Monocyte Chemotactic Protein-1 (MCP1) Accumulation in Human Osteoclast Precursor Cultures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Culture of Human CD14+ Mononuclear Cells from Peripheral Blood
2.2. Real-Time Polymerase Chain Reaction (RT-PCR)
2.3. Microscopy
2.4. Statistical Analysis
3. Results
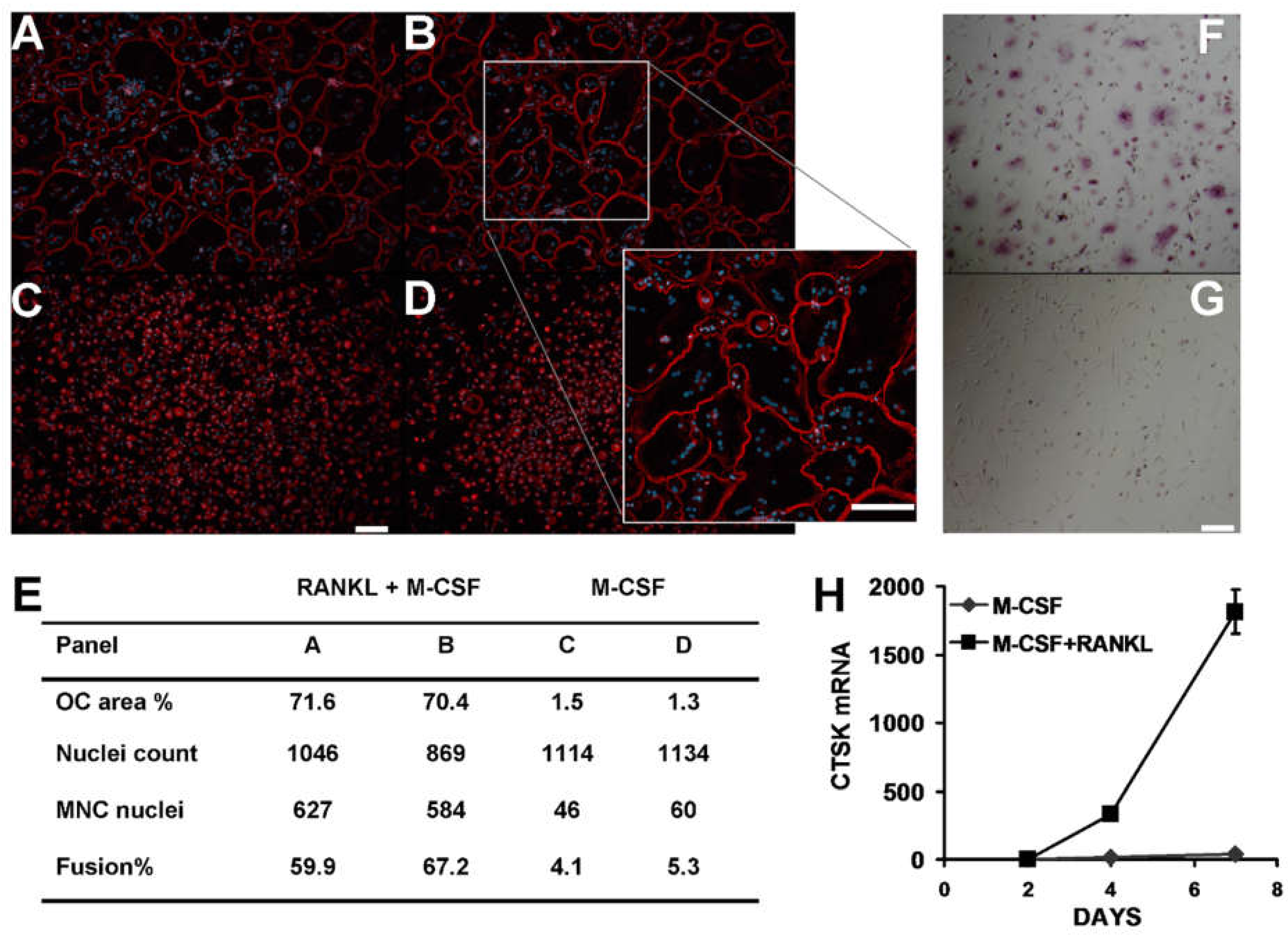
3.1. Osteoclast-like Cells in Culture Derived from Human CD14+ Mononuclear Cells
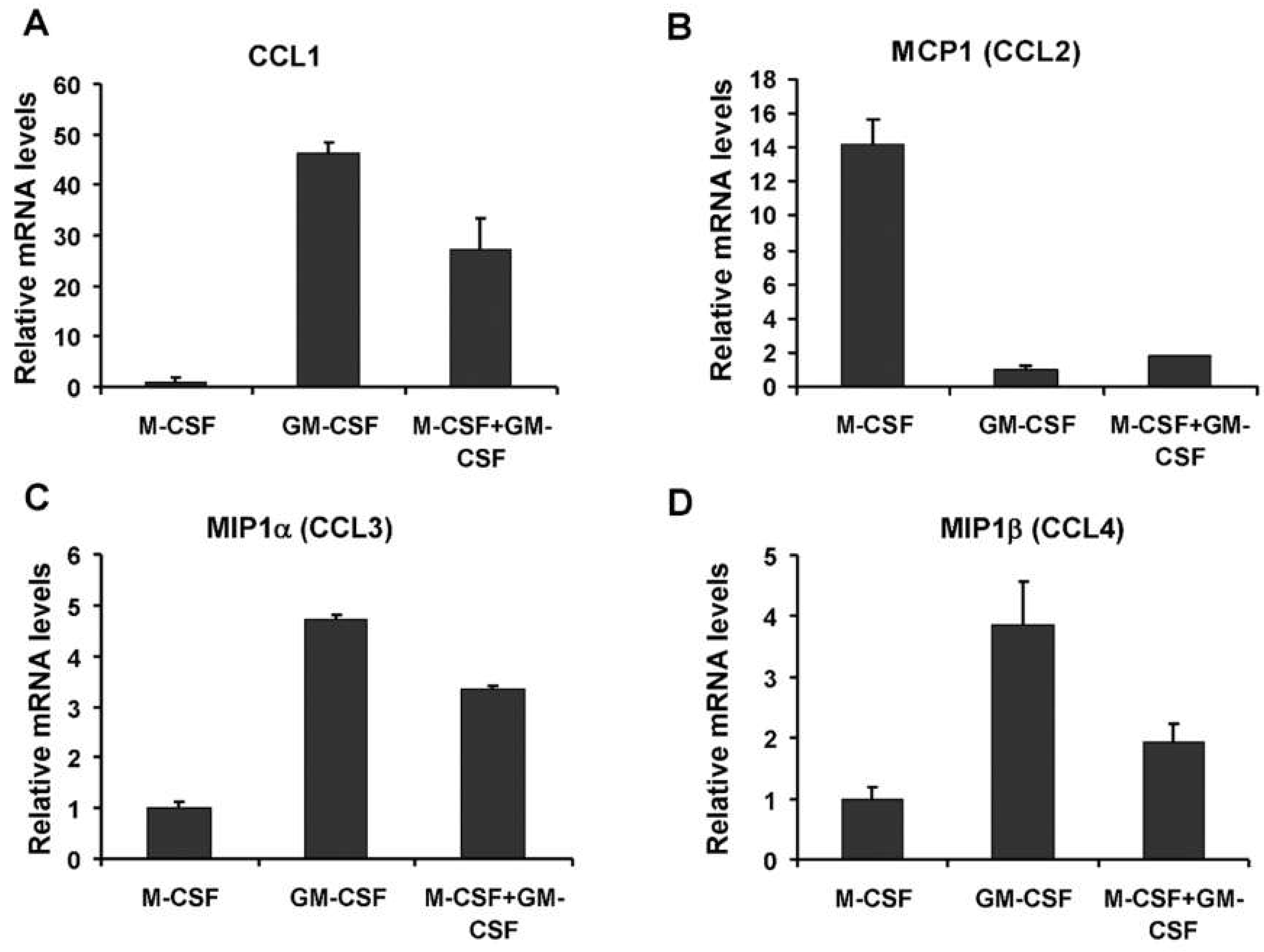
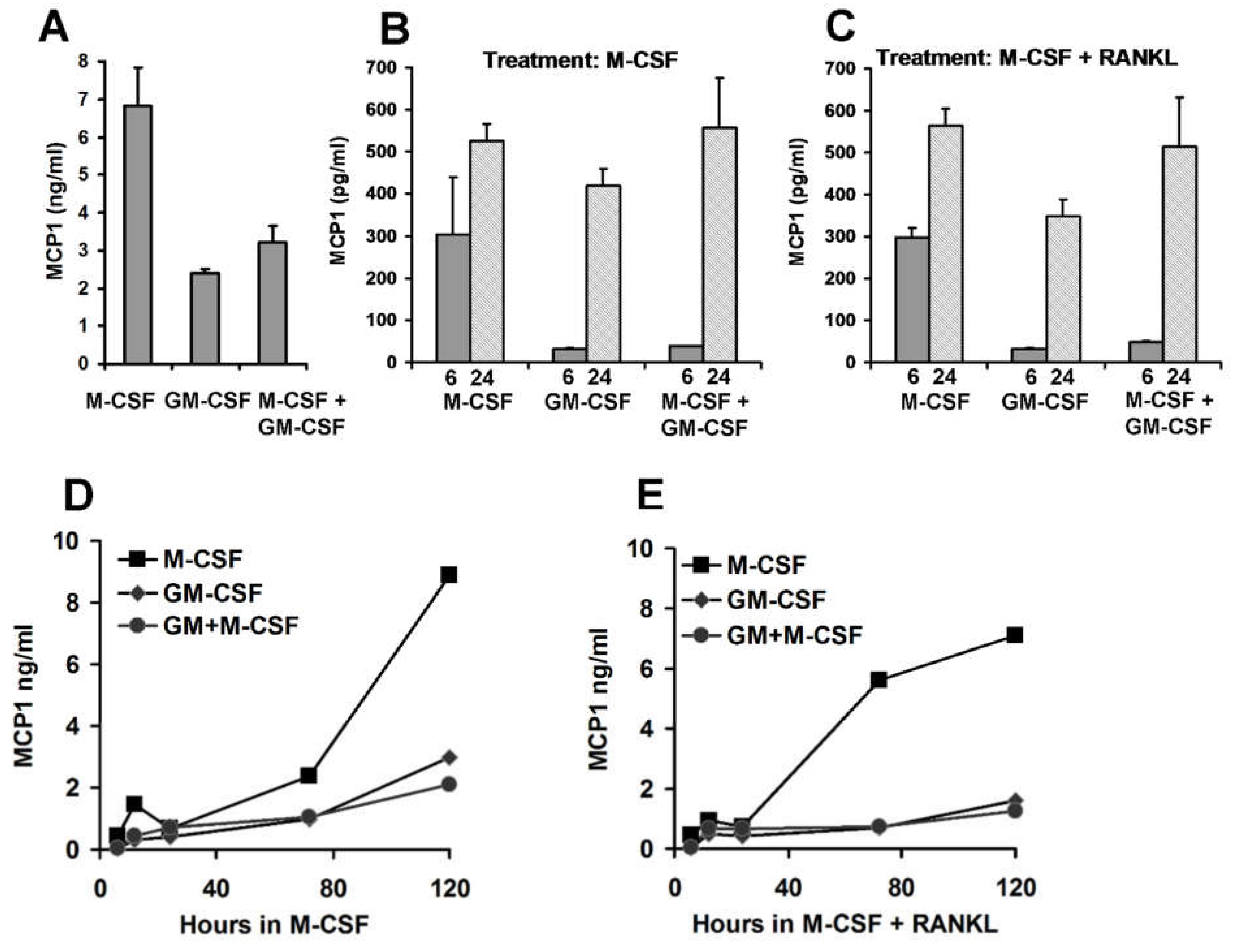
3.2. Effect of M-CSF and GM-CSF on MCP1 Abundance in CD14+ Mononuclear Cells
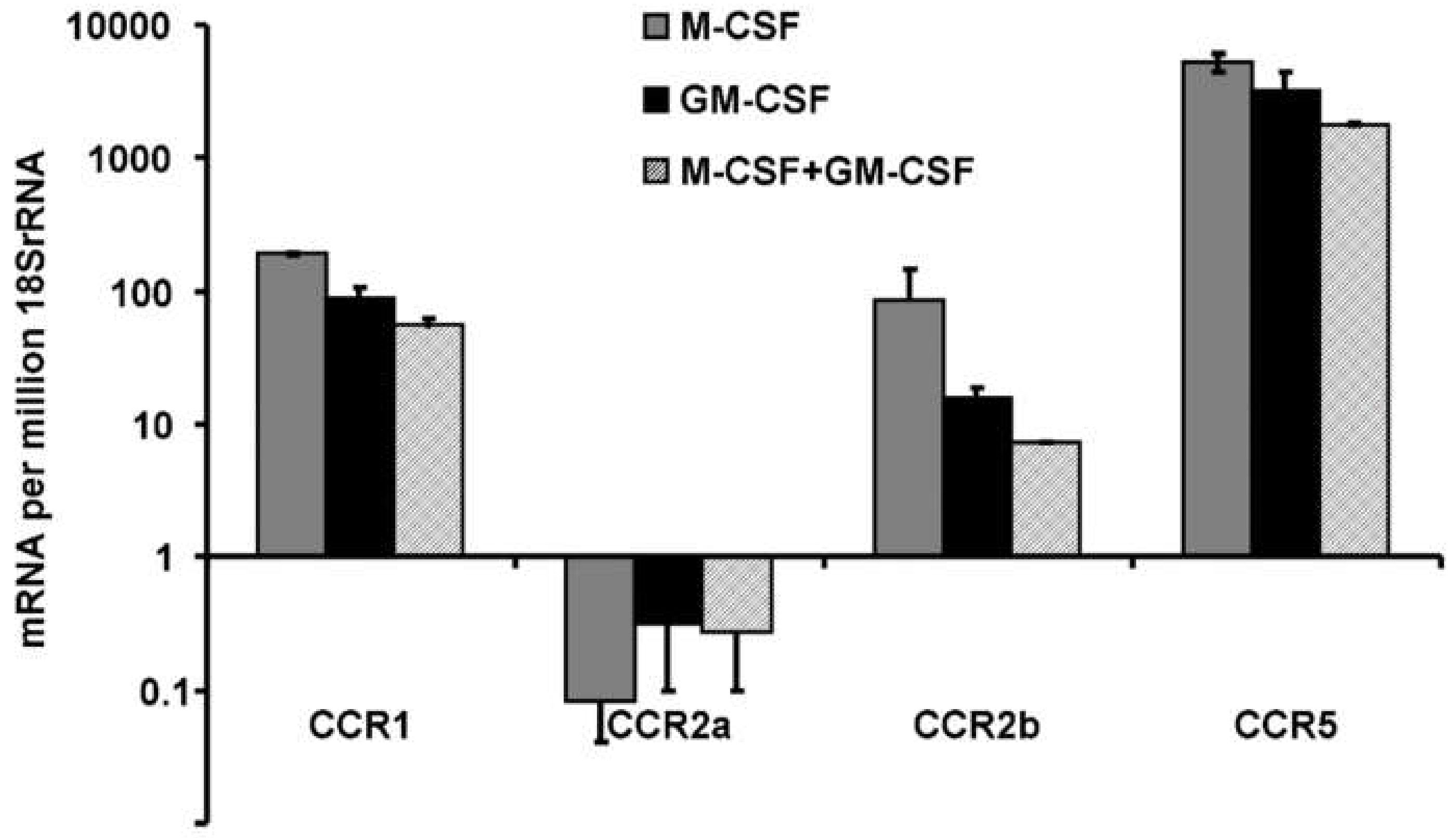
3.3. Expression of Chemokine Receptors
3.4. Pre-Treatment and Post-Treatment Culture Conditions and Chemokine mRNA Levels
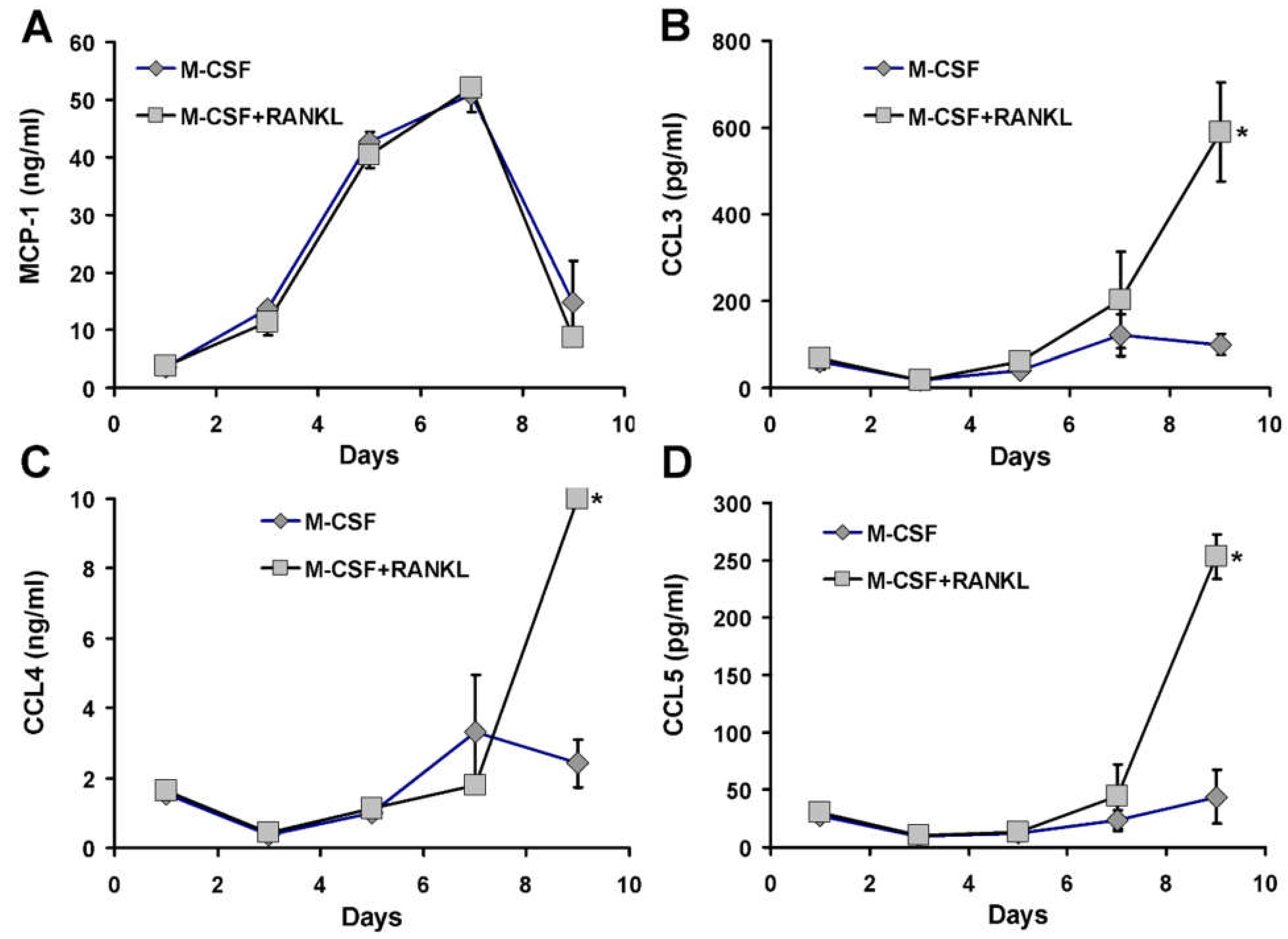
3.5. Pre-Treatment Culture Conditions and MCP1 Protein Levels
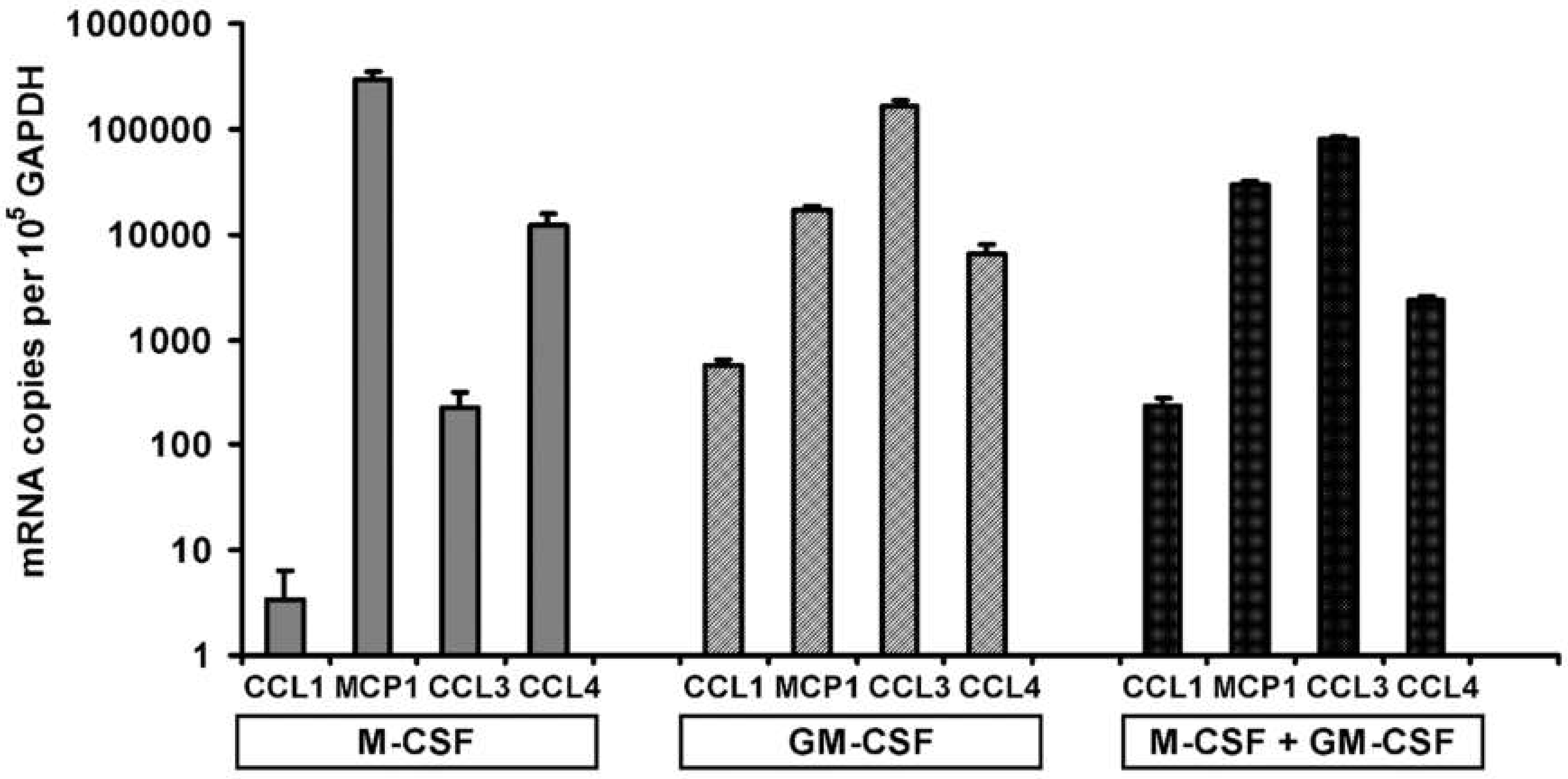
3.6. Chemokine Accumulation in Media of CD14+ Human Mononuclear Cells in Culture
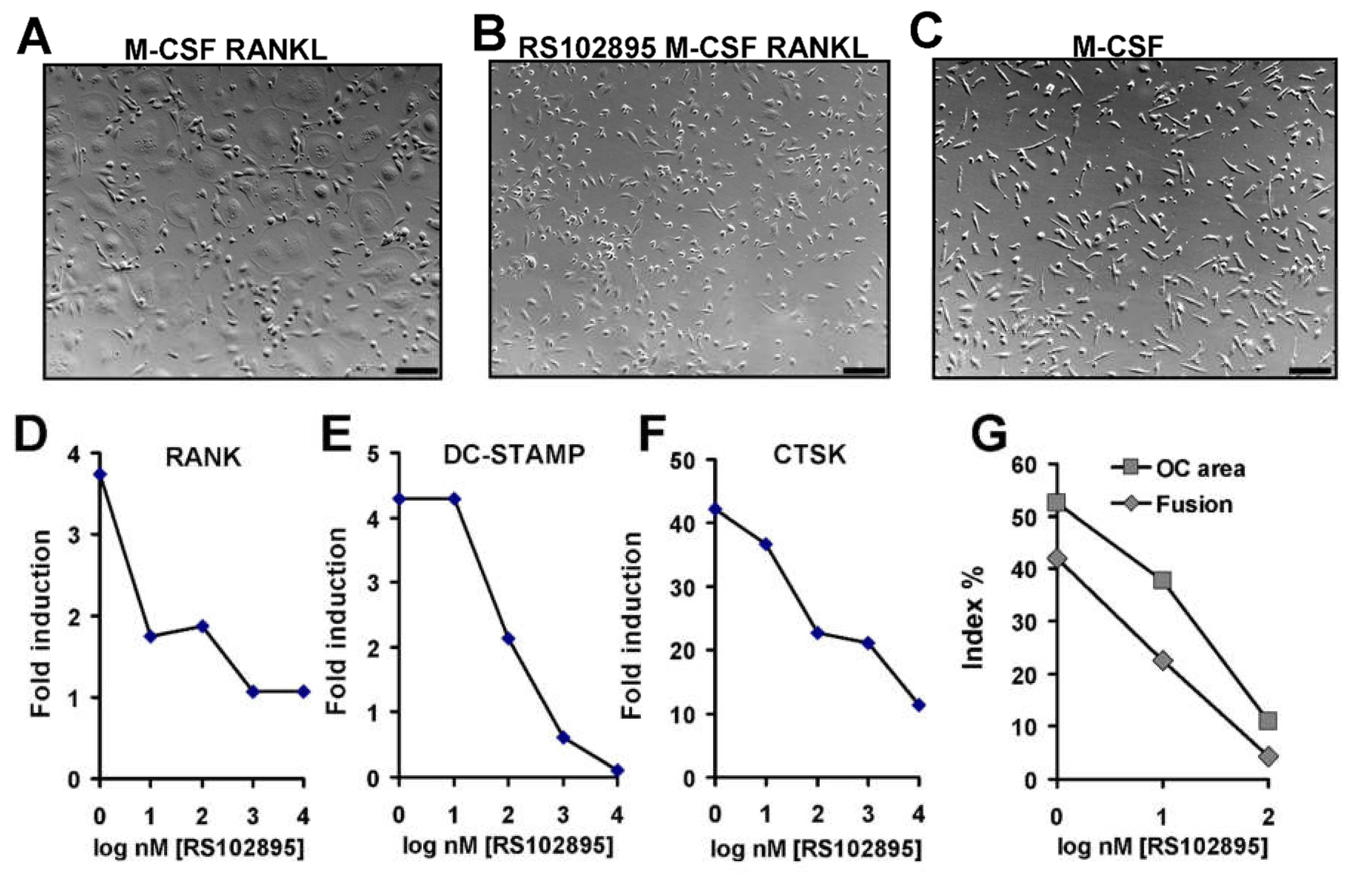
3.7. Effect of CCR2 Antagonist on Human Osteoclast Formation and Function
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Murphy, P.M.; Baggiolini, M.; Charo, I.F.; Hébert, C.A.; Horuk, R.; Matsushima, K.; Miller, L.H.; Oppenheim, J.J.; Power, C.A. International Union of Pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol. Rev. 2000, 52, 145–176. [Google Scholar] [PubMed]
- Miller, M.; Mayo, K. Chemokines from a structural perspective. Int. J. Mol. Sci. 2017, 18, 2088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, C.E.; Nibbs, R.J. A guide to chemokines and their receptors. FEBS J. 2018, 285, 2944–2971. [Google Scholar] [CrossRef] [PubMed]
- Mulholland, B.S.; Forwood, M.R.; Morrison, N.A. Monocyte chemoattractant protein-1(MCP-1/CCL2) drives activation of bone remodelling and skeletal metastasis. Curr. Osteoporos. Rep. 2019, 17, 538–547. [Google Scholar] [CrossRef] [Green Version]
- Gschwandtner, M.; Derler, R.; Midwood, K.S. More than just attractive: How CCL2 influences myeloid cell behavior beyond chemotaxis. Front. Immunol. 2019, 10, 2759–2788. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, S.; Takahashi, N.; Udagawa, N.; Tamura, T.; Akatsu, T.; Stanley, E.R.; Kurokawa, T.; Suda, T. Macrophage colony-stimulating factor is indispensable for both proliferation and differentiation of osteoclast progenitors. J. Clin. Investig. 1993, 91, 257–263. [Google Scholar] [CrossRef] [Green Version]
- Suda, T.; Takahashi, N.; Udagawa, N.; Jimi, E.; Gillespie, M.T.; Martin, T.J. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr. Rev. 1999, 20, 345–357. [Google Scholar] [CrossRef]
- Day, C.J.; Kim, M.S.; Stephens, S.R.; Simcock, W.E.; Aitken, C.J.; Nicholson, G.C.; Morrison, N.A. Gene array identification of osteoclast genes: Differential inhibition of osteoclastogenesis by cyclosporin A and granulocyte macrophage colony stimulating factor. J. Cell. Biochem. 2004, 91, 303–315. [Google Scholar] [CrossRef]
- Kim, M.S.; Day, C.J.; Morrison, N.A. MCP-1 is induced by receptor activator of nuclear factor-{kappa}B ligand, promotes human osteoclast fusion, and rescues granulocyte macrophage colony-stimulating factor suppression of osteoclast formation. J. Boil. Chem. 2005, 280, 16163–16169. [Google Scholar] [CrossRef] [Green Version]
- Morrison, N.A.; Day, C.J.; Nicholson, G.C. Dominant negative MCP-1 blocks human osteoclast differentiation. J. Cell. Biochem. 2014, 115, 303–312. [Google Scholar] [CrossRef]
- Quan, J.; Morrison, N.A.; Johnson, N.W.; Gao, J. MCP-1 as a potential target to inhibit the bone invasion by oral squamous cell carcinoma. J. Cell. Biochem. 2014, 115, 1787–1798. [Google Scholar] [CrossRef] [PubMed]
- Sul, O.J.; Ke, K.; Kim, W.K.; Kim, S.H.; Lee, S.C.; Kim, H.J.; Kim, S.Y.; Suh, J.H.; Choi, H.S. Absence of MCP-1 leads to elevated bone mass via impaired actin ring formation. J. Cell. Physiol. 2012, 227, 1619–1627. [Google Scholar] [CrossRef] [PubMed]
- Binder, N.B.; Niederreiter, B.; Hoffmann, O.; Stange, R.; Pap, T.; Stulnig, T.M.; Mack, M.; Erben, R.G.; Smolen, J.S.; Redlich, K. Estrogen-dependent and C-C chemokine receptor-2-dependent pathways determine osteoclast behavior in osteoporosis. Nat. Med. 2009, 15, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, H.; Qin, L.; Tamasi, J.; Bergenstock, M.; Shapses, S.; Feyen, J.H.; Notterman, D.A.; Partridge, N.C. Determination of parathyroid hormone’s dual effects on skeletal gene expression in vivo by microarray and network analysis. J. Biol. Chem. 2007, 282, 33086–33097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamasi, J.A.; Vasilov, A.; Shimizu, E.; Benton, N.; Johnson, J.; Bitel, C.L.; Morrison, N.; Partridge, N.C. Monocyte chemoattractant protein-1 is a mediator of the anabolic action of parathyroid hormone on bone. J. Bone Miner. Res. 2013, 28, 1975–1986. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, J.A.; Johnson, J.; Le Henaff, C.; Bitel, C.L.; Tamasi, J.A.; Partridge, N.C. Catabolic effects of human PTH (1–34) on bone: Requirement of monocyte chemoattractant protein-1 in murine model of hyperparathyroidism. Sci. Rep. 2017, 7, 15300. [Google Scholar] [CrossRef] [Green Version]
- Khan, U.A.; Hashimi, S.M.; Khan, S.; Quan, J.; Bakr, M.M.; Forwood, M.R.; Morrison, N.A. Differential expression of chemokines, chemokine receptors and proteinases by foreign body giant cells (FBGCs) and osteoclasts. J. Cell. Biochem. 2014, 115, 1290–1298. [Google Scholar] [CrossRef] [Green Version]
- Khan, U.A.; Hashimi, S.M.; Bakr, M.M.; Forwood, M.R.; Morrison, N.A. CCL2 and CCR2 are essential for the formation of osteoclasts and foreign body giant cells. J. Cell. Biochem. 2016, 117, 382–389. [Google Scholar] [CrossRef] [Green Version]
- Lean, J.M.; Murphy, C.; Fuller, K.; Chambers, T.J. CCL9/MIP-1gamma and its receptor CCR1 are the major chemokine ligand/receptor species expressed by osteoclasts. J. Cell. Biochem. 2002, 87, 386–393. [Google Scholar] [CrossRef]
- Okamatsu, Y.; Kim, D.; Battaglino, R.; Sasaki, H.; Späte, U.; Stashenko, P. MIP-1 gamma promotes receptor-activator-of-NF-kappa-B-ligand-induced osteoclast formation and survival. J. Immunol. 2004, 173, 2084–2090. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Mailhot, G.; MacKay, C.A.; Mason-Savas, A.; Aubin, J.; Odgren, P.R. Chemokine and chemokine receptor expression during colony stimulating factor-1-induced osteoclast differentiation in the toothless osteopetrotic rat: A key role for CCL9 (MIP-1gamma) in osteoclastogenesis in vivo and in vitro. Blood 2006, 107, 2262–2270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houser, B. Bio-Rad’s Bio-Plex® suspension array system, xMAP technology overview. Arch. Physiol. Biochem. 2012, 118, 192–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirzadegan, T.; Diehl, F.; Ebi, B.; Bhakta, S.; Polsky, I.; McCarley, D.; Mulkins, M.; Weatherhead, G.S.; Lapierre, J.M.; Dankwardt, J.; et al. Identification of the binding site for a novel class of CCR2b chemokine receptor antagonists: Binding to a common chemokine receptor motif within the helical bundle. J. Biol. Chem. 2000, 275, 25562–25571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.S.; Magno, C.L.; Day, C.J.; Morrison, N.A. Induction of chemokines and chemokine receptors CCR2b and CCR4 in authentic human osteoclasts differentiated with RANKL and osteoclast like cells differentiated by MCP-1 and RANTES. J. Cell. Biochem. 2006, 97, 512–518. [Google Scholar] [CrossRef]
- Granfar, R.M.; Day, C.J.; Kim, M.S.; Morrison, N.A. Optimised real-time quantitative PCR assays for RANKL regulated genes. Mol. Cell. Probes 2005, 19, 119–126. [Google Scholar] [CrossRef]
- Stephens, A.S.; Stephens, S.R.; Morrison, N.A. Internal control genes for quantitative RT-PCR expression analysis in mouse osteoblasts, osteoclasts and macrophages. BMC Res. Notes 2011, 4, 410. [Google Scholar] [CrossRef] [Green Version]
- Hodge, J.M.; Kirkland, M.A.; Aitken, C.J.; Waugh, C.M.; Myers, D.E.; Lopez, C.M.; Adams, B.E.; Nicholson, G.C. Osteoclastic potential of human CFU-GM: Biphasic effect of GM-CSF. J. Bone Miner. Res. 2004, 19, 190–199. [Google Scholar] [CrossRef]
- Ishikawa, M.; Ito, H.; Kitaori, T.; Murata, K.; Shibuya, H.; Furu, M.; Yoshitomi, H.; Fujii, T.; Yamamoto, K.; Matsuda, S. MCP/CCR2 signaling is essential for recruitment of mesenchymal progenitor cells during the early phase of fracture healing. PLoS ONE 2014, 9, e104954. [Google Scholar] [CrossRef] [Green Version]
| Gene | Forward | Reverse | Size | Reference |
|---|---|---|---|---|
| CCL1 | GACACAGTTGGATGGGTTCA | CCTCTGTGACCTAGCAAAAG | 174 | This study |
| MCP1(CCL2) | TCGCGAGCTATAGAAGAATCA | TGTTCAAGTCTTCGGAGTTTG | 161 | [9] |
| CCL3 | CTATGGACTGGTTGTTGCCA | AGGGGAACTCTCAGAGCAAA | 140 | [24] |
| CCL4 | TCCATGAGACACATCTCCTC | GCAACAGCAGAGAAACAGTG | 186 | This study |
| CCL5 | GAGCTTCTGAGGCGCTGCT | TCTAGAGGCATGCTGACTTC | 112 | [24] |
| CCR1 | TTCCTGTTCACCCATGAGTG | AAGGGGAGCCATTTAACCAG | 191 | [24] |
| CCR2a | CATAGCTCTTGGCTGTAGGA | GTGAAGCCAGACGTGTGATT | 224 | [24] |
| CCR2b | AACAAACACGCCTTCCACTG | GTCAAAGTCTCTACCCACAG | 259 | [24] |
| CCR3 | ACCCTACAATGTGGCTATCC | TTCATGCAGCAGTGGGAGTA | 132 | This study |
| CCR4 | CTTATGGGGTCATCACCAGT | AGTAGGTATGGTTGCGCTCA | 105 | [24] |
| CCR5 | ACCAAGCTATGCAGGTGACA | GAACAGCATTTGCAGAAGCG | 148 | [25] |
| CCR6 | GGACAGCTGGAATTATGCTG | CCCATGACAGTACCTTCCTA | 208 | This study |
| CCR7 | ACGCAACTTTGAGCGCAACA | TTGCTTACTGAGCTCACAGG | 151 | This study |
| CCR8 | GTCCCATTCAACGTGGTTCT | AGCTCTCCCTAGGCATTTGT | 250 | This study |
| CTSK | TGAGGCTTCTCTTGGTGTCCATAC | AAAGGGTGTCATTACTGCGGG | 134 | [8] |
| TRAP | GACCACCTTGGCAATGTCTCTG | TGGCTGAGGAAGTCATCTGAGTTG | 176 | [8] |
| RANK | CAGAACTAAGCTCAGTATGTGA | GAATGCCAAGCTGCAGCAAC | 121 | [9] |
| DC-STAMP | AGACCTGGGTTCCTCTCAGTGTTAT | GTTGGTGCGATGTGGCTGAGG | 326 | This study |
| GAPDH | ACAGTCCATGCCATCACTGCC | GCCTGCTTCACCACCTTCTTG | 266 | [26] |
| 18S rRNA | CTTAGAGGGACAAGTGGCG | ACGCTGAGCCAGTCAGTGTA | 107 | [9] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morrison, N.A.; Forwood, M.R. Monocyte Chemotactic Protein-1 (MCP1) Accumulation in Human Osteoclast Precursor Cultures. Life 2022, 12, 789. https://doi.org/10.3390/life12060789
Morrison NA, Forwood MR. Monocyte Chemotactic Protein-1 (MCP1) Accumulation in Human Osteoclast Precursor Cultures. Life. 2022; 12(6):789. https://doi.org/10.3390/life12060789
Chicago/Turabian StyleMorrison, Nigel A., and Mark R. Forwood. 2022. "Monocyte Chemotactic Protein-1 (MCP1) Accumulation in Human Osteoclast Precursor Cultures" Life 12, no. 6: 789. https://doi.org/10.3390/life12060789
APA StyleMorrison, N. A., & Forwood, M. R. (2022). Monocyte Chemotactic Protein-1 (MCP1) Accumulation in Human Osteoclast Precursor Cultures. Life, 12(6), 789. https://doi.org/10.3390/life12060789





