Effects of Heat Stress and Exogenous Salicylic Acid on Secondary Metabolites Biosynthesis in Pleurotus ostreatus (Jacq.) P. Kumm
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Medium
2.2. Salicylic Acid Treatment
2.3. Heat Stress Treatment
2.4. Metabolome Analysis
2.5. Transcriptome Analysis
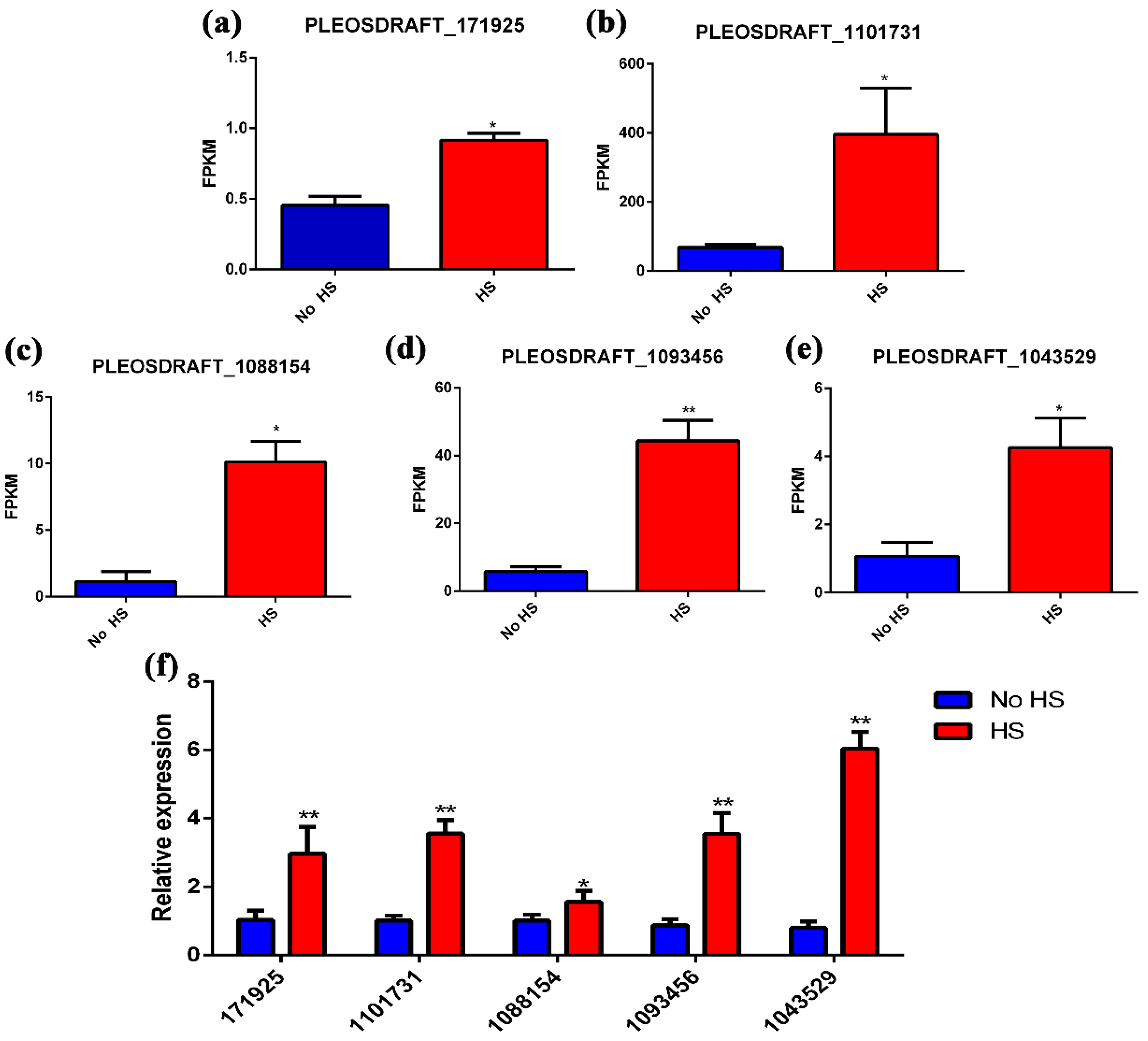
2.6. Gene Expression Analysis
2.7. Significance Analysis
3. Results
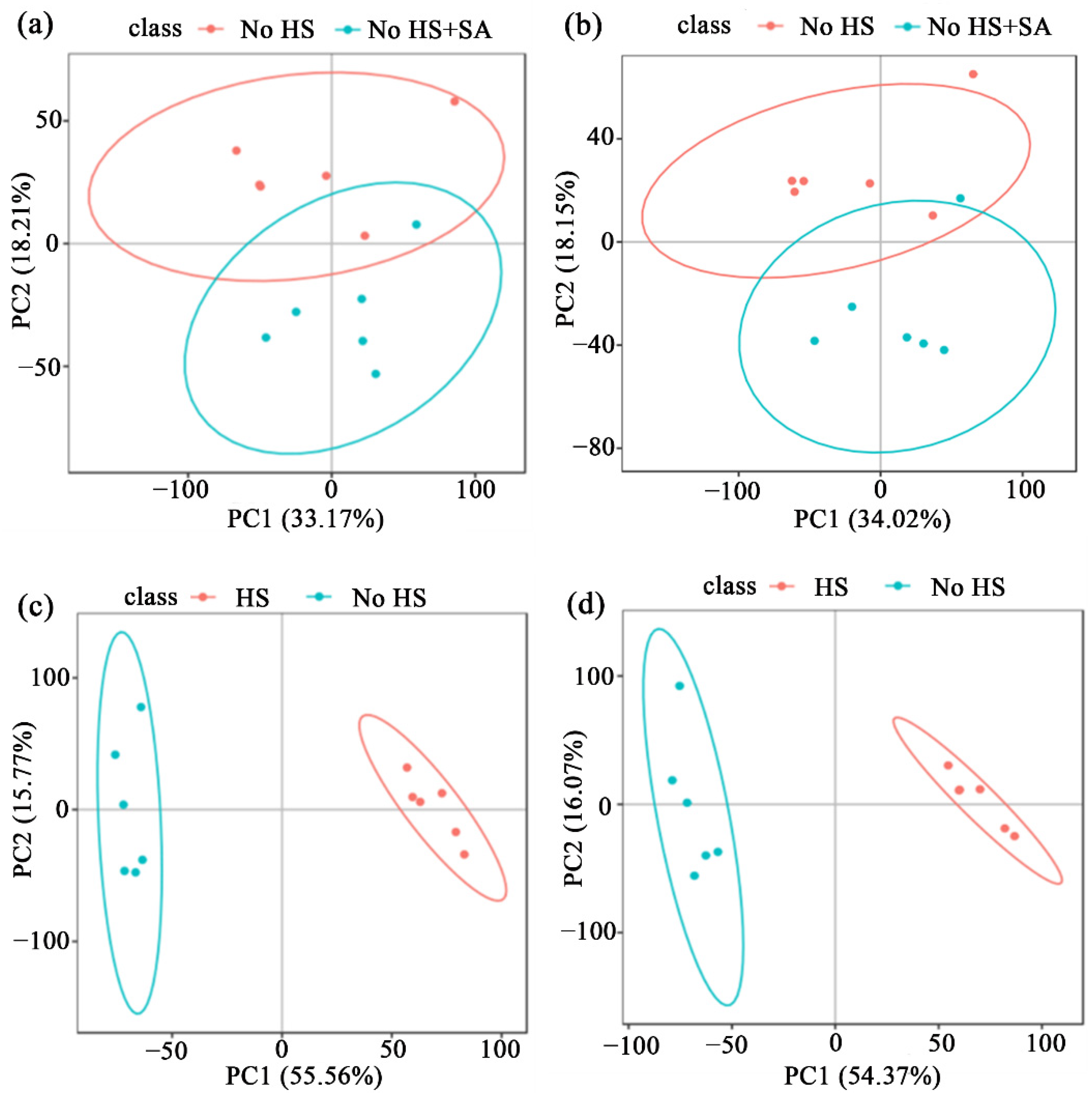
3.1. Metabolome Analysis
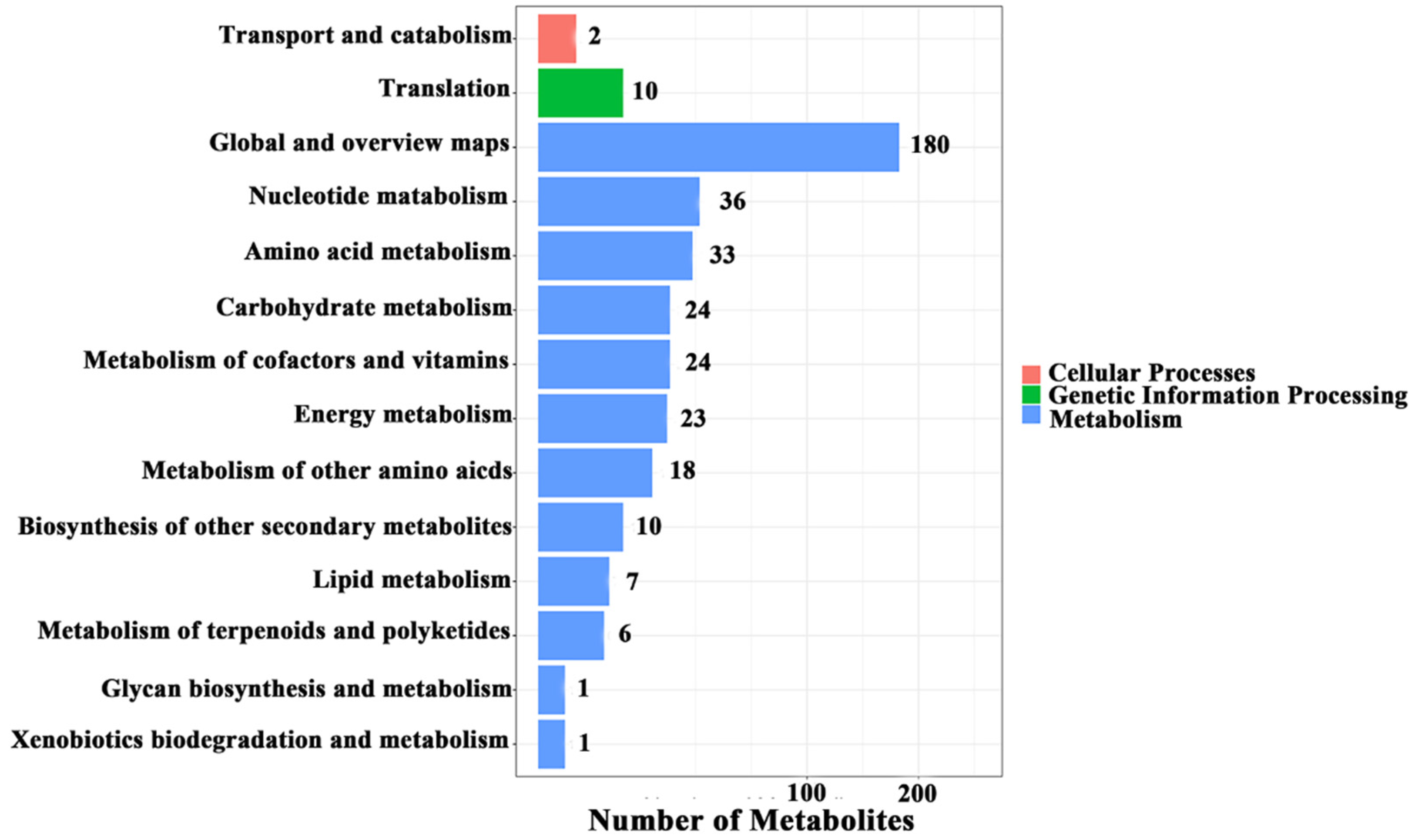
3.2. Metabolic Changes and the Metabolic Pathway Analysis
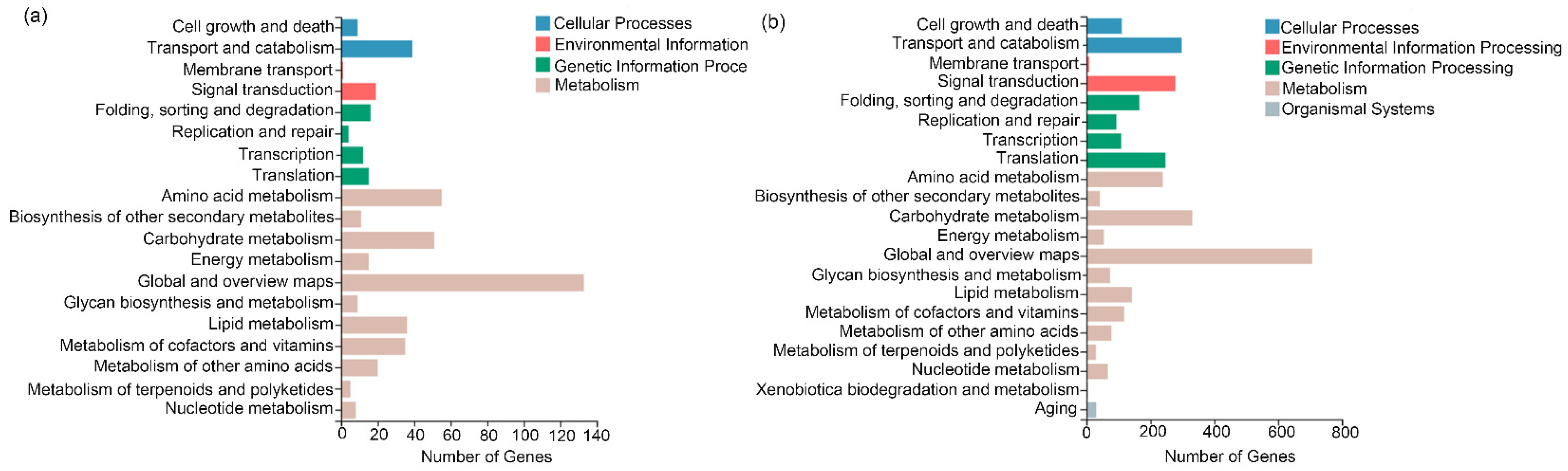
3.3. Transcriptome Analysis
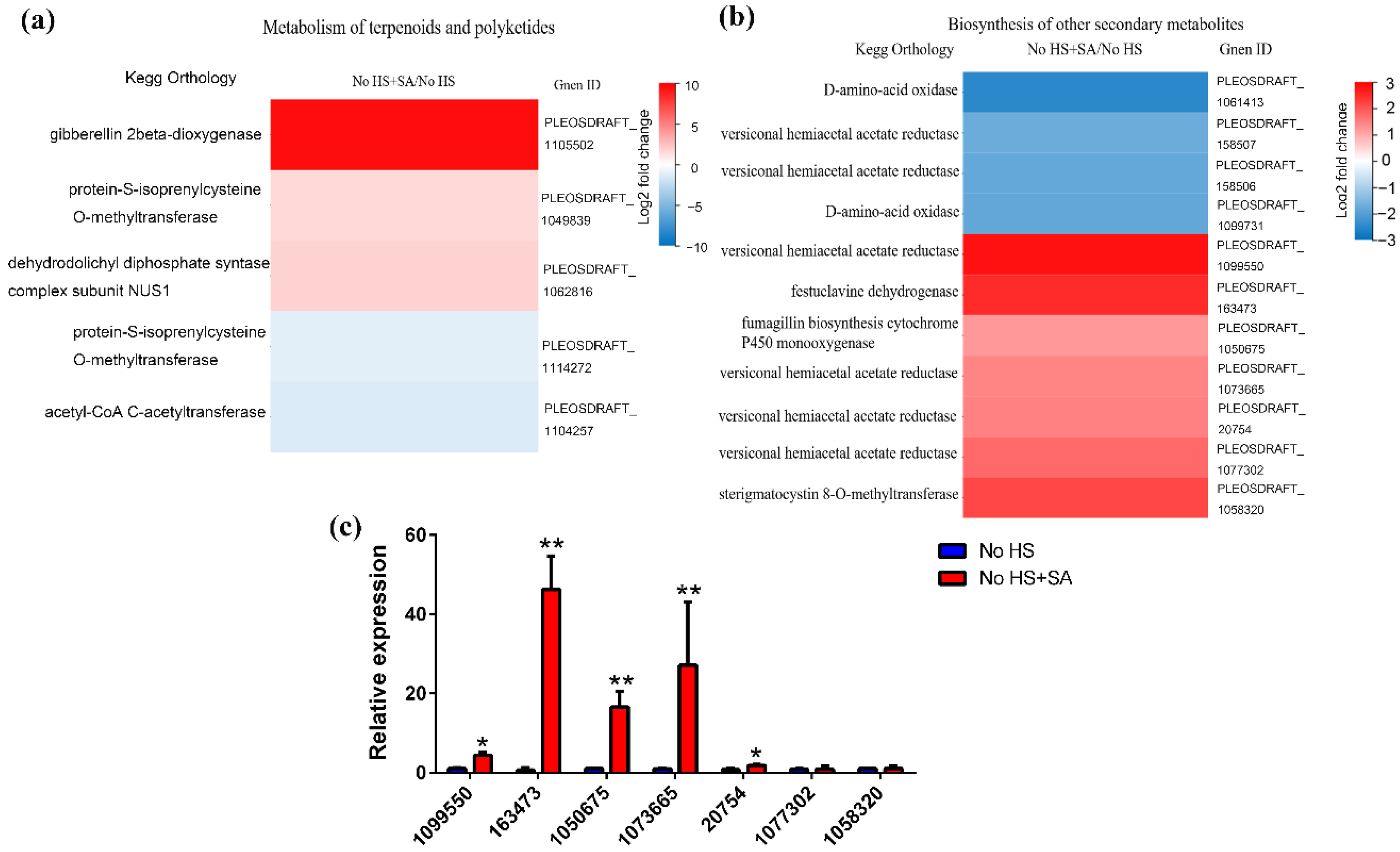
3.4. Putative Secondary Metabolic Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32. [Google Scholar] [CrossRef]
- Parola, S.; Chiodaroli, L.; Orlandi, V.; Vannin, C.; Panno, L. Lentinula edodes and Pleurotus ostreatus: Functional food with antioxidant-antimicrobial activity and an important source of Vitamin D and medicinal compounds. Funct. Foods Health Dis. 2017, 7, 773–794. [Google Scholar] [CrossRef] [Green Version]
- Elkhateeb, W.A.; Daba, G.M.; Thomas, P.W.; Wen, T.C. Medicinal mushrooms as a new source of natural therapeutic bioactive compounds. Egypt. Pharm. J. 2019, 18, 88–101. [Google Scholar]
- McLean, T.C.; Wilkinson, B.; Hutchings, M.I.; Devine, R. Dissolution of the disparate: Co-ordinate regulation in antibiotic biosynthesis. Antibiotics 2019, 8, 83. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.T.; Mo, X.H.; Zhu, L.P.; Tan, L.L.; Du, F.Y.; Wang, Q.W.; Zhou, Y.M.; Yuan, X.J.; Qiao, B.; Yang, S. Unusual and highly bioactive sesterterpenes synthesized by Pleurotus ostreatus during coculture with Trametes robiniophila Murr. Appl. Environ. Microbiol. 2019, 85, e00293-19. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Ren, A.; Li, M.J.; Cao, P.F.; Chen, T.X.; Zhang, G.; Shi, L.; Jiang, A.L.; Zhao, M.W. Heat stress modulates mycelium growth, heat shock protein expression, ganoderic acid biosynthesis, and hyphal branching of Ganoderma lucidum via cytosolic Ca2+. Appl. Environ. Microbiol. 2016, 82, 4112–4125. [Google Scholar] [CrossRef] [Green Version]
- Rana, M.; Dahot, M.U. Optimization of culture conditions to produce secondary metabolites by Pleurotus ostreatus. Pak. J. Biotechnol. 2017, 14, 251–256. [Google Scholar]
- Cao, P.F.; Wu, C.G.; Dang, Z.H.; Jiang, A.L.; Ren, A.; Zhao, M.W. Effects of exogenous salicylic acid on ganoderic acid biosynthesis and the expression of key genes in the ganoderic acid biosynthesis pathway in the ling zhi or reishi medicinal mushroom, Ganoderma lucidum (Agaricomycetes). Int. J. Med. Mushrooms 2017, 19, 65–73. [Google Scholar] [CrossRef]
- Zhang, G.; Ren, A.; Wu, F.L.; Yu, H.S.; Shi, L.; Zhao, M.W. Ethylene promotes mycelial growth and ganoderic acid biosynthesis in Ganoderma lucidum. Biotechnol. Lett. 2017, 39, 269–275. [Google Scholar] [CrossRef]
- Hu, Y.; Ahmed, S.; Li, J.W.; Luo, B.B.; Gao, Z.Y.; Zhang, Q.Y.; Li, X.H.; Hu, X.B. Improved ganoderic acids production in Ganoderma lucidum by wood decaying components. Sci. Rep. 2017, 7, 46623. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Xu, J.W.; Zhong, J.J. Enhanced production of ganoderic acids in static liquid culture of Ganoderma lucidum under nitrogen-limiting conditions. Bioresour. Technol. 2011, 102, 8185–8190. [Google Scholar] [CrossRef]
- Liu, G.Q.; Wang, X.L.; Han, W.J.; Lin, Q.L. Improving the fermentation production of the individual key triterpene ganoderic acid me by the medicinal fungus Ganoderma lucidum in submerged culture. Molecules 2012, 17, 12575–12586. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, C. Cultivation of Pleurotus ostreatus and other edible mushrooms. Appl. Microbiol. Biotechnol. 2010, 85, 1321–1337. [Google Scholar] [CrossRef]
- Reisa, F.S.; Lillian Barros, L.; Martins, A.; Ferreira, I.C.F.R. Chemical composition and nutritional value of the most widely appreciated cultivated mushrooms: An inter-species comparative study. Food Chem. Toxicol. 2012, 50, 191–197. [Google Scholar] [CrossRef] [Green Version]
- Baraza, L.D.; Neser, W.; Jackson, K.C.; Fredrick, J.B.; Dennis, O.; Wairimu, K.R.; Keya, A.O.; Heydenreich, M. Antimicrobial coumarins from the oyster culinary-medicinal mushroom, Pleurotus ostreatus (Agaricomycetes), from Kenya. Int. J. Med. Mushrooms 2016, 18, 905–913. [Google Scholar] [CrossRef]
- Annang, F.; Pérez-Victoria, I.; Appiah, T.; Pérez-Moreno, G.; Domingo, E.; Martín, J.; Mackenzie, T.; Ruiz-Pérez, L.; González-Pacanowska, D.; Genilloud, O.; et al. Antiprotozoan sesterterpenes and triterpenes isolated from two Ghanaian mushrooms. Fitoterapia 2018, 127, 341–348. [Google Scholar] [CrossRef]
- Cai, X.; Yu, Y.; Li, Q.; Chen, B.K.; Huang, Y.; Zou, X.W.; Tang, J.T.; Huang, B.S. Asperpyrone F, a new dimeric naphtho-γ-pyrone from the edible fungus Pleurotus ostreatus. Nat. Prod. Res. 2018, 16, 1953–1960. [Google Scholar] [CrossRef]
- Yan, Z.; Zhao, M.; Wu, X.; Zhang, J. Metabolic response of Pleurotus ostreatus to continuous heat stress. Front. Microbiol. 2020, 10, 3148. [Google Scholar] [CrossRef]
- Hu, Y.R.; Wang, Y.; Chen, Y.J.; Chai, Q.Q.; Dong, H.Z.; Shen, J.W.; Qi, Y.C.; Wang, F.Q.; Wen, Q. Salicylic acid enhances heat stress resistance of Pleurotus ostreatus (Jacq.) P. Kumm through metabolic rearrangement. Antioxidants 2022, 11, 968. [Google Scholar] [CrossRef]
- Qi, Y.; Chen, H.; Zhang, M.; Wen, Q.; Qiu, L.Y.; Shen, J.W. Identification and expression analysis of Pofst3 suggests a role during Pleurotus ostreatus primordia formation. Fungal Biol. 2019, 123, 200–208. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wang, C.S.; Lu, L.; Yin, W.B.; Zhang, K.Q. From taxonomy and industry to genetics: Fungal Biology in China. Fungal Genet. Biol. 2015, 81, 110–112. [Google Scholar] [CrossRef]
- Ren, A.; Qin, L.; Shi, L.; Dong, X.; Li, Y.X.; Zhao, M.W. Methyl jasmonate induces ganoderic acid biosynthesis in the basidiomycetous fungus Ganoderma lucidum. Bioresour. Technol. 2010, 101, 6785–6790. [Google Scholar] [CrossRef]
- Liang, C.X.; Li, Y.B.; Xu, J.W.; Wang, J.L.; Miao, X.L.; Tang, Y.J.; Gu, T.Y.; Zhong, J.J. Enhanced biosynthetic gene expressions and production of ganoderic acids in static liquid culture of Ganoderma lucidum under phenobarbital induction. Appl. Microbiol. Biot. 2010, 86, 1367–1374. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, S.; Wu, J.; Sun, X.; Ma, A. Heat stress in macrofungi: Effects and response mechanisms. Appl. Microbiol. Biot. 2021, 105, 7567–7576. [Google Scholar] [CrossRef]
- Devine, R.; Hutchings, M.; Holmes, N. Future directions for the discovery of antibiotics from actinomycete bacteria. Emerg. Top. Life Sci. 2017, 1, 1–12. [Google Scholar]
- Künzler, M. How fungi defend themselves against microbial competitors and animal predators. PLoS Pathog. 2018, 14, e1007184. [Google Scholar] [CrossRef]
- Hertweck, C.; Luzhetskyy, A.; Rebets, Y.; Bechthold, A. Type II polyketide synthases: Gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 2007, 24, 162–190. [Google Scholar] [CrossRef]
- Rawlings, B.J. Type I polyketide biosynthesis in bacteria (Part B). Nat. Prod. Rep. 2001, 18, 231–281. [Google Scholar] [CrossRef]
- Shao, L.; Chen, J.; Wang, C.; Li, J.A.; Tang, Y.; Chen, D.; Liu, W. Characterization of a key aminoglycoside phosphotransferase in gentamicin biosynthesis. Bioorg. Med. Chem. Lett. 2013, 23, 1438–1441. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, S.S.; Isbrandt, T.; Kirkegaard, M.; Buijs, Y.; Strube, M.L.; Sonnenschein, E.C.; Larsen, T.O.; Gram, L. Production of the antimicrobial compound tetrabromopyrrole and the Pseudomonas quinolone system precursor, 2-heptyl-4-quinolone, by a novel marine species Pseudoalteromonas galatheae sp. nov. Sci. Rep. 2020, 10, 21630. [Google Scholar] [CrossRef] [PubMed]
- Honda, M.; Katsuki, T.; Yamaguchi, M. Total synthesis of protomycinolide iv. Tetrahedron Lett. 1984, 25, 3857–3860. [Google Scholar] [CrossRef]
- Chang, P.K.; Yabe, K.; Yu, J. The Aspergillus parasiticus estA-encoded esterase converts versiconal hemiacetal acetate to versiconal and versiconol acetate to versiconol in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004, 70, 3593–3599. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Chang, P.K.; Ehrlich, K.C.; Cary, J.W.; Bhatnagar, D.; Cleveland, T.E.; Payne, G.A.; Linz, J.E.; Woloshuk, C.P.; Bennett, J.W. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004, 70, 1253–1262. [Google Scholar] [CrossRef] [Green Version]
- Bilovol, Y.; Panaccione, D.G. Functional analysis of the gene controlling hydroxylation of festuclavine in the ergot alkaloid pathway of Neosartorya fumigata. Curr. Genet. 2016, 62, 853–860. [Google Scholar] [CrossRef] [Green Version]
- Urlacher, V.B.; Eiben, S. Cytochrome P450 monooxygenases: Perspectives for synthetic application. Trends Biotechnol. 2006, 24, 324–330. [Google Scholar] [CrossRef]
- Mycroft-West, C.J.; Su, D.; Pagani, I.; Rudd, T.R.; Elli, S.; Guimond, S.E.; Miller, G.J.; Meneghetti, M.C.Z.; Nader, H.B.; Li, Y.; et al. Heparin inhibits cellular invasion by SARS-CoV-2: Structural dependence of the interaction of the surface protein (spike) S1 receptor binding domain with heparin. Thromb. Haemost. 2020, 120, 1700–1715. [Google Scholar]
- Baytas, S.N.; Linhardt, R.J. Advances in the preparation and synthesis of heparin and related products. Drug Discov. Today 2020, 25, 2095–2109. [Google Scholar] [CrossRef]
- Selles, B.; Dhalleine, T.; Hériché, M.; Rouhier, N.; Couturier, J. The cytosolic Arabidopsis thaliana cysteine desulfurase ABA3 delivers sulfur to the sulfurtransferase STR18. J. Biol. Chem. 2022, 298, 101749. [Google Scholar] [CrossRef]
- Pecora, F.; Gualeni, B.; Forlino, A.; Superti-Furga, A.; Tenni, R.; Cetta, G.; Rossi, A. In vivo contribution of amino acid sulfur to cartilage proteoglycan sulfation. Biochem. J. 2006, 398, 509–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Álvarez-Cao, M.E.; Rico-Díaz, A.; Cerdán, M.E.; Becerra, M.; González-Siso, M.I. Valuation of agro-industrial wastes as substrates for heterologous production of α-galactosidase. Microb. Cell Fact. 2018, 17, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Y.; Chai, Q.; Wang, Y.; Chen, Y.; Dong, H.; Shen, J.; Qi, Y.; Yu, H.; Wang, F.; Wen, Q. Effects of Heat Stress and Exogenous Salicylic Acid on Secondary Metabolites Biosynthesis in Pleurotus ostreatus (Jacq.) P. Kumm. Life 2022, 12, 915. https://doi.org/10.3390/life12060915
Hu Y, Chai Q, Wang Y, Chen Y, Dong H, Shen J, Qi Y, Yu H, Wang F, Wen Q. Effects of Heat Stress and Exogenous Salicylic Acid on Secondary Metabolites Biosynthesis in Pleurotus ostreatus (Jacq.) P. Kumm. Life. 2022; 12(6):915. https://doi.org/10.3390/life12060915
Chicago/Turabian StyleHu, Yanru, Qianqian Chai, Yue Wang, Yujie Chen, Haozhe Dong, Jinwen Shen, Yuancheng Qi, Haiyou Yu, Fengqin Wang, and Qing Wen. 2022. "Effects of Heat Stress and Exogenous Salicylic Acid on Secondary Metabolites Biosynthesis in Pleurotus ostreatus (Jacq.) P. Kumm" Life 12, no. 6: 915. https://doi.org/10.3390/life12060915
APA StyleHu, Y., Chai, Q., Wang, Y., Chen, Y., Dong, H., Shen, J., Qi, Y., Yu, H., Wang, F., & Wen, Q. (2022). Effects of Heat Stress and Exogenous Salicylic Acid on Secondary Metabolites Biosynthesis in Pleurotus ostreatus (Jacq.) P. Kumm. Life, 12(6), 915. https://doi.org/10.3390/life12060915






