Abstract
Immune checkpoint inhibitors (ICIs) have a major impact on cancer treatment. However, the therapeutic efficacy of ICIs is only effective in some patients. Programmed death ligand 1 (PD-L1), tumor mutation burden (TMB), and high-frequency microsatellite instability (MSI-high) are markers that predict the efficacy of ICIs but are not universally used in many carcinomas. The gut microbiota has received much attention recently because of its potential to have a significant impact on immune cells in the cancer microenvironment. Metabolites of the gut microbiota modulate immunity and have a strong influence on the therapeutic efficacy of ICI. It has been suggested that the gut microbiota may serve as a novel marker to predict the therapeutic efficacy of ICI. Therefore, there is an urgent need to develop biomarkers that can predict anti-tumor effects and adverse events, and the study of the gut microbiota is essential in this regard.
1. Introduction
Immune checkpoint inhibitors (ICIs) have a significant impact on cancer treatment. However, further improvement in the efficacy of ICIs is required. For this reason, there is an urgent need for biomarkers of high efficacy and prognostic value. Therefore, it is important to investigate the conditions of the cancer microenvironment in which the efficacy of ICIs can be demonstrated. The expression of programmed death ligand 1 (PD-L1) on tumor cells, tumor mutation burden (TMB), and high-frequency microsatellite instability (MSI-high) have been reported as biomarkers for the efficacy of anti-PD-1/PD-L1 antibodies. However, the expression of PD-L1 has limitations as a biomarker because of its heterogeneity within and between tumors. Additionally, PD-L1 expression itself is transient and changes with the immune response over time and space. In the IMpower 133 trial, the efficacy of carboplatin and etoposide with atezolizumab, an anti-PD-L1 antibody, was analyzed based on the PD-L1 expression on tumor cells, but there was no difference in the efficacy between patients with PD-L1 expression and those without [1]. There is no clear biomarker for anti-PD-L1 combination chemotherapy for small cell lung cancer [1]. In addition, intestinal bacteria have been attracting attention in recent years because they may have a significant impact on the immune cells in the cancer microenvironment.
In addition to monotherapy with anti-PD-1/PD-L1 antibodies, combination therapy with chemotherapy and molecular target therapy as well as combination therapy with anti-PD-1 and anti-cytotoxic T lymphocyte antigen-4 (anti-CTLA-4) antibodies is now widely used in cancer treatment with ICIs [2]. As a result, there is an urgent need to develop biomarkers that can predict the anti-tumor response and adverse events, and research on intestinal bacteria is essential in this regard.
2. Mechanism of Action of Anti-PD-1/PD-L1 and Anti-CTLA-4 Antibodies
The immunosuppressive immune checkpoints pathway is an important immune resistance mechanism in cancer [3]. Programmed cell death 1 (PD-1) is one of the critical immune checkpoint receptors that are primarily expressed on a variety of immune cells, including activated T, B, regulatory T (Treg), and dendritic cells (DC) [4]. PD-1 and PD-L1 are immune checkpoints that regulate adaptive immune responses to prevent excessive activation and maintain immune homeostasis [3,4]. Anti-PD-1/PD-L1 antibodies mainly suppress the negative regulatory mechanisms between tumors and T cells. This is referred to as the effector phase [5]. The anti-PD-1 antibodies pembrolizumab and nivolumab showed promising results in non-small cell lung cancer (NSCLC) and melanoma patients [6,7,8]. Subsequently, their efficacy was demonstrated in many cancers, including urothelial bladder cancer, renal cell carcinoma, gastric cancer, Hodgkin’s lymphoma, and head and neck squamous cell carcinoma [9,10,11,12,13,14]. Atezolizumab was the first PD-L1 antibody to be approved by the FDA for non-small cell lung cancer patients [15]. Avelumab showed efficacy against Merkel cell carcinoma [16].
Furthermore, atetzolizumab administered after adjuvant chemotherapy in patients with resected stage IB-IIIA non-small cell lung cancer statistically significantly prolonged disease-free survival compared to best supportive care after adjuvant chemotherapy [17].
In contrast, anti-CTLA-4 antibodies maintain T-cell activation by blocking inhibitory signals from dendritic cells in the lymph nodes [18]. This phase is called the priming phase [5]. The primary mechanism by which CTLA-4 suppresses T-cell activation is through transendocytosis, which traps the ligand CD86 from DCs and sends a negative signal to T cells. Transendocytosis can be prevented by the anti-CTLA-4 antibody, which maintains the expression of the CD86 ligand [19]. Transendocytosis of CD86 on Tregs results in the removal of the ligand from antigen-presenting cells (APCs) and prevents normal T-cell activation, resulting in an antitumor effect [20]. Ipilimumab, a CTLA-4 inhibitor, was the first approved ICI for patients with advanced melanoma [21,22] (Table 1).

Table 1.
Types of cancer effective for ICI (monotherapy).
3. PD-L1 Expression in Tumor Cells
PD-L1 is a ligand for PD-1 [4]. PD-L1 has been used as a biomarker to predict clinical response to PD-1 or PD-L1 antibody therapy [26]. From 2011 through April 2019, only some immune checkpoint inhibitors approved by the US Food and Drug Administration (FDA) approved PD-L1 as a companion diagnostic [27]. During the above time period, FDA-approved immune checkpoint inhibitor carcinomas included non-small cell lung cancer (NSCLC), melanoma, bladder, renal, head and neck, colon, hepatocellular, small cell lung cancer, Merkel cell carcinoma, squamous cell carcinoma of the skin, Hodgkin lymphoma, breast, gastric/gastroesophageal junction (GEJ), primary mediastinal B-cell lymphoma, and MSI-high/mismatch repair-deficient status [27]. Among the above, PD-L1 is FDA-approved as a companion diagnostic for NSCLC, bladder cancer, triple-negative breast cancer, cervical cancer, and gastric/GEJ cancer [27].
However, the type of PD-L1-expressing cells to be measured differs depending on the carcinoma. The immunohistochemistry for PD-L1 is based on PD-L1 staining on tumor cells for NSCLC. In contrast, triple-negative breast cancer was approved based on tumor-infiltrating immune cells, and cervical cancer was approved based on a combined tumor and immune cell percentage score [27,28,29].
The problem is the heterogeneity of PD-L1 expression within and between tumors, and PD-L1 expression itself is transient and changes over time and space due to the immune response [30] (Table 2).

Table 2.
Predictive biomarkers for treatment of immune checkpoint inhibitors.
4. Tumor Mutation Burden
Genetic mutations in tumors can generate immunogenic neoplastic antigens, which have been investigated as predictive biomarkers of the response to ICIs [33]. The KEYNOTE-158 study showed that pembrolizumab was effective in treating tumors of anal, bile duct, cervical, endometrial, mesothelioma, neuroendocrine, salivary, small cell lung, thyroid, and vulvar origins that had high oncogene mutation scores (TMB-high, ≥10 mutations per megabase). Pembrolizumab was also effective in tumors of salivary, small-cell lung, thyroid, and vulvar origin with high oncogene mutation scores (TMB-high) [23]. The FDA approved pembrolizumab monotherapy in 2020 for the treatment of adult and pediatric patients with TMB-high, previously unresectable, or metastatic solid tumors [31,32]. However, TMB-high has not been demonstrated in all cancers; glioma, prostate cancer, and breast cancer were not included in the KEYNOTE-158 study. McGrail et al. divided TMB-high tumors into two groups, those in which the neoantigen levels correlated with the number of CD8 T cells in the tumor and those in which the number of CD8 T cells did not correlate, and analyzed the effect of treatment with ICIs [48]. Some patients with TMB do not respond to PD-L1 antibodies or anti-CTLA4 [49,50]. Their findings suggest that TMB-high does not uniformly predict the efficacy of ICI therapy (Table 2).
5. High-Frequency Microsatellite Instability and DNA Mismatch Repair
Microsatellites are repeats of a few nucleotides in DNA. In the case of defective DNA mismatch repair (MMR), the number of microsatellite repeats differs from the normal number of DNA repeats because of the inability to repair mistakes during DNA replication [51]. MSI is usually caused by germline mutations in components of mismatch repair (MSH2, MSH6, MLH1, PMS2) in patients with Lynch syndrome or somatic hypermethylation of the MLH1 promoter [34]. This is called MSI-high [52]. It is more frequent in gastrointestinal and gynecological cancers [53]. MSI-high colorectal cancer (CRC) has shown a higher mutation load than microsatellite stable CRC, and a high somatic mutation load was associated with prolonged progression-free survival (PFS) [54]. A phase III randomized controlled trial (KEYNOTE-177) was conducted in patients with unresectable or metastatic MSI-H/dMMR CRC [24]. Patients with MSI-H/dMMR metastatic CRC who were treated with pembrolizumab as a first-line therapy demonstrated a significantly longer PFS than those treated with chemotherapy. Pembrolizumab has become the first-line treatment for unresectable or metastatic MSI-H/dMMR CRC [24]. Pembrolizumab was also approved by FDA in 2017 for the treatment of patients with unresectable or metastatic, microsatellite instability-high (MSI-H), or mismatch repair deficient (dMMR) solid tumors [25]. MSI-high is a predictive biomarker for response to ICI [33,34].
Dostarlimab, an anti-PD-1 monoclonal antibody, was administered for 6 months as neoadjuvant therapy to patients with deficient mismatch repair (dMMR)–stage II or III rectal adenocarcinoma. After 6 months of treatment, twelve patients showed clinically complete responses [55]. This result suggests that dMMR is a predictive marker for the efficacy of preoperative immunotherapy for rectal cancer (Table 2).
6. Intestinal Bacteria
Approximately 100 trillion different bacteria coexist in the human intestine, forming intestinal microflora (also called the intestinal flora) weighing 1.5 to 2 kg [56]. However, with advancements in technology, the number of intestinal bacteria in the body is now being disputed, and this number may be revised in the future. How these intestinal bacteria coexist with humans is not known. However, it is an extremely important relationship, and it has become known in recent years that the disruption of the relationship with the intestinal microflora causes inflammatory bowel disease, rheumatic diseases, obesity, diabetes, atopy, and allergies. This disordered state of the intestinal microflora is called dysbiosis, which means a breakdown in the composition of the intestinal bacteria [57].
Rapid progress in the analysis of gut microbiota began with the advent of next-generation sequencing. In other words, it came about as a result of the instantaneous availability of large amounts of genetic analysis techniques. Before the full-scale introduction of next-generation sequencing, it was known that the bacterial genome, which consists of several million base pairs, contains a polymorphic region of approximately 1600 base pairs called the 16S ribosomal RNA region. The 16S ribosomal RNA region has nine hypervariable regions consisting of tens to hundreds of base pairs, which have characteristic sequences depending on the type of bacteria. The sequences of these hypervariable regions are conserved among the same bacterial species, and it is believed that the bacterial species can be identified by reading and analyzing the full or partial length of the 16S region without sequencing the entire bacterial genome.
In the analysis of intestinal microflora by 16S metagenomics, the DNA of enterobacteria in feces is extracted and purified as a template, and the gene is amplified using primers designed to amplify a part of the 16S region. The gene sequences contained in the library are read using a next-generation sequencer and matched with a database to identify the type of bacteria. Using this genetic testing method, it is possible to detect DNA fragments of living bacteria in feces and DNA fragments of dead bacteria that cannot be detected by the culture method, and it is believed to provide accurate information regarding the intestinal microflora. The process of gene extraction and purification from feces, gene amplification, purification and quantification of amplified products, library preparation, sequencing, identification, and calculation of the percentage of bacteria using analysis software can be completed in about 3 days. In the future, it is expected that the information obtained will be successively compiled into databases (Figure 1). Recent technological innovations have led to the accumulation of a considerable amount of information on intestinal bacteria and diseases, especially cancer [58,59].
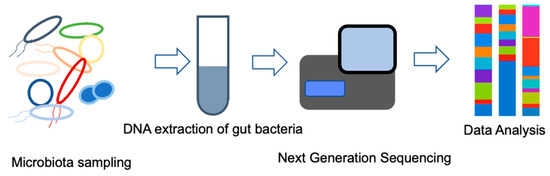
Figure 1.
Microbiome identification by 16S rRNA gene analysis.
7. Composition of the Intestinal Microflora
The formation of human gut microbiota begins immediately after birth. The gut flora formed in the neonatal period is not constant throughout life, and the composition of the constituent bacteria changes with age [58]. The formation process of the intestinal flora can vary and is affected by various environmental factors such as the duration of fetal life, mode of delivery, and mode of lactation [59]. According to the research on dysbiosis described above, it is important to have a good balance between the so-called good and bad bacteria in the intestinal microbiota, and it has often been reported that oligotrophic anaerobic bacteria (fermentative bacteria) are good bacteria, whereas commensal anaerobic bacteria are bad bacteria. However, with recent advances in the study of dysbiosis, either this rule does not necessarily apply, or the terms “good” and “bad” bacteria themselves are being less commonly used [36,37,38,60,61,62,63].
The effect of gut bacteria on ICI therapy has been reported by research groups in the USA and in France [35,36,37,38]. They have claimed that certain gut bacteria may modulate the clinical effects of anti-PD-1 antibodies. However, the gut microbiota reported by each group was different and has not been identified yet. In addition, the pattern of gut microbiota differs among countries and diets (Table 2).
8. Microbiota Is a Potential Biomarker of ICIs
Various studies have been conducted to explore how intestinal bacteria act on the immune system. It has been shown that the involvement of single-chain fatty acids is a major mechanism of action. It is believed that the actions of single-chain fatty acids may change due to differences in their receptors, but the details need to be clarified in future studies [64].
It is important to note that many researchers, including us, are currently conducting research aimed at identifying the immune states that are likely to cause immune-related adverse events (irAEs) and those that are likely to result in effective immunotherapy by analyzing the intestinal bacteria. Research is underway to induce a state in which irAEs are less likely to occur, and immunotherapy is more likely to be effective by using various therapies, including the modification of the intestinal microbiota. If the optimal immune conditions can be estimated using biomarkers, treatment strategies, especially the management of adverse events, will become easier.
9. Attenuation of ICI Efficacy Via the Effects of Antibiotics on Gut Bacteria
In the field of cancer research, data are emerging that gut microbiota may be highly correlated with the therapeutic effects of cancer immunotherapy [65,66,67]. It is also believed that intestinal bacteria may be involved in cancers of the esophagus, stomach, and many other organs [68].
Interestingly, a growing body of evidence suggests that antibiotics have a strongly negative impact on gut bacteria [69]. In addition, there are some interesting data on the negative effects of antibiotics during treatment with ICIs.
We compared NSCLC patients receiving anti-PD-1 antibody treatment with and without antibiotics and found that both the PFS and overall survival (OS) were significantly lower in the group that used some antibiotics within 3 weeks before and after the start of anti-PD-1 antibody treatment than those in the group that did not use antibiotics. In other words, antibiotic use hampers the efficacy of ICIs [70]. Using antibiotics is known to cause dysbiosis of the gut microbiota [71], suggesting that dysbiosis could reduce the efficacy of PD-1 antibody therapy.
10. Metabolites of Gut Bacteria Affect Immunity
The above-mentioned ectopic anaerobic bacteria produce short-chain fatty acids (SCFAs) by fermentation using dietary fiber as a nutrient source [72,73,74,75]. These SCFAs include propionate, acetate, and butyrate, all of which have fewer than six carbons and are believed to influence immune activity and regulation. Certain microbiota, such as Akkermansia muciniphila, modulate the activation of monocytes, dendritic cells, and macrophages in the cancer microenvironment via bacterially derived substances. Thereby, anti-tumor immunity is activated, and the efficacy of anti-PD-1 immunotherapy is enhanced [76] (Figure 2). In any case, SCFAs play important roles in human immunity and homeostasis, such as the induction of Tregs and type 1 helper T cells and the maintenance of intestinal epithelial cell proliferation [64]. However, SCFAs have not been shown to be effective against tumors, and there are still many unresolved questions on the relationship between dietary fiber and anti-tumor effects. SCFAs certainly play a vital role, and research on the importance of fiber in the diet and the effects of each SCFA on immunity is becoming increasingly important [77,78]. Recently, in addition to SCFAs, metabolites produced by intestinal bacteria have been extensively studied. It is known that commensal anaerobes have a low expression of digestive enzymes that digest dietary fiber [79]. It is also known that they utilize nutrient sources that are abundant in Westernized diets, such as monosaccharides, disaccharides, fats, proteins, and alcohol, rather than dietary fiber.
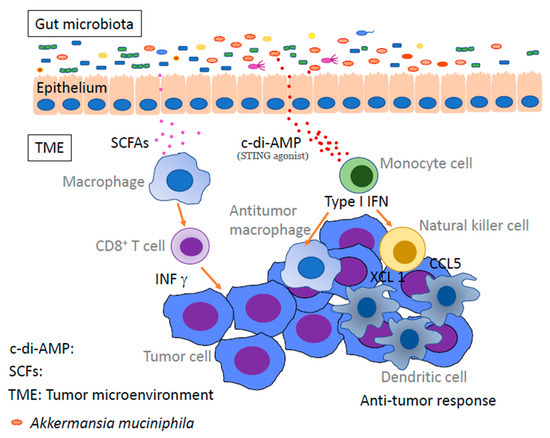
Figure 2.
Gut microbiota enhances antitumor immunity via microbial metabolites. Microbe-derived short-chain fatty acids and STING agonists enhance anti-tumor immunity by activating monocytes, dendritic cells, anti-tumor macrophages, and CD8 T cells in the tumor microenvironment. TME: tumor microenvironment, SCFAs: short-chain fatty acids, c-di-AMP: c-di-adenosine monophosphate.
These SCFAs are generally believed to increase anti-tumor activity, but there are also data that they may inhibit some types of cancer immunity or in certain conditions. A representative example of this is a mouse study that demonstrated that sodium butyrate inhibited anti-CTLA-4-induced dendritic cell (DC) maturation and T-cell priming [40]. Therefore, further studies are needed to elucidate the mechanism underlying the effects of individual SCFAs on cancer immunity.
11. Other Potential Biomarkers for ICIs
The analysis of immune cells in peripheral blood is a non-invasive method of predicting the outcome of immunotherapy treatments [80]. In patients with stage IV melanoma, the high frequency of CD14+ CD16− HLA-DRhi monocytes in peripheral blood samples before anti-PD-1 immunotherapy was a strong predictor of PFS and OS in response to PD-1 immunotherapy [44]. In patients with metastatic melanoma, a higher fraction of activated CD4 effector memory T cells, which lack expression of CCR7 and SELL (CD62L), is associated with a higher incidence of severe irAEs within 3 months [81]. In patients with NSCLC and gastric cancer, significance was found to correlate with decreased soluble PD-L1 levels and tumor regression after four cycles of PD-1 Ab [41]. Changes in plasma soluble PD-1 concentrations before and after two and four cycles of anti-PD-1 antibody treatment were significantly correlated with tumor size progression [42]. PD-L1+ CD14+ monocytes in peripheral blood are correlated with shorter survival in patients treated with anti-PD-1 antibodies, including NSCLC, gastric cancer, melanoma, parotid cancer, and bladder cancer [43]. The neutrophil-to-lymphocyte ratio (NLR) has the potential to predict treatment outcomes in patients receiving ICIs [45,46,82]. Furthermore, the probability of obtaining an ICI benefit is significantly higher in the NLR low/TMB high group compared to the NLR high/TMB low group (OR = 3.22; 95% CI, 2.26–4.58; p < 0.001) [47]. CXCL13 in the tumor microenvironment of many different types of cancer, and CXCL13 is associated with favorable outcomes in cancer patients treated with ICIs [39].
12. Conclusions
Although PD-L1, TMB, and MSI-high are predictive markers for the efficacy of ICIs, they are not common in many carcinomas. Recently, it has been recognized that the gut microbiota functions as a single organ. It is becoming clear that the gut microbiota influences many diseases, including cancer, through its metabolism. Metabolites of the gut microbiota have been shown to modulate immunity and have a strong effect on the therapeutic efficacy of ICIs. Accumulating evidence suggests that the gut microbiota may act as a novel predictive marker for the efficacy of ICIs.
Author Contributions
Writing—original draft preparation, K.H. and K.Y.; writing—review and editing, K.H., K.Y. and T.T.; funding acquisition, K.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Liu, S.V.; Reck, M.; Mansfield, A.S.; Mok, T.; Scherpereel, A.; Reinmuth, N.; Garassino, M.C.; De Castro Carpeno, J.; Califano, R.; Nishio, M.; et al. Updated Overall Survival and PD-L1 Subgroup Analysis of Patients with Extensive-Stage Small-Cell Lung Cancer Treated With Atezolizumab, Carboplatin, and Etoposide (IMpower133). J. Clin. Oncol. 2021, 39, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Robert, C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat. Commun. 2020, 11, 3801. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Freeman, G.J.; Long, A.J.; Iwai, Y.; Bourque, K.; Chernova, T.; Nishimura, H.; Fitz, L.J.; Malenkovich, N.; Okazaki, T.; Byrne, M.C.; et al. Engagement of the Pd-1 Immunoinhibitory Receptor by a Novel B7 Family Member Leads to Negative Regulation of Lymphocyte Activation. J. Exp. Med. 2000, 192, 1027–1034. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Oncology Meets Immunology: The Cancer-Immunity Cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef]
- Garon, E.B.; Rizvi, N.A.; Hui, R.; Leighl, N.; Balmanoukian, A.S.; Eder, J.P.; Patnaik, A.; Aggarwal, C.; Gubens, M.; Horn, L.; et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 2015, 372, 2018–2028. [Google Scholar] [CrossRef]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Cowey, C.L.; Lao, C.D.; Schadendorf, D.; Dummer, R.; Smylie, M.; Rutkowski, P.; et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med. 2015, 373, 23–34. [Google Scholar] [CrossRef]
- Cheng, W.; Fu, D.; Xu, F.; Zhang, Z. Unwrapping the genomic characteristics of urothelial bladder cancer and successes with immune checkpoint blockade therapy. Oncogenesis 2018, 7, 2. [Google Scholar] [CrossRef]
- Ansell, S.M.; Lesokhin, A.M.; Borrello, I.; Halwani, A.; Scott, E.C.; Gutierrez, M.; Schuster, S.J.; Millenson, M.M.; Cattry, D.; Freeman, G.J.; et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N. Engl. J. Med. 2015, 372, 311–319. [Google Scholar] [CrossRef]
- Kang, Y.K.; Boku, N.; Satoh, T.; Ryu, M.H.; Chao, Y.; Kato, K.; Chung, H.C.; Chen, J.S.; Muro, K.; Kang, W.K.; et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017, 390, 2461–2471. [Google Scholar] [CrossRef]
- Motzer, R.J.; Escudier, B.; McDermott, D.F.; George, S.; Hammers, H.J.; Srinivas, S.; Tykodi, S.S.; Sosman, J.A.; Procopio, G.; Plimack, E.R.; et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2015, 373, 1803–1813. [Google Scholar] [CrossRef] [PubMed]
- Bellmunt, J.; de Wit, R.; Vaughn, D.J.; Fradet, Y.; Lee, J.L.; Fong, L.; Vogelzang, N.J.; Climent, M.A.; Petrylak, D.P.; Choueiri, T.K.; et al. Pembrolizumab as Second-Line Therapy for Advanced Urothelial Carcinoma. N. Engl. J. Med. 2017, 376, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Burtness, B.; Harrington, K.J.; Greil, R.; Soulières, D.; Tahara, M.; de Castro, G., Jr.; Psyrri, A.; Basté, N.; Neupane, P.; Bratland, Å.; et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): A randomised, open-label, phase 3 study. Lancet 2019, 394, 1915–1928. [Google Scholar] [CrossRef]
- Rittmeyer, A.; Barlesi, F.; Waterkamp, D.; Park, K.; Ciardiello, F.; von Pawel, J.; Gadgeel, S.M.; Hida, T.; Kowalski, D.M.; Dols, M.C.; et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): A phase 3, open-label, multicentre randomised controlled trial. Lancet 2017, 389, 255–265. [Google Scholar] [CrossRef]
- Kaufman, H.L.; Russell, J.; Hamid, O.; Bhatia, S.; Terheyden, P.; D’Angelo, S.P.; Shih, K.C.; Lebbé, C.; Linette, G.P.; Milella, M.; et al. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: A multicentre, single-group, open-label, phase 2 trial. Lancet Oncol. 2016, 17, 1374–1385. [Google Scholar] [CrossRef]
- Felip, E.; Altorki, N.; Zhou, C.; Csőszi, T.; Vynnychenko, I.; Goloborodko, O.; Luft, A.; Akopov, A.; Martinez-Marti, A.; Kenmotsu, H.; et al. Adjuvant atezolizumab after adjuvant chemotherapy in resected stage IB-IIIA non-small-cell lung cancer (IMpower010): A randomised, multicentre, open-label, phase 3 trial. Lancet 2021, 398, 1344–1357. [Google Scholar] [CrossRef]
- Ribas, A. Tumor immunotherapy directed at PD-1. N. Engl. J. Med. 2012, 366, 2517–2519. [Google Scholar] [CrossRef]
- Qureshi, O.S.; Zheng, Y.; Nakamura, K.; Attridge, K.; Manzotti, C.; Schmidt, E.M.; Baker, J.; Jeffery, L.E.; Kaur, S.; Briggs, Z.; et al. Trans-endocytosis of CD80 and CD86: A molecular basis for the cell-extrinsic function of CTLA-4. Science 2011, 332, 600–603. [Google Scholar] [CrossRef]
- Soskic, B.; Qureshi, O.S.; Hou, T.; Sansom, D.M. A transendocytosis perspective on the CD28/CTLA-4 pathway. Adv. Immunol. 2014, 124, 95–136. [Google Scholar] [CrossRef]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Thomas, L.; Bondarenko, I.; O’Day, S.; Weber, J.; Garbe, C.; Lebbe, C.; Baurain, J.F.; Testori, A.; Grob, J.J.; et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N. Engl. J. Med. 2011, 364, 2517–2526. [Google Scholar] [CrossRef] [PubMed]
- Marabelle, A.; Fakih, M.; Lopez, J.; Shah, M.; Shapira-Frommer, R.; Nakagawa, K.; Chung, H.C.; Kindler, H.L.; Lopez-Martin, J.A.; Miller, W.H., Jr.; et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: Prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020, 21, 1353–1365. [Google Scholar] [CrossRef]
- André, T.; Shiu, K.K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab in Microsatellite-Instability-High Advanced Colorectal Cancer. N. Engl. J. Med. 2020, 383, 2207–2218. [Google Scholar] [CrossRef] [PubMed]
- Marcus, L.; Lemery, S.J.; Keegan, P.; Pazdur, R. FDA Approval Summary: Pembrolizumab for the Treatment of Microsatellite Instability-High Solid Tumors. Clin. Cancer Res. 2019, 25, 3753–3758. [Google Scholar] [CrossRef]
- Tsao, M.S.; Kerr, K.M.; Kockx, M.; Beasley, M.B.; Borczuk, A.C.; Botling, J.; Bubendorf, L.; Chirieac, L.; Chen, G.; Chou, T.Y.; et al. PD-L1 Immunohistochemistry Comparability Study in Real-Life Clinical Samples: Results of Blueprint Phase 2 Project. J. Thorac. Oncol. 2018, 13, 1302–1311. [Google Scholar] [CrossRef]
- Davis, A.A.; Patel, V.G. The role of PD-L1 expression as a predictive biomarker: An analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J. Immunother Cancer 2019, 7, 278. [Google Scholar] [CrossRef]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Hegg, R.; Im, S.A.; Shaw Wright, G.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef]
- Colombo, N.; Dubot, C.; Lorusso, D.; Caceres, M.V.; Hasegawa, K.; Shapira-Frommer, R.; Tewari, K.S.; Salman, P.; Hoyos Usta, E.; Yañez, E.; et al. Pembrolizumab for Persistent, Recurrent, or Metastatic Cervical Cancer. N. Engl. J. Med. 2021, 385, 1856–1867. [Google Scholar] [CrossRef]
- Shklovskaya, E.; Rizos, H. Spatial and Temporal Changes in PD-L1 Expression in Cancer: The Role of Genetic Drivers, Tumor Microenvironment and Resistance to Therapy. Int. J. Mol. Sci. 2020, 21, 7139. [Google Scholar] [CrossRef]
- Marcus, L.; Fashoyin-Aje, L.A.; Donoghue, M.; Yuan, M.; Rodriguez, L.; Gallagher, P.S.; Philip, R.; Ghosh, S.; Theoret, M.R.; Beaver, J.A.; et al. FDA Approval Summary: Pembrolizumab for the Treatment of Tumor Mutational Burden–High Solid Tumors. Clin. Cancer Res. 2021, 27, 4685–4689. [Google Scholar] [CrossRef] [PubMed]
- Yarchoan, M.; Hopkins, A.; Jaffee, E.M. Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 2017, 377, 2500–2501. [Google Scholar] [CrossRef]
- Rizzo, A.; Ricci, A.D.; Brandi, G. PD-L1, TMB, MSI, and Other Predictors of Response to Immune Checkpoint Inhibitors in Biliary Tract Cancer. Cancers 2021, 13, 558. [Google Scholar] [CrossRef] [PubMed]
- Dudley, J.C.; Lin, M.T.; Le, D.T.; Eshleman, J.R. Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin. Cancer Res. 2016, 22, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Chaput, N.; Lepage, P.; Coutzac, C.; Soularue, E.; Le Roux, K.; Monot, C.; Boselli, L.; Routier, E.; Cassard, L.; Collins, M.; et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017, 28, 1368–1379. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.-L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Hsieh, C.H.; Jian, C.Z.; Lin, L.I.; Low, G.S.; Ou, P.Y.; Hsu, C.; Ou, D.L. Potential Role of CXCL13/CXCR5 Signaling in Immune Checkpoint Inhibitor Treatment in Cancer. Cancers 2022, 14, 294. [Google Scholar] [CrossRef]
- Coutzac, C.; Jouniaux, J.-M.; Paci, A.; Schmidt, J.; Mallardo, D.; Seck, A.; Asvatourian, V.; Cassard, L.; Saulnier, P.; Lacroix, L.; et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat. Commun. 2020, 11, 2168. [Google Scholar] [CrossRef]
- Ando, K.; Hamada, K.; Watanabe, M.; Ohkuma, R.; Shida, M.; Onoue, R.; Kubota, Y.; Matsui, H.; Ishiguro, T.; Hirasawa, Y.; et al. Plasma Levels of Soluble PD-L1 Correlate with Tumor Regression in Patients with Lung and Gastric Cancer Treated with Immune Checkpoint Inhibitors. Anticancer Res. 2019, 39, 5195–5201. [Google Scholar] [CrossRef]
- Ohkuma, R.; Ieguchi, K.; Watanabe, M.; Takayanagi, D.; Goshima, T.; Onoue, R.; Hamada, K.; Kubota, Y.; Horiike, A.; Ishiguro, T.; et al. Increased Plasma Soluble PD-1 Concentration Correlates with Disease Progression in Patients with Cancer Treated with Anti-PD-1 Antibodies. Biomedicines 2021, 9, 1929. [Google Scholar] [CrossRef] [PubMed]
- Ando, K.; Hamada, K.; Shida, M.; Ohkuma, R.; Kubota, Y.; Horiike, A.; Matsui, H.; Ishiguro, T.; Hirasawa, Y.; Ariizumi, H.; et al. A high number of PD-L1(+) CD14(+) monocytes in peripheral blood is correlated with shorter survival in patients receiving immune checkpoint inhibitors. Cancer Immunol. Immunother. 2021, 70, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Krieg, C.; Nowicka, M.; Guglietta, S.; Schindler, S.; Hartmann, F.J.; Weber, L.M.; Dummer, R.; Robinson, M.D.; Levesque, M.P.; Becher, B. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat. Med. 2018, 24, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Ferrucci, P.; Gandini, S.; Battaglia, A.; Alfieri, S.; Di Giacomo, A.; Giannarelli, D.; Cappellini, G.; De Galitiis, F.; Marchetti, P.; Amato, G. Baseline neutrophil-to-lymphocyte ratio is associated with outcome of ipilimumab-treated metastatic melanoma patients. Br. J. Cancer 2015, 112, 1904–1910. [Google Scholar] [CrossRef] [PubMed]
- Capone, M.; Giannarelli, D.; Mallardo, D.; Madonna, G.; Festino, L.; Grimaldi, A.M.; Vanella, V.; Simeone, E.; Paone, M.; Palmieri, G. Baseline neutrophil-to-lymphocyte ratio (NLR) and derived NLR could predict overall survival in patients with advanced melanoma treated with nivolumab. J. Immunother. Cancer 2018, 6, 74. [Google Scholar] [CrossRef]
- Valero, C.; Lee, M.; Hoen, D.; Weiss, K.; Kelly, D.W.; Adusumilli, P.S.; Paik, P.K.; Plitas, G.; Ladanyi, M.; Postow, M.A.; et al. Pretreatment neutrophil-to-lymphocyte ratio and mutational burden as biomarkers of tumor response to immune checkpoint inhibitors. Nat. Commun. 2021, 12, 729. [Google Scholar] [CrossRef]
- McGrail, D.J.; Pilié, P.G.; Rashid, N.U.; Voorwerk, L.; Slagter, M.; Kok, M.; Jonasch, E.; Khasraw, M.; Heimberger, A.B.; Lim, B.; et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann. Oncol. 2021, 32, 661–672. [Google Scholar] [CrossRef]
- Chan, T.A.; Wolchok, J.D.; Snyder, A. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N. Engl. J. Med. 2015, 373, 1984. [Google Scholar] [CrossRef]
- Rosenberg, J.E.; Hoffman-Censits, J.; Powles, T.; van der Heijden, M.S.; Balar, A.V.; Necchi, A.; Dawson, N.; O’Donnell, P.H.; Balmanoukian, A.; Loriot, Y.; et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: A single-arm, multicentre, phase 2 trial. Lancet 2016, 387, 1909–1920. [Google Scholar] [CrossRef]
- Luchini, C.; Bibeau, F.; Ligtenberg, M.J.L.; Singh, N.; Nottegar, A.; Bosse, T.; Miller, R.; Riaz, N.; Douillard, J.Y.; Andre, F.; et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: A systematic review-based approach. Ann. Oncol. 2019, 30, 1232–1243. [Google Scholar] [CrossRef] [PubMed]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087.e2073. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef]
- Cercek, A.; Lumish, M.; Sinopoli, J.; Weiss, J.; Shia, J.; Lamendola-Essel, M.; El Dika, I.H.; Segal, N.; Shcherba, M.; Sugarman, R.; et al. PD-1 Blockade in Mismatch Repair–Deficient, Locally Advanced Rectal Cancer. N. Engl. J. Med. 2022, 386, 2363–2376. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Cani, P.D. Gut microbiota—At the intersection of everything? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 321–322. [Google Scholar] [CrossRef]
- Arboleya, S.; Binetti, A.; Salazar, N.; Fernández, N.; Solís, G.; Hernández-Barranco, A.; Margolles, A.; de Los Reyes-Gavilán, C.G.; Gueimonde, M. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol. Ecol. 2012, 79, 763–772. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 852. [Google Scholar] [CrossRef]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef]
- Ahn, J.; Sinha, R.; Pei, Z.; Dominianni, C.; Wu, J.; Shi, J.; Goedert, J.J.; Hayes, R.B.; Yang, L. Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 1907–1911. [Google Scholar] [CrossRef]
- Lepage, P.; Häsler, R.; Spehlmann, M.E.; Rehman, A.; Zvirbliene, A.; Begun, A.; Ott, S.; Kupcinskas, L.; Doré, J.; Raedler, A.; et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 2011, 141, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Dubin, K.; Callahan, M.K.; Ren, B.; Khanin, R.; Viale, A.; Ling, L.; No, D.; Gobourne, A.; Littmann, E.; Huttenhower, C.; et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun. 2016, 7, 10391. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Wu, W.; Liu, Z.; Cong, Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J. Gastroenterol. 2017, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.M.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.-L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Wang, Y.; Wiesnoski, D.H.; Helmink, B.A.; Gopalakrishnan, V.; Choi, K.; DuPont, H.L.; Jiang, Z.-D.; Abu-Sbeih, H.; Sanchez, C.A.; Chang, C.-C.; et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat. Med. 2018, 24, 1804–1808. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef]
- Derosa, L.; Hellmann, M.D.; Spaziano, M.; Halpenny, D.; Fidelle, M.; Rizvi, H.; Long, N.; Plodkowski, A.J.; Arbour, K.C.; Chaft, J.E.; et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann. Oncol. 2018, 29, 1437–1444. [Google Scholar] [CrossRef]
- Hamada, K.; Yoshimura, K.; Hirasawa, Y.; Hosonuma, M.; Murayama, M.; Narikawa, Y.; Ariizumi, H.; Ohkuma, R.; Shida, M.; Kubota, Y.; et al. Antibiotic Usage Reduced Overall Survival by over 70% in Non-small Cell Lung Cancer Patients on Anti-PD-1 Immunotherapy. Anticancer Res. 2021, 41, 4985–4993. [Google Scholar] [CrossRef]
- Dethlefsen, L.; Relman, D.A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4554–4561. [Google Scholar] [CrossRef] [PubMed]
- Tanes, C.; Bittinger, K.; Gao, Y.; Friedman, E.S.; Nessel, L.; Paladhi, U.R.; Chau, L.; Panfen, E.; Fischbach, M.A.; Braun, J.; et al. Role of dietary fiber in the recovery of the human gut microbiome and its metabolome. Cell Host Microbe 2021, 29, 394–407.e395. [Google Scholar] [CrossRef] [PubMed]
- Myhrstad, M.C.W.; Tunsjø, H.; Charnock, C.; Telle-Hansen, V.H. Dietary Fiber, Gut Microbiota, and Metabolic Regulation-Current Status in Human Randomized Trials. Nutrients 2020, 12, 859. [Google Scholar] [CrossRef] [PubMed]
- Makki, K.; Deehan, E.C.; Walter, J.; Bäckhed, F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef]
- Kasubuchi, M.; Hasegawa, S.; Hiramatsu, T.; Ichimura, A.; Kimura, I. Dietary Gut Microbial Metabolites, Short-chain Fatty Acids, and Host Metabolic Regulation. Nutrients 2015, 7, 2839–2849. [Google Scholar] [CrossRef]
- Lam, K.C.; Araya, R.E.; Huang, A.; Chen, Q.; Di Modica, M.; Rodrigues, R.R.; Lopès, A.; Johnson, S.B.; Schwarz, B.; Bohrnsen, E.; et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell 2021, 184, 5338–5356.e5321. [Google Scholar] [CrossRef]
- Sivaprakasam, S.; Prasad, P.D.; Singh, N. Benefits of short-chain fatty acids and their receptors in inflammation and carcinogenesis. Pharmacol. Ther. 2016, 164, 144–151. [Google Scholar] [CrossRef]
- Thiruvengadam, M.; Subramanian, U.; Venkidasamy, B.; Thirupathi, P.; Samynathan, R.; Shariati, M.A.; Rebezov, M.; Chung, I.M.; Rengasamy, K.R.R. Emerging role of nutritional short-chain fatty acids (SCFAs) against cancer via modulation of hematopoiesis. Crit. Rev. Food Sci. Nutr. 2021, 1–18. [Google Scholar] [CrossRef]
- Skelly, A.N.; Sato, Y.; Kearney, S.; Honda, K. Mining the microbiota for microbial and metabolite-based immunotherapies. Nat. Rev. Immunol. 2019, 19, 305–323. [Google Scholar] [CrossRef]
- Gascón, M.; Isla, D.; Cruellas, M.; Gálvez, E.M.; Lastra, R.; Ocáriz, M.; Paño, J.R.; Ramírez, A.; Sesma, A.; Torres-Ramón, I.; et al. Intratumoral versus Circulating Lymphoid Cells as Predictive Biomarkers in Lung Cancer Patients Treated with Immune Checkpoint Inhibitors: Is the Easiest Path the Best One? Cells 2020, 9, 1525. [Google Scholar] [CrossRef]
- Lozano, A.X.; Chaudhuri, A.A.; Nene, A.; Bacchiocchi, A.; Earland, N.; Vesely, M.D.; Usmani, A.; Turner, B.E.; Steen, C.B.; Luca, B.A.; et al. T cell characteristics associated with toxicity to immune checkpoint blockade in patients with melanoma. Nat. Med. 2022, 28, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Joshi, I.; Peravali, M.; Geng, X.; Rao, S.; Chen, K.Y.; Veytsman, I.; Giaccone, G.; Liu, S.V.; Kim, C. Impact of Baseline Clinical Biomarkers on Treatment Outcomes in Patients with Advanced NSCLC Receiving First-line Pembrolizumab-Based Therapy. Clin. Lung Cancer 2022, 23, 438–445. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).