google Scholar]
- Maury, C.P.J. Self-Propagating β-Sheet Polypeptide Structures as Prebiotic Informational Molecular Entities: The Amyloid World. Orig. Life Evol. Biosph. 2009, 39, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Riesner, D. Biochemistry and structure of PrPC and PrPSc. Br. Med. Bull. 2003, 66, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Baskakov, I.V. Autocatalytic Conversion of Recombinant Prion Proteins Displays a Species Barrier. J. Biol. Chem. 2004, 279, 7671–7677. [Google Scholar] [CrossRef]
- Portillo, A.; Hashemi, M.; Zhang, Y.; Breydo, L.; Uversky, V.N.; Lyubchenko, Y.L. Role of monomer arrangement in the amyloid self-assembly. Biochim. Biophys. Acta 2015, 1854, 218–228. [Google Scholar]
- Hegde, R.S.; Mastrianni, J.A.; Scott, M.R.; DeFea, K.A.; Tremblay, P.; Torchia, M.; DeArmond, S.J.; Prusiner, S.B.; Lingappa, V.R. A Transmembrane Form of the Prion Protein in Neurodegenerative Disease. Science 1998, 279, 827–834. [Google Scholar] [CrossRef]
- Cronin, J.R.; Pizzarello, S. Amino Acids in Meteorites. Space Res. 1983, 3, 5–18. [Google Scholar] [CrossRef]
- Flissi, A.; Ricart, E.; Campart, C.; Chevalier, M.; Dufresne, Y.; Michalik, J.; Jacques, P.; Flahaut, C.; Lisacek, F.; Leclère, V.; et al. Norine: Update of the nonribosomal peptide resource. Nucleic Acids Res. 2020, 48, D465–D469. [Google Scholar] [CrossRef]
- German, C.; Seyfriedm, W.E., Jr. Hydrothermal Processes. In Treatise on Geochemistry, 2nd ed.; Holland, H.D., Turekian, K.K., Eds.; Oxford Elsevier: Oxford, UK, 2014; pp. 191–233. [Google Scholar]
- Fuchida, S.; Mizuno, Y.; Masuda, H.; Toki, T.; Makita, H. Concentrations and distributions of amino acids in black and white smoker fluids at temperatures over 200 °C. Org. Geochem. 2014, 66, 98–106. [Google Scholar] [CrossRef]
- Iadanza, M.G.; Jackson, M.P.; Hewitt, E.W.; Ranson, N.A.; Radford, S.E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 755–773. [Google Scholar] [CrossRef]
- Halley, J.W. How Likely Is Extraterrestrial Life; SpringerBriefs in Astronomy Series; Hillebrandt, W., Ratcliffe, M., Eds.; Springer: Berlin/Heidelberg, Germany; Dordrecht, The Netherlands; London, UK; New York, NY, USA, 2012. [Google Scholar]
- Popa, R. Between Necessity and Probability: Searching for the Definition and Origin of Life; Springer: Berlin, Germany, 2004. [Google Scholar]
- Pross, A. Toward a general theory of evolution: Extending Darwinian theory to inanimate matter. J. Syst. Chem. 2011, 2, 1. [Google Scholar] [CrossRef] [Green Version]
- Dyson, F. Origins of Life; Cambridge University Press: Cambridge, UK, 1999. [Google Scholar]
- Cairns-Smith, A.G. The origin of life and the nature of the primitive gene. J. Theor. Biol. 1966, 10, 53–88. [Google Scholar] [CrossRef]
- Zaia, D.A.; Zaia, C.T.B. A few experimental suggestions using minerals to obtain peptides with a high concentration of L-amino acids and protein amino acids. Symmetry 2020, 12, 2046. [Google Scholar] [CrossRef]
- Kitadai, N.; Nishiuchi, K.; Takahagi, W. Thermodynamic Impact of Mineral Surfaces on Amino Acid Polymerization: Aspartate Dimerization on Two-Line Ferrihydrite, Anatase, and γ-Alumina. Minerals 2021, 11, 234. [Google Scholar] [CrossRef]
- Intoy, B.F.; Wynveen, A.; Halley, J.W. Effects of spatial diffusion on nonequilibrium steady states in a model for prebiotic evolution. Phys. Rev. E 2016, 94, 042424. [Google Scholar] [CrossRef]
- Matsuno, K. Hydrothermal Vent Origin of Life Models. In Encyclopedia of Astrobiology; Gargaud, M., Irvine, W., Amils, R., Cleaves, H.J., II, Pinti, D.L., Quintanilla, J.C., Rouan, D., Spohn, T., Tirard, S., Viso, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2020; pp. 1162–1166. [Google Scholar] [CrossRef]
- Nejdl, L.; Petera, L.; Šponer, J.; Zemánková, K.; Pavelicová, K.; Knížek, A.; Adam, V.; Vaculovičová, M.; Ivanek, O. Ferus, M Quantum Dots in Peroxidase-like Chemistry and Formamide-Based Hot Spring Synthesis of Nucleobases. Astrobiology 2022, 5, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Taran, O.; Chen, C.; Omosun, T.O.; Hsieh, M.C.; Rha, A.; Goodwin, J.T.; Mehta, A.K.; Grover, M.A.; Lynn, D.G. Expanding the informational chemistries of life: Peptide/RNA networks. Phys. Trans. R. Soc. A 2017, 375, 20160356. [Google Scholar] [CrossRef] [PubMed]
- Müller-Plathe, F. Coarse-graining in polymer simulation: From the atomistic to the mesoscopic scale and back. Chem. Phys. Chem. 2002, 3, 754–769. [Google Scholar] [CrossRef]
- Anderson, P.W. More is Different. Science 1972, 177, 393. [Google Scholar] [CrossRef]
- Kivelson, S.; Kivelson, S. Defining emergence in physics. Quantum Mater. 2016, 1, 16024. [Google Scholar] [CrossRef]
- Kauffman, S.A. The Origins of Order; Oxford University Press: New York, NY, USA, 1993; Chapter 7. [Google Scholar]
- Farmer, J.D.; Kauffman, S.A.; Packard, N.H. Autocatalytic eplication of polymers. Physica 1986, 220, 50–67. [Google Scholar]
- Bagley, R.; Farmer, J.D. Spontaneous Emergence of a Metabolism. In Artificial Life II; Langton, G.D., Taylor, C., Farmer, J.D., Rasmussen, S., Eds.; Addison Wesley: Redwood City, CA, USA, 1991; p. 93. [Google Scholar]
- Wynveen, A.; Fedorov, I.; Halley, J.W. Nonequilibrium steady states in a model for prebiotic evolution. Phys. Rev. E 2014, 89, 022725. [Google Scholar] [CrossRef] [PubMed]
- Intoy, B.F.; Halley, J.W. Energetics in a model of prebiotic evolution. Phys. Rev. E 2017, 96, 062402. [Google Scholar] [CrossRef] [PubMed]
- Intoy, B.F.; Halley, J.W. Some generic measures of the extent of chemical disequilibrium applied to living and abiotic systems. Phys. Rev. E 2019, 99, 062419. [Google Scholar] [CrossRef] [PubMed]
- Baum, D. The origin and early evolution of life in chemical complexity space. J. Theor. Biol. 2018, 456, 295–304. [Google Scholar] [CrossRef]
- Marín-Moreno, A.; Fernández-Borges, N.; Espinosa, J.C.; Andréoletti, O.; Torres, J.M. Transmission and Replication of Prions. Prog. Mol. Biol. Transl. Sci. 2017, 150, 181–201. [Google Scholar]
- Sheng, Q.; Intoy, B.F.; Halley, J.W. Quenching to fix metastable states in models of Prebiotic Chemistry. Phys. Rev. E 2020, 102, 062412. [Google Scholar] [CrossRef]
- Sheng, Q.; Intoy, B.F.; Halley, J.W. Effects of activation barriers on quenching to stabilize prebiotic chemical systems. Phys. Rev. E 2022, in press. [Google Scholar]
- Lee, N.; Foustoukos, D.I.; Sverjensky, D.A.; Hazen, R.M.; Cody, G. D Hydrogen enhances the stability of glutamic acid in hydrothermal environments. Chem. Geol. 2014, 386, 184–189. [Google Scholar] [CrossRef]
- Radzicka, A.; Wolfenden, R. Rates of uncatalyzed peptide bond hydrolysis in neutral solution and the transition state affinities of proteases. J. Am. Chem. Soc. 1996, 118, 6105–6109. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclope- dia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27. [Google Scholar] [CrossRef] [PubMed]
- Tolman, R.C. The Principles of Statistical Mechanics; Oxford University Press: Oxford, UK, 1938; pp. 58–59. [Google Scholar]
- Kohn, J.E.; Millett, I.S.; Jacob, J.; Zagrovic, B.; Dillon, T.M.; Cingel, N.; Dothager, R.S.; Seifert, S.; Thiyagarajan, P.; Sosnick, T.R.; et al. Random-coil Behavior and the Dimensions of Chemically Unfolded Proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 12491–12496, Erratum in Proc. Natl. Acad. Sci. USA 2005, 102, 14475. [Google Scholar] [CrossRef] [PubMed]
- Gautschi, W. Error Function and Fresnel Integrals. In Handbook of Mathematical Functions; Abramowitz, M., Stegun, I., Eds.; Dover Publications: New York, NY, USA, 1965; pp. 295–296. [Google Scholar]
- Liu, L.; Zhai, S. Basic mathematical model for the normal black smoker system and the hydrothermal megaplume formation. Acta Oceanol. Sin. 2007, 26, 30–40. [Google Scholar]
- Imai, E.I.; Honda, H.; Hatori, K.; Brack, A.; Matsuno, K. Elongation of Oligopeptides in a Simulated Submarine Hydrothermal System. Science 1999, 283, 831. [Google Scholar] [CrossRef] [PubMed]
- Ogata, Y.; Imai, E.I.; Honda, H.; Hatori, K.; Matsuno, K. Hydrothermla Circulation of Seawater through hot vents and contribution of Interface Chemistry to Prebiotic Synthesis. Orig. Life Evol. Biosph. 2000, 30, 527–537. [Google Scholar] [CrossRef]
- Tsukahara, H. Prebiotic oligomerization on or inside lipid vesicles in hydrothermal environments. Orig. Life Evol. Biosph. 2002, 32, 13–21. [Google Scholar] [CrossRef]
- Sibilska, I.K.; Chen, B.; Li, L.; Yin, J. Effects of trimetaphosphate on abiotic formation and hydrolysis of peptides. Life 2017, 7, 50. [Google Scholar] [CrossRef]
- Sibilska, I.; Feng, Y.; Li, L.; Yin, J. Trimetaphosphate Activates Prebiotic Peptide Synthesis across a Wide Range of Temperature and pH. Orig. Life Evol. Biosph. 2018, 48, 277–287. [Google Scholar] [CrossRef]
- Sibilska-Kaminski, I.K.; Yin, J. Toward Molecular Cooperation by De Novo Peptides. Orig. Life Evol. Biosph. 2021, 51, 71–82. [Google Scholar] [CrossRef]
- Larralde, R.; Robertson, M.P.; Miller, S.L. Rates of decomposition of ribose and other sugars: Implications for chemical evolution. Proc. Natl. Acad. Sci. USA 1995, 92, 8158–8160. [Google Scholar] [CrossRef]
- Caetano-Anollés, G.; Caetano-Anollés, D. Computing the origin and evolution of the ribosome from its structure—Uncovering processes of macromolecular accretion benefiting synthetic biology. Comput. Struct. Biotechnol. J. 2015, 13, 427–447. [Google Scholar] [CrossRef] [PubMed]
- Baaske, P.; Weinert, F.M.; Duhr, S.; Lemke, K.H.; Russell, M.J.; Braun, D. Extreme accumulation of nucleotides in simulated hydrothermal pore systems. Proc. Natl. Acad. Sci. USA 2007, 104, 9346–9351. [Google Scholar] [CrossRef] [PubMed]
- Navrotsky, A.; Hervig, R.; Lyons, J.; Seo, D.-K.; Shock, E.; Voskanyan, A. Cooperative formation of porous silica and peptides on the prebiotic Earth. Proc. Natl. Acad. Sci. USA 2021, 118, e2021117118. [Google Scholar] [CrossRef]
- Damm, V. Chemistry of submarine hydrothermal solutions at Guaymas Basin, Gulf of California. Gechemica Cosmochim. AGU 1985, 49, 2221–2237. [Google Scholar] [CrossRef]
- Potter, R.J.; Barnes, H.L. The Kuroko and Related Volcanogenic Massive Sulfide Deposits; Ohmoto, H., Skinner, B.J., Eds.; Society of Economic Geologists: Littleton, CO, USA, 1983; pp. 198–223. [Google Scholar]
- Liu, C.Z.; Dick, H.J.; Mitchell, R.N.; Wei, W.; Zhang, Z.Y.; Hofmann, A.W.; Yang, J.F.; Li, Y. Archean cratonic mantle recycled at a mid-ocean ridge. Sci. Adv. 2022, 8, eabn6749. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Halley, J.W. Some Factors from Theory, Simulation, Experiment and Proteomes in the Current Biosphere Supporting Deep Oceans as the Location of the Origin of Terrestrial Life. Life 2022, 12, 1330. https://doi.org/10.3390/life12091330
Halley JW. Some Factors from Theory, Simulation, Experiment and Proteomes in the Current Biosphere Supporting Deep Oceans as the Location of the Origin of Terrestrial Life. Life. 2022; 12(9):1330. https://doi.org/10.3390/life12091330
Chicago/Turabian StyleHalley, J. W. 2022. "Some Factors from Theory, Simulation, Experiment and Proteomes in the Current Biosphere Supporting Deep Oceans as the Location of the Origin of Terrestrial Life" Life 12, no. 9: 1330. https://doi.org/10.3390/life12091330
APA StyleHalley, J. W. (2022). Some Factors from Theory, Simulation, Experiment and Proteomes in the Current Biosphere Supporting Deep Oceans as the Location of the Origin of Terrestrial Life. Life, 12(9), 1330. https://doi.org/10.3390/life12091330