Comparative Characteristics and Pathogenic Potential of Escherichia coli Isolates Originating from Poultry Farms, Retail Meat, and Human Urinary Tract Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Escherichia coli Isolates
2.2. Phylogenetic Groups of E. coli
2.3. Enterobacterial Repetitive Intergenic Consensus Polymerase Chain Reaction (ERIC-PCR)
2.4. Multilocus Sequence Typing (MLST)
2.5. Detection of Selected Virulence Genes (VGs)
2.6. Determination of E. coli Susceptibility to Antibiotics and Chemotherapeutics
2.7. Statistical Analysis
3. Results
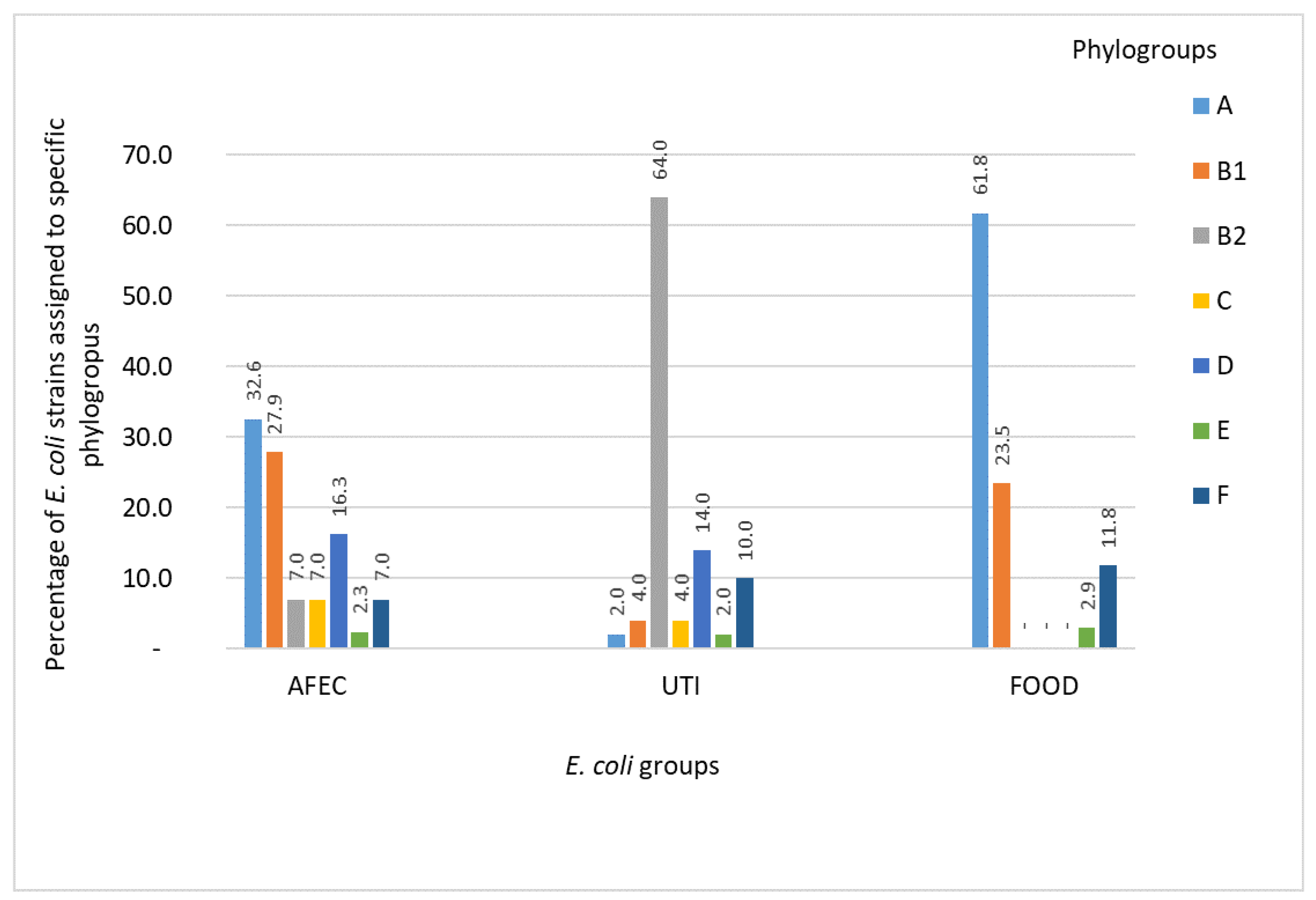
3.1. Phylogenetic Groups of E. coli
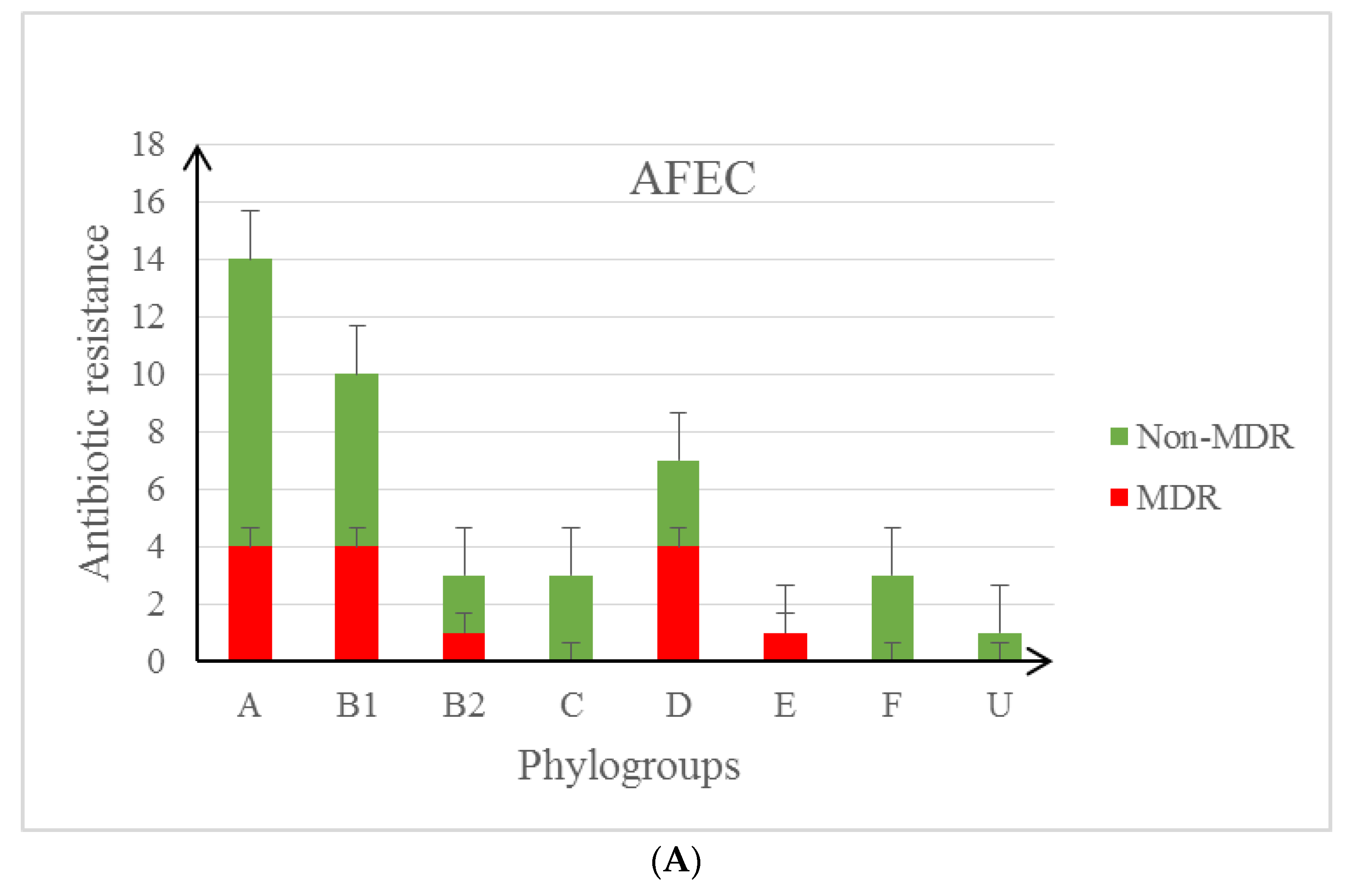
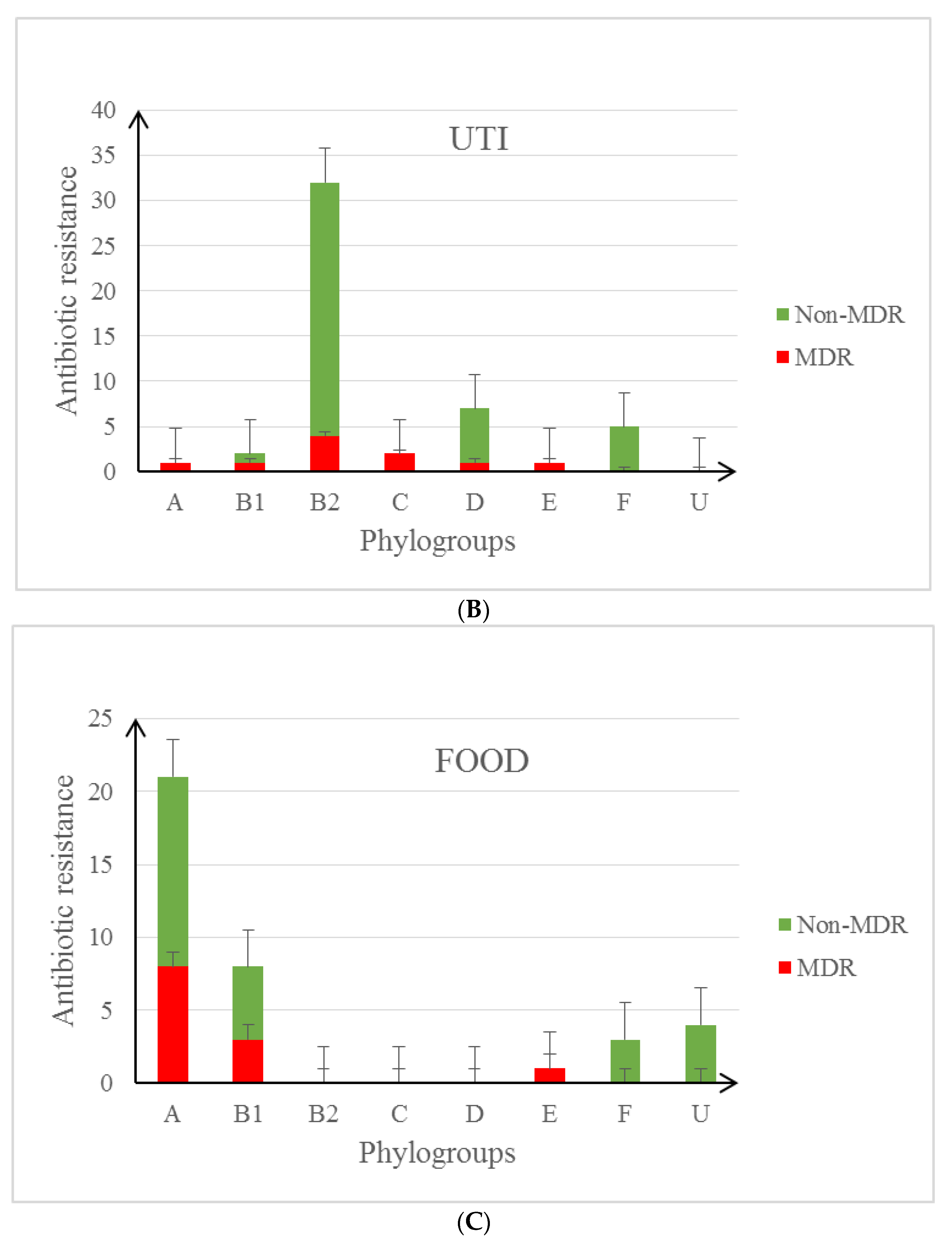
3.2. Characteristics of the Studied E. coli Isolates in the Context of Their Antibiotic Resistance Patterns and Virulence Profile
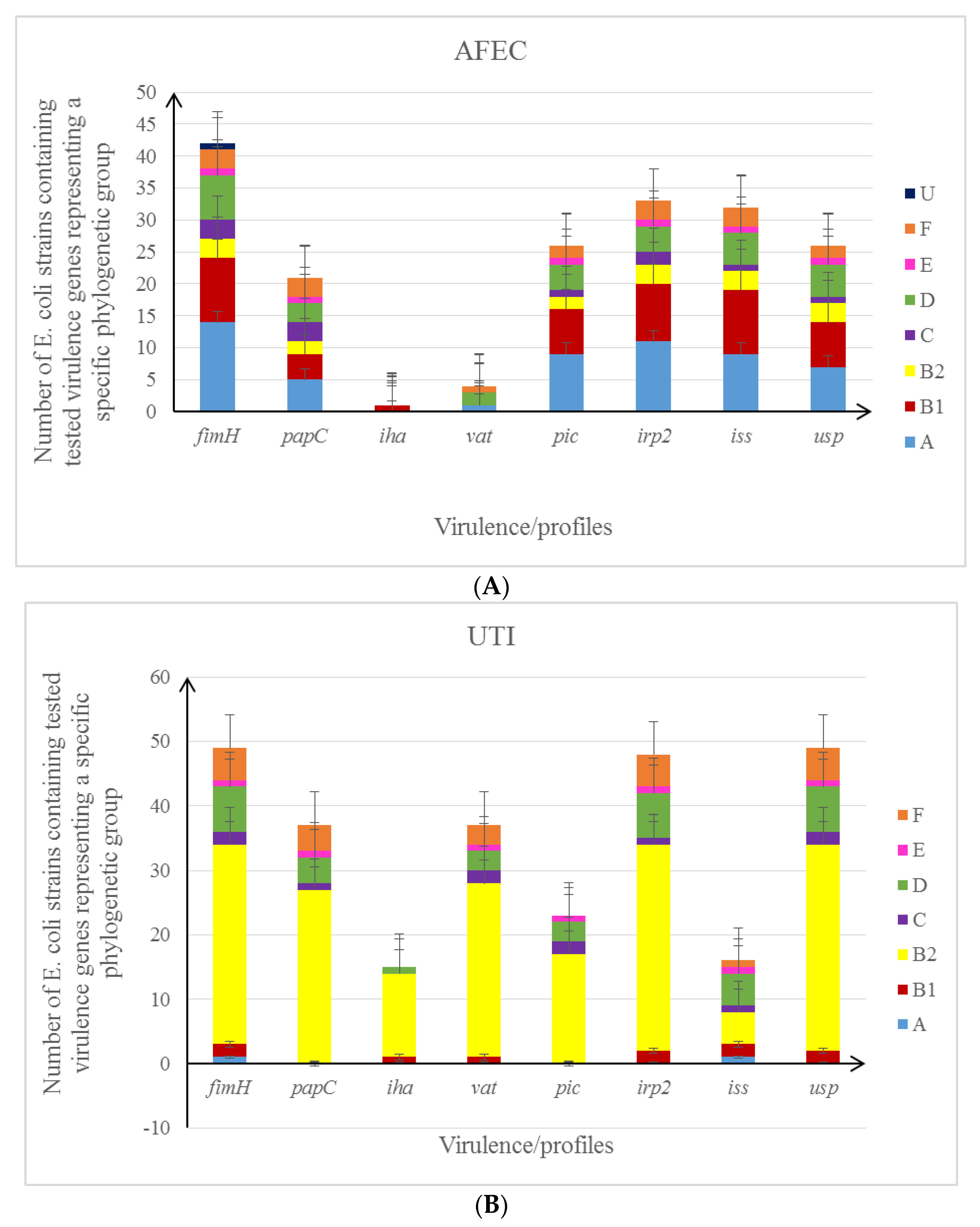
3.3. The Genotyping Analysis of E. coli Strains
3.4. Statistical Analysis of the Obtained Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Köhler, C.D.; Dobrindt, U. What defines extraintestinal pathogenic Escherichia coli? Int. J. Med. Microbiol. 2011, 301, 642–647. [Google Scholar] [CrossRef]
- Bogema, D.R.; McKinnon, J.; Liu, M.; Hitchick, N.; Miller, N.; Venturini, C.; Iredell, J.; Darling, A.E.; Chowdury, P.R.; Djordjevic, S.P. Wholegenome analysis of extraintestinal Escherichia coli sequence type 73 from a single hospital over a 2 year period identified different circulating clonal groups. Microb. Genom. 2020, 6, e000255. [Google Scholar]
- Hussain, A.; Shaik, S.; Ranjan, A.; Nandanwar, N.; Tiwari, S.K.; Majid, M.; Baddam, R.; Qureshi, I.A.; Semmler, T.; Wieler, L.H.; et al. Risk of transmission of antimicrobial resistant Escherichia coli from commercial broiler and free-range retail chicken in India. Front. in Microbiol. 2017, 13, 2120. [Google Scholar] [CrossRef]
- European Commission. A European One Health Action Plan against Antimicrobial Resistance (AMR). 2017. Available online: https://ec.europa.eu/health/amr/sites/amr/files/amr_action_plan_2017_en.pdf (accessed on 13 September 2018).
- Gootz, T.D. The global problem of antibiotic resistance. Crit. Rev. Immunol. 2010, 30, 79–93. [Google Scholar] [CrossRef] [PubMed]
- García, A.; Fox, J.G. A One Health Perspective for Defining and Deciphering Escherichia coli Pathogenic Potential in Multiple Hosts. Comp. Med. 2021, 71, 3–45. [Google Scholar] [CrossRef]
- Vincent, C.; Boerlin, P.; Daignault, D.; Dozois, C.M.; Dutil, L.; Galanakis, C.; Reid-Smith, R.J.; Tellier, P.P.; Tellis, P.A.; Ziebell, K.; et al. Food reservoir for Escherichia coli causing urinary tract infections. Emerg. Infect. Dis. 2010, 16, 88–95. [Google Scholar] [CrossRef]
- Mitchell, N.M.; Johnson, J.R.; Johnston, B.; Curtiss, R.; Mellata, M. Zoonotic potential of Escherichia coli isolates from retail chicken meat products and eggs. Appl. Environ. Microbiol. 2015, 81, 1177–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sora, V.M.; Meroni, G.; Martino, P.A.; Soggiu, A.; Bonizzi, L.; Zecconi, A. Extraintestinal Pathogenic Escherichia coli: Virulence Factors and Antibiotic Resistance. Pathogens 2021, 10, 1355. [Google Scholar] [CrossRef]
- Castellanos, L.R.; Donado-Godoy, P.; León, M.; Clavijo, V.; Arevalo, A.; Bernal, J.F.; Timmerman, A.J.; Mevius, D.J.; Wagenaar, J.A.; Hordijk, J. High Heterogeneity of Escherichia coli Sequence Types Harbouring ESBL/AmpC Genes on IncI1 Plasmids in the Colombian Poultry Chain. PLoS ONE 2017, 12, e0170777. [Google Scholar] [CrossRef]
- Stromberg, Z.R.; Johnson, J.R.; Fairbrother, J.M.; Kilbourne, J.; Van Goor, A.; Curtiss, R., 3rd; Mellata, M. Evaluation of Escherichia coli isolates from healthy chickens to determine their potential risk to poultry and human health. PLoS ONE 2017, 12, e0180599. [Google Scholar] [CrossRef] [Green Version]
- Magray, S.; Wani, S.; Kashoo, Z.; Bhat, M.; Adil, S.; Farooq, S. Serological diversity, molecular characterisation and antimicrobial sensitivity of avian pathogenic Escherichia coli (APEC) isolates from broiler chickens in Kashmir, India. Anim. Prod. Sci. 2019, 59, 338–346. [Google Scholar] [CrossRef]
- Johnson, T.J.; Wannemuehler, Y.; Doetkott, C.; Johnson, S.J.; Rosenberger, S.C.; Nolan, L.K. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J. Clin. Microbiol. 2008, 46, 3987–3996. [Google Scholar] [CrossRef] [Green Version]
- Awad, A.; Arafat, N.; Elhadidy, M. Genetic elements associated with antimicrobial resistance among avian pathogenic Escherichia coli. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 59. [Google Scholar] [CrossRef] [Green Version]
- Wiles, T.J.; Kulesus, R.R.; Mulvey, M.A. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp. Mol. Pathol. 2008, 85, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; Fan, Y.; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Gen. Res. 2020, 30, 138–152. [Google Scholar] [CrossRef] [Green Version]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wel. Open. Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Nascimento, M.; Sousa, A.; Ramirez, M.; Francisco, A.P.; Carriço, J.A.; Vaz, C. PHYLOViZ 2.0: Providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 2017, 33, 128–129. [Google Scholar] [CrossRef]
- Yun, K.W.; Kim, D.S.; Kim, W.; Lim, I.S. Molecular typing of uropathogenic Escherichia coli isolated from Korean children with urinary tract infection. Korean J. Ped. 2015, 58, 20–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paniagua-Contreras, G.L.; Hernández-Jaimes, T.; Monroy-Pérez, E.; Vaca-Paniagua, F.; Díaz-Velásquez, C.; Uribe-García, A.; Vaca, S. Comprehensive expression analysis of pathogenicity genes in uropathogenic Escherichia coli strains. Microb Path. 2017, 103, 1–7. [Google Scholar] [CrossRef]
- Ewers, C.; Li, G.; Wilking, H.; Kiessling, S.; Alt, K.; Antáo, E.M.; Laturnus, C.; Diehl, I.; Glodde, S.; Homeier, T.; et al. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: How closely related are they? Int. J. Med. Microbiol. 2007, 297, 163–176. [Google Scholar] [CrossRef]
- Rodriguez-Siek, K.E.; Giddings, C.W.; Doetkott, C.; Johnson, T.J.; Fakhr, M.K.; Nolan, L.K. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 2005, 151, 2097–2110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, A.; Kanamaru, S.; Kurazono, H.; Kunishima, Y.; Tsukamoto, T.; Ogawa, O.; Yamamoto, S. Escherichia coli isolates associated with uncomplicated and complicated cystitis and asymptomatic bacteriuria possess similar phylogenies, virulence genes, and O-serogroup profiles. J. Clin. Microbiol. 2006, 44, 4589–4592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 7.1. 2017. Available online: http://www.eucast.org/documents/rd/ (accessed on 31 December 2017).
- Economou, V.; Gousia, P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect. Drug Resist. 2015, 1, 49–61. [Google Scholar] [CrossRef] [Green Version]
- Manges, A.R. Escherichia coli and urinary tract infections: The role of poultry-meat”. Clin. Microbiol. Infect. 2016, 22, 122–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Medicines Agency. European Surveillance of Veterinary Antimicrobial Consumption, 2018. Sales of Veterinary Antimicrobial Agents in 30 European Countries in 2016. (EMA/275982/2018); European Medicines Agency: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Casella, T.; Nogueira, M.; Saras, E. High prevalence of ESBLs in retail chicken meat despite reduced use of antimicrobials in chicken production, France. Int J Food Microbiol. 2017, 18, 271–275. [Google Scholar] [CrossRef] [Green Version]
- Reich, F.; Atanassova, V.; Klein, G. Extended-spectrum β-lactamase- and AmpC-producing enterobacteria in healthy broiler chickens, Germany. Emerg. Infect. Dis. 2013, 19, 1253–1259. [Google Scholar] [CrossRef]
- Melendez, D.; Roberts, M.C.; Greninger, A.L.; Weissman, S.; No, D.; Rabinowitz, P.; Wasser, S. Whole-genome analysis of extraintestinal pathogenic Escherichia coli (ExPEC) MDR ST73 and ST127 isolated from endangered southern resident killer whales (Orcinus orca). J. Antimicrob. Chemother. 2019, 74, 2176–2180. [Google Scholar] [CrossRef]
- Clermont, O.; Olier, M.; Hoede, C.; Diancourt, L.; Brisse, S.; Keroudean, M.; Glodt, J.; Picard, B.; Oswald, E.; Denamur, E. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect. Genet. Evol. 2011, 11, 654–662. [Google Scholar] [CrossRef]
- Mellata, M. Human and avian extraintestinal pathogenic Escherichia coli: Infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog. Dis. 2013, 10, 916–932. [Google Scholar] [CrossRef] [Green Version]
- Singer, R.S. Urinary tract infections attributed to diverse ExPEC strains in food animals: Evidence and data gaps. Front. Microbiol. 2015, 6, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Najafi, S.; Rahimi, M.; Nikousefat, Z. Extra-intestinal pathogenic Escherichia coli from human and avian origin: Detection of the most common virulence-encoding genes. Vet. Res. Forum. 2019, 10, 43–49. [Google Scholar]
- Pesciaroli, M.; Magistrali, C.F.; Filippini, G.; Epifanio, E.M.; Lovito, C.; Marchi, L.; Maresca, C.; Massacci, F.R.; Orsini, S.; Scoccia, E.; et al. Antibiotic-resistant commensal Escherichia coli are less frequently isolated from poultry raised using non-conventional management systems than from conventional broiler. Int. J. Food Microbiol. 2020, 314, 108–391. [Google Scholar] [CrossRef] [PubMed]
- Xiaojing, X.; Quing, S.; Lixiang, Z. Virulence factors and antibiotic resistance of avian pathogenic Escherichia coli in eastern China. J. Vet. Res. 2019, 63, 317–320. [Google Scholar]
- Hung, C.; Zhou, Y.; Pinkner, J.S.; Dodson, K.W.; Crowley, J.R.; Heuser, J.; Chapman, M.R.; Hadjifrangiskou, M.; Henderson, J.P.; Hultgren, S.J. Escherichia coli biofilms have an organized and complex extracellular matrix structure. MBio 2013, 4, e00645-13. [Google Scholar] [CrossRef] [Green Version]
- Gunther, N.W.; Lockatell, V.; Johnson, D.E.; Mobley, H.L. In vivo dynamics of type 1 fimbria regulation in uropathogenic Escherichia coli during experimental urinary tract infection. Infect. Immun. 2001, 69, 2838–2846. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Banda, D.A.; Carrillo-Casas, E.M.; Leyva-Leyva, M.; Orozco-Hoyuela, G.; Manjarrez-Hernandez, A.H.; Arroyo-Escalante, S. Identification of virulence factors genes in Escherichia coli isolates from women with urinary tract infection in Mexico. Biomed Res Int. 2014, 2014, 959206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ewers, C.; Antao, E.M.; Diehl, I.; Philipp, H.C.; Wieler, L.H. Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Appl. Environ. Microbiol. 2009, 75, 184–192. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.R.; Russo, T.A.; Tarr, P.I.; Carlino, U.; Bilge, S.S.; Vary, J.C.; Stell., A.L. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroNE. coli, among Escherichia coli isolates from patients with urosepsis. Infect. Immun. 2000, 68, 3040–3047. [Google Scholar] [CrossRef] [Green Version]
- Kanamura, S.; Kurazono, H.; Ishitoya, S.; Terai, A.; Habuchi, T.; Nakano, M.; Ogawa, O.; Yamamoto, S. Distribution and genetic association of putative uropathogenic virulence factors iroN, iha, kpsMT, ompT and usp in Escherichia coli isolated from urinary tract infections in Japan. J. Urol. 2003, 170, 2490–2493. [Google Scholar] [CrossRef]
- Kurazono, H.; Yamamoto, S.; Nakano, M.; Nair, G.B.; Terai, A.; Chaicumpa, W.; Hayashi, H. Characterization of a putative virulence island in the chromosome of uropathogenic Escherichia coli possessing a gene encoding a uropathogenic-specific protein. Microb. Pathog. 2000, 28, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Clermont, O.; Menard, M.; Kuskowski, M.A.; Picard, B.; Denamur, E. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J. Infect. Dis. 2006, 194, 1141–1150. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Li, Y.J.; Fan, C. Different loci and mRNA copy number of the increased serum survival gene of Escherichia coli. Can. J. Microbiol. 2018, 64, 147–154. [Google Scholar] [CrossRef] [Green Version]
- McPeake, S.J.; Smyth, J.A.; Ball, H.J. Characterization of avian pathogenic Escherichia coli (APEC) associated with colisepticaemia compared to faecal isolates from healthy birds. Vet. Microbiol. 2005, 110, 245–253. [Google Scholar] [CrossRef]
- Navarro-Garcia, F.; Gutierrez-Jimenez, J.; Garcia-Tovar, C. Pic, an autotransporter protein secreted by different pathogens in the Enterobacteriaceae family, is a potent mucus secretagogue. Infect. Immun. 2010, 78, 4101–4109. [Google Scholar] [CrossRef] [Green Version]
- Nichols, K.B.; Totsika, M.; Moriel, D.G. Molecular Characterization of the Vacuolating Autotransporter Toxin in Uropathogenic Escherichia coli. J. Bacteriol. 2016, 198, 1487–1498. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.R.; Porter, S.; Johnston, B.; Kuskowski, M.A.; Spurbeck, R.R.; Mobley, H.L.; Williamson, D.A. Host Characteristics and Bacterial Traits Predict Experimental Virulence for Escherichia coli Bloodstream Isolates from Patients With Urosepsis. Open Forum Infect. Dis. 2015, 2, ofv083. [Google Scholar] [CrossRef]
- Ewers, C.; Bethe, A.; Semmler, T.; Guenther, S.; Wieler, L.H. Extended-spectrum beta-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 2012, 18, 646–655. [Google Scholar] [CrossRef] [Green Version]
- Rogers, B.A.; Sidjabat, H.E.; Paterson, D.L. Escherichia coli O25b- -ST131: A pandemic, multiresistant, community associated strain. J. Antimicrob. Chemother. 2011, 66, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, J.R.; Porter, S.B.; Johnston, B.; Thuras, P.; Clock, S.; Crupain, M.; Rangan, U. Extraintestinal pathogenic and antimicrobial-resistant Escherichia coli, including sequence Type 131 (ST131), from retail chicken breasts in the United States in 2013. Appl. Environ. Microbiol. 2017, 83, e02956-16. [Google Scholar] [CrossRef] [Green Version]
- Koga, L.V.; Maluta, R.P.; da Silveira, W.D.; Ribeiro, R.A.; Hungria, M.; Vespero, E.C.; Nakazato, G.; Kobayashi, R.K.T. Characterization of CMY-2-type beta-lactamase-producing Escherichia coli isolated from chicken carcasses and human infection in a city of South Brazil. BMC Microbiol. 2019, 19, 174. [Google Scholar] [CrossRef] [Green Version]
- Vangchhia, B.; Abraham, S.; Bell, J.M.; Collignon, P.; Gibson, J.S.; Ingram, P.R.; Johnson, J.R.; Kennedy, K.; Trott, D.J.; Turnidge, J.D.; et al. Phylogenetic diversity, antimicrobial susceptibility and virulence characteristics of phylogroup F Escherichia coli in Australia. Microbiology 2016, 162, 1904–1912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toval, F.; Köhler, C.D.; Vogel, U.; Wagenlehner, F.; Mellmann, A.; Fruth, A.; Schmidt, M.A.; Karch, H.; Bielaszewska, M.; Dobrindta, U. Characterization of Escherichia coli Isolates from Hospital Inpatients or Outpatients with Urinary Tract Infection. J. Clin. Microbiol. 2014, 52, 407–418. [Google Scholar] [CrossRef] [Green Version]
- Johnson, T.J.; Kariyawasam, S.; Wannemuehler, Y.; Mangiamele, P.; Johnson, S.J.; Doetkott, C.; Skyberg, J.A.; Lynne, A.M.; Johnson, J.R.; Nolan, L.K. The genome sequence of avian pathogenic Escherichia coli strain O1: K1: H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 2007, 189, 3228–3236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Functions | Gene | Starter Forward (5′-3′) Starter Reverse (3′-5′) | Product Size (bp) |
|---|---|---|---|
| Adhesins | fimH—gene encoding for type 1 fimbria adhesin; | TGCAGAACGGATAAGCCGTGG GCAGTCACCTGCCCTCCGGTA | 508 [21] |
| papC—gene encoding for adhesin, an initiator of the formation of P-fimbria; | GTGGCAGTATGAGTAATGACCGTTA ATATCCTTTCTGCAGGGATGCAATA | 205 [22] | |
| iha—gene encoding for adhesin homologous to the Vibrio cholerae receptor (IrgA); | CTGGCGGAGGCTCTGAGATCA TCCTTAAGCTCCCGCGGCTGA | 827 [22] | |
| Miscellaneous | irp2—gene encoding the protein responsible for iron acquisition, protectin; | AAGGATTCGCTGTTACCGGAC TCGTCGGGCAGCGTTTCTTCT | 287 [23] |
| iss—gene encoding protectin (increased serum survival gene); | CAGCAACCCGAACCACCTGATG AGCATTGCCAGAGCGGCAGAA | 323 [24] | |
| usp—gene encoding a toxin which is a homologue of the Vibrio cholerae toxin; | CGGCTCTTACATCGGTGCGTTG GACATATCCAGCCAGCGAGTTC | 615 [25] | |
| Toxins | vat—gene encoding for the cytotoxin responsible for E. coli infection; | TCCTGGGACATAATGGTCAG GTGTCAGAACGGAATTGTC | 981 [23] |
| pic—gene encoding serine protease, toxin; | ACTGGATCTTAAGGCTCAGG TGGAATATCAGGGTGCCACT | 409 [23] |
| Functional Cathegory VG | Number (%) of E. coli Isolates with VGs | |||
|---|---|---|---|---|
| UTI (N = 50) | FOOD (N = 38) | Poultry Farms (N = 44) | p-Value UTI/Food (U-F) UTI/Poultry Farms (U-P) Food/Poultry Farms (F-P) | |
| Adhesins | ||||
| fimH | 49 (98.0) | 34 (89.5) | 42 (95.4) | N.S. |
| papC | 37 (74.0) | 28 (73.7) | 23 (52.3) | 0.97 U-F 0.03 U-P * 0.04 F-P * |
| iha | 14 (28.0) | 5 (13.1) | 1 (2.3) | 0.14 U-F <0.01 U-P * 0.05 F-P * |
| Miscellaneous | ||||
| irp2 | 48 (96.0) | 22 (57.9) | 33 (75.0) | <0.01 U-F * 0.01 U-P * 0.05 F-P * |
| iss | 16 (32.0) | 27 (71.1) | 32 (72.7) | <0.01 U-F * <0.01 U-P * 0.87 F-P |
| usp | 49 (98.0) | 23 (60.5) | 26 (59.1) | <0.01 U-F * <0.01 U-P * 0.87 F-P |
| Toxins | ||||
| vat | 37 (74.0) | 4 (10.5) | 4 (9.1) | <0.01 U-F * <0.01 U-P * 0.86 F-P |
| pic | 23 (46.0) | 28 (73.7) | 25 (56.8) | 0.01 U-F * 0.28 U-P * 0.12 F-P |
| Number (%) of E.coli Isolates with VGs within Resistance Groups | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Functional Category VG | UTI (N = 50) | FOOD (N = 38) | Poultry Farms (N = 44) | ||||||
| Non-MDR (n = 40) | MDR (n = 10) | p | Non-MDR (n = 24) | MDR (n = 14) | p | Non-MDR (n = 30) | MDR (n = 14) | p | |
| Adhesins | |||||||||
| fimH | 40 (100.0) * | 9 (90.0) * | 0.04 * | 21 (87.0) | 13 (92.9) | 0.98 | 29 (96.7) | 13 (92.9) | 0.83 |
| papC | 31 (77.5) | 6 (60.0) | 0.26 | 16 (66.7) | 12 (85.7) | 0.37 | 18 (60.0) | 5 (35.7) | 0.13 |
| iha | 12 (30.0) | 3 (30.0) | 1.0 | 3 (12.5) | 4 (28.6) | 0.42 | 1 (3.3) | 0 (0.0) | 0.69 |
| Miscellaneous | |||||||||
| irp2 | 40 (100.0) * | 8 (80.0) * | 0.04 | 15 (62.5) | 7 (50.0) | 0.45 | 21 (70.0) | 12 (85.7) | 0.26 |
| iss | 10 (25.0) * | 6 (60.0) * | 0.03 * | 13 (54.2) * | 14 (100.0) * | 0.01 * | 19 (63.3) * | 13 (92.8) * | 0.04 * |
| usp | 40 (100.0) | 9 (90.0) | 0.04 * | 18 (75.0) * | 5 (35.7) * | 0.02 * | 17 (56.7) | 9 (64.3) | 0.63 |
| Toxins | |||||||||
| vat | 29 (72.5) | 8 (80.0) | 0.63 | 3 (12.5) | 1 (7.1) | 0.69 | 2 (6.7) | 2 (14.3) | 0.80 |
| pic | 18 (45.0) | 5 (50.0) | 0.78 | 16 (66.7) | 12 (85.7) | 0.20 | 16 (53.3) | 9 (64.3) | 0.49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarowska, J.; Olszak, T.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Futoma-Koloch, B.; Gawel, A.; Drulis-Kawa, Z.; Choroszy-Krol, I. Comparative Characteristics and Pathogenic Potential of Escherichia coli Isolates Originating from Poultry Farms, Retail Meat, and Human Urinary Tract Infection. Life 2022, 12, 845. https://doi.org/10.3390/life12060845
Sarowska J, Olszak T, Jama-Kmiecik A, Frej-Madrzak M, Futoma-Koloch B, Gawel A, Drulis-Kawa Z, Choroszy-Krol I. Comparative Characteristics and Pathogenic Potential of Escherichia coli Isolates Originating from Poultry Farms, Retail Meat, and Human Urinary Tract Infection. Life. 2022; 12(6):845. https://doi.org/10.3390/life12060845
Chicago/Turabian StyleSarowska, Jolanta, Tomasz Olszak, Agnieszka Jama-Kmiecik, Magdalena Frej-Madrzak, Bozena Futoma-Koloch, Andrzej Gawel, Zuzanna Drulis-Kawa, and Irena Choroszy-Krol. 2022. "Comparative Characteristics and Pathogenic Potential of Escherichia coli Isolates Originating from Poultry Farms, Retail Meat, and Human Urinary Tract Infection" Life 12, no. 6: 845. https://doi.org/10.3390/life12060845
APA StyleSarowska, J., Olszak, T., Jama-Kmiecik, A., Frej-Madrzak, M., Futoma-Koloch, B., Gawel, A., Drulis-Kawa, Z., & Choroszy-Krol, I. (2022). Comparative Characteristics and Pathogenic Potential of Escherichia coli Isolates Originating from Poultry Farms, Retail Meat, and Human Urinary Tract Infection. Life, 12(6), 845. https://doi.org/10.3390/life12060845






