Chronic Exposure to the Food Additive tBHQ Modulates Expression of Genes Related to SARS-CoV-2 and Influenza Viruses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Transcriptome Analysis
2.3. Statistical Analysis
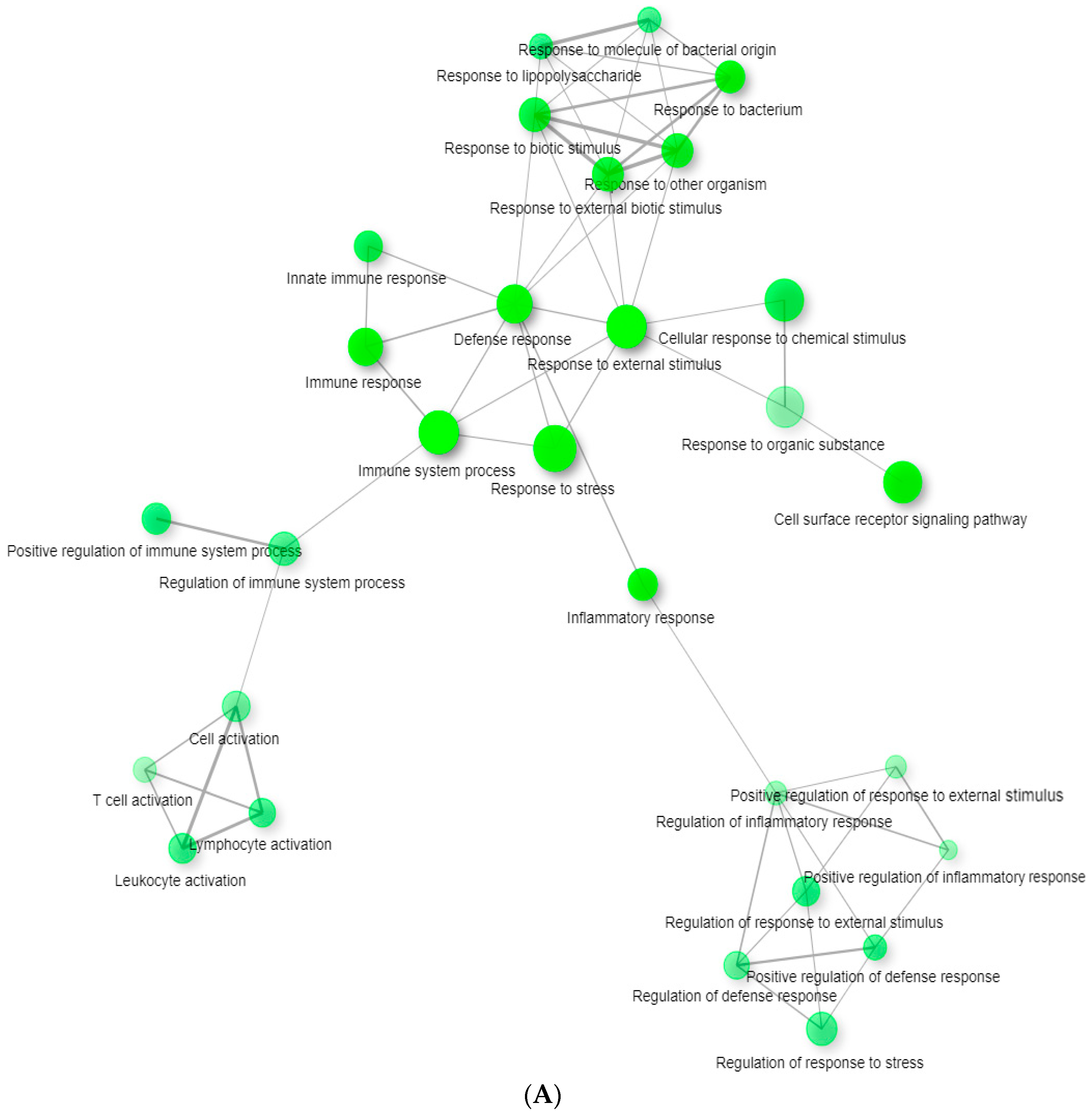
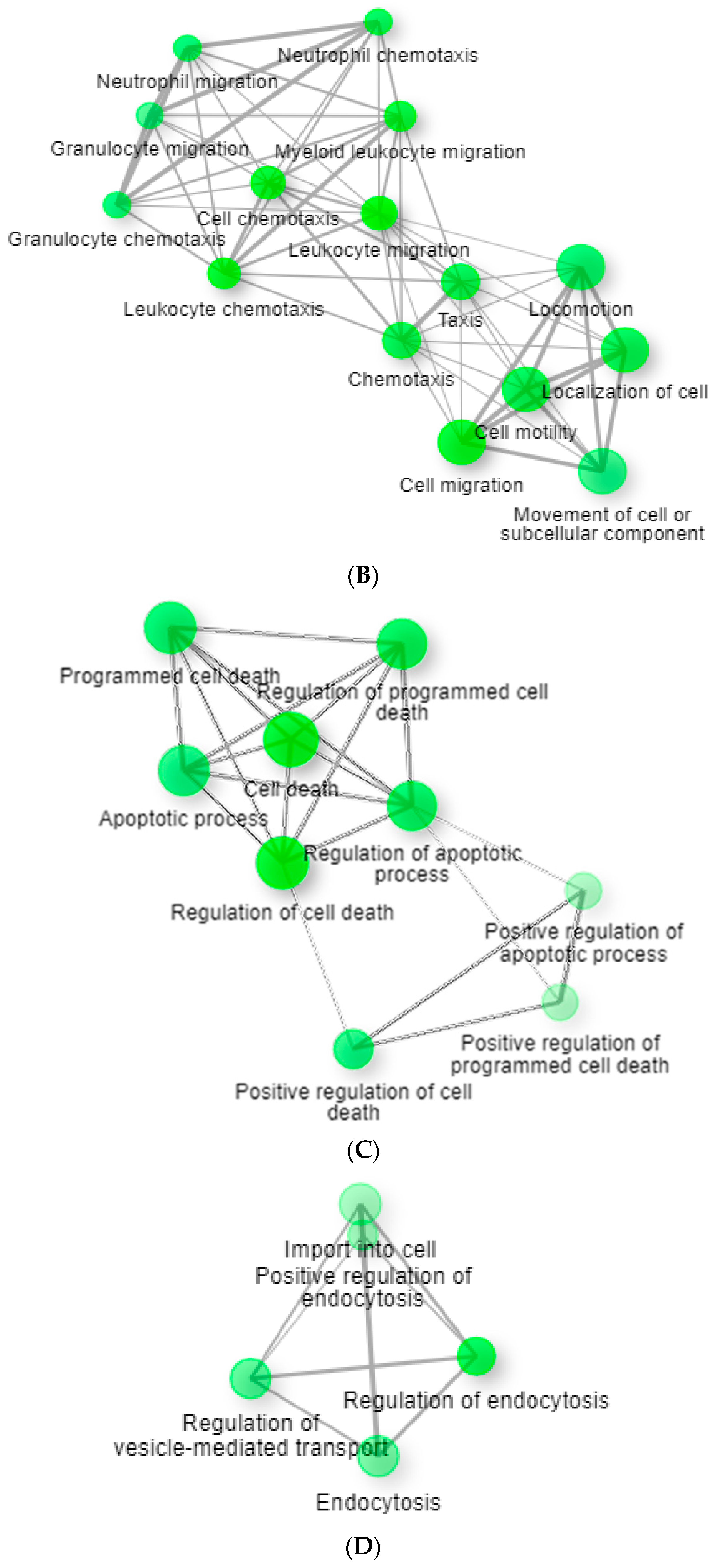
3. Results
3.1. Profiling mRNA Expression after tBHQ Treatment
3.2. Network Analysis
4. Discussion
4.1. tBHQ Exposure and Influenza Virus Infection
4.2. tBHQ Exposure and SARS-CoV-2 Virus Infection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hazardous Substances Data Bank. Bethesda (MD): National Library of Medicine (US); t-Butylhydroquinone; CASRN: 1948-33-0; Hazardous Substances Data Bank: Bethesda, MD, USA, 2022. Available online: https://toxnet.nlm.nih.gov/cgi-bin/sis/search/a?dbs+hsdb:@term+@DOCNO+838 (accessed on 15 April 2022).
- European Food Safety Authority (EFSA). Opinion of the Scientific Panel on Food Additives, Flavourings, Processing Aids and Materials in Contact with Food on a request from the Commission related to tertiary-Butylhydroquinone (TBHQ) Question number EFSA-Q-2003-141. EFSA J. 2004, 2, 84. [Google Scholar]
- Le Coz, C.J.; Schneider, G.A. Contact dermatitis from tertiary butylhydroquinone in a hair dye with cross sensitivity to BHA and BHT. Contact Dermat. 1987, 17, 257–258. [Google Scholar] [CrossRef]
- Osburn, W.O.; Yates, M.S.; Dolan, P.D.; Chen, S.; Liby, K.T.; Sporn, M.B.; Taguchi, K.; Yamamoto, M.; Kensler, T.W. Genetic or pharmacologic amplification of nrf2 signaling inhibits acute inflammatory liver injury in mice. Toxicol. Sci. 2008, 104, 218–227. [Google Scholar] [CrossRef] [PubMed]
- van Esch, G.J. Toxicology of tert-butylhydroquinone (TBHQ). Food Chem. Toxicol. 1986, 24, 1063–1065. [Google Scholar] [CrossRef]
- Rockwell, C.E.; Zhang, M.; Fields, P.E.; Klaassen, C.D. Th2 skewing by activation of Nrf2 in CD4(+) T cells. J. Immunol. 2012, 188, 1630–1637. [Google Scholar] [CrossRef] [Green Version]
- Zagorski, J.W.; Turley, A.E.; Dover, H.E.; Van Den Berg, K.R.; Compton, J.R.; Rockwell, C.E. The Nrf2 activator, tBHQ, differentially affects early events following stimulation of Jurkat cells. Toxicol. Sci. 2013, 136, 63–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turley, A.E.; Zagorski, J.W.; Rockwell, C.E. The Nrf2 activator tBHQ inhibits T cell activation of primary human CD4 T cells. Cytokine 2015, 71, 289–295. [Google Scholar] [CrossRef] [Green Version]
- Freeborn, R.A.; Boss, A.P.; Liu, S.; Jin, Y.; Brocke, S.; Turley, A.E.; Zagordki, J.W.; Kennedy, R.C.; Gardner, E.M.; Rockwell, C.E. The Immune Response to Influenza is Suppressed by the Synthetic Food Additive and Nrf2 Activator, tert-butylhydroquinone (tBHQ). FASEB J. 2019, 33, 505-3. [Google Scholar] [CrossRef]
- Ito, N.; Hirose, M.; Fukushima, S.; Tsuda, H.; Shirai, T.; Tatematsu, M. Studies on antioxidants: Their carcinogenic and modifying effects on chemical carcinogenesis. Food Chem. Toxicol. 1986, 24, 1071–1082. [Google Scholar] [CrossRef]
- Petschner, P.; Bagdy, G.; Tóthfalusi, L. The problem of small “n” and big “P” in neuropsycho-pharmacology, or how to keep the rate of false discoveries under control. Neuropsychopharmacol. Hung. 2015, 17, 23–30. [Google Scholar]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A Free, Open-Source System for Microarray Data Management and Analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saeed, A.I.; Bhagabati, N.K.; Braisted, J.C.; Liang, W.; Sharov, V.; Howe, E.A.; Li, J.; Thiagarajan, M.; White, J.A.; Quackenbush, J. [9] TM4 Microarray Software Suite. Methods Enzymol. 2006, 411, 134–193. [Google Scholar] [CrossRef] [PubMed]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Wu, C.; Jin, X.; Tsueng, G.; Afrasiabi, C.; Su, A.I. BioGPS: Building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 2016, 44, D313–D316. [Google Scholar] [CrossRef] [Green Version]
- Zerbino, D.R.; Wilder, S.P.; Johnson, N.; Juettemann, T.; Flicek, P.R. The Ensembl Regulatory Build. Genome Biol. 2015, 16, 56. [Google Scholar] [CrossRef] [Green Version]
- Andersson, R.; Gebhard, C.; Miguel-Escalada, I.; Hoof, I.; Bornholdt, J.; Boyd, M.; Chen, Y.; Zhao, X.; Schmidl, C.; Suzuki, T.; et al. An atlas of active enhancers across human cell types and tissues. Nature 2014, 507, 455–461. [Google Scholar] [CrossRef]
- Visel, A.; Minovitsky, S.; Dubchak, I.; Pennacchio, L.A. VISTA Enhancer Browser—A database of tissue-specific human enhancers. Nucleic Acids Res. 2006, 35, D88–D92. [Google Scholar] [CrossRef]
- Khan, A.; Zhang, X. DbSUPER: A database of super-enhancers in mouse and human genome. Nucleic Acids Res. 2016, 44, D164–D171. [Google Scholar] [CrossRef] [Green Version]
- Dreos, R.; Ambrosini, G.; Périer, R.C.; Bucher, P. EPD and EPDnew, high-quality promoter resources in the next-generation sequencing era. Nucleic Acids Res. 2013, 41, D157–D164. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-W.; Liang, J.; Yan, J.-X.; Ye, Y.-C.; Wang, J.-J.; Chen, C.; Sun, H.-T.; Chen, F.; Tu, Y.; Li, X.-H. TBHQ improved neurological recovery after traumatic brain injury by inhibiting the overactivation of astrocytes. Brain Res. 2020, 1739, 146818. [Google Scholar] [CrossRef] [PubMed]
- Kesic, M.J.; Simmons, S.O.; Bauer, R.; Jaspers, I. Nrf2 expression modifies influenza A entry and replication in nasal epithelial cells. Free Radic. Biol. Med. 2011, 51, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Olagnier, D.; Farahani, E.; Thyrsted, J.; Blay-Cadanet, J.; Herengt, A.; Idorn, M.; Hait, A.; Hernaez, B.; Knudsen, A.; Iversen, M.B.; et al. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat. Commun. 2020, 11, 4938. [Google Scholar] [CrossRef] [PubMed]
- Herengt, A.; Thyrsted, J.; Holm, C.K. NRF2 in Viral Infection. Antioxidants 2021, 10, 1491. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, F.; Li, Z.; Garcia, C.C.; Quintana, F.J. A potential role for AHR in SARS-CoV-2 pathology. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Dimitrieva, S.; Bucher, P. UCNEbase—A database of ultraconserved non-coding elements and genomic regulatory blocks. Nucleic Acids Res. 2012, 41, D101–D109. [Google Scholar] [CrossRef]
- Antanasijevic, A.; Cheng, H.; Wardrop, D.J.; Rong, L.; Caffrey, M. Inhibition of Influenza H7 Hemagglutinin-Mediated Entry. PLoS ONE 2013, 8, e76363. [Google Scholar] [CrossRef] [Green Version]
- Vanderlinden, E.; Göktaş, F.; Cesur, Z.; Froeyen, M.; Reed, M.L.; Russell, C.J.; Cesur, N.; Naesens, L. Novel Inhibitors of Influenza Virus Fusion: Structure-Activity Relationship and Interaction with the Viral Hemagglutinin. J. Virol. 2010, 84, 4277–4288. [Google Scholar] [CrossRef] [Green Version]
- Russell, R.J.; Kerry, P.S.; Stevens, D.J.; Steinhauer, D.A.; Martin, S.R.; Gamblin, S.J.; Skehel, J.J. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc. Natl. Acad. Sci. USA 2008, 105, 17736–17741. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Zhang, X.; Liu, S. Novel hemagglutinin-based influenza virus inhibitors. J. Thorac. Dis. 2013, 5, S149–S159. [Google Scholar] [CrossRef]
- Ng, W.C.; Liong, S.; Tate, M.D.; Irimura, T.; Denda-Nagai, K.; Brooks, A.G.; Londrigan, S.L.; Reading, P.C. The Macrophage Galactose-Type Lectin Can Function as an Attachment and Entry Receptor for Influenza Virus. J. Virol. 2014, 88, 1659–1672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauligk, C.; Nain, M.; Reiling, N.; Gemsa, D.; Kaufmann, A. CD14 is required for influenza A virus-induced cytokine and chemokine production. Immunobiology 2004, 209, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.R.; Skeate, J.; Kast, W.M. Annexin A2 in Virus Infection. Front. Microbiol. 2018, 9, 2954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ampomah, P.B.; Kong, W.T.; Zharkova, O.; Chua, S.C.J.H.; Samy, R.P.; Lim, L.H.K. Annexins in Influenza Virus Replication and Pathogenesis. Front. Pharmacol. 2018, 9, 1282. [Google Scholar] [CrossRef]
- Alessi, M.-C.; Cenac, N.; Si-Tahar, M.; Riteau, B. FPR2: A Novel Promising Target for the Treatment of Influenza. Front. Microbiol. 2017, 8, 1719. [Google Scholar] [CrossRef]
- Tran, A.T.; Rahim, M.N.; Ranadheera, C.; Kroeker, A.; Cortens, J.P.; Opanubi, K.J.; Wilkins, J.A.; Coombs, K.M. Knockdown of specific host factors protects against influenza virus-induced cell death. Cell Death Dis. 2013, 4, e769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, B.; Schuster, S.; Zysset, D.; Rihs, S.; Dickgreber, N.; Schurch, C.; Riether, C.; Siegrist, M.; Schneider, C.; Pawelski, H.; et al. TREM-1 Deficiency Can Attenuate Disease Severity without Affecting Pathogen Clearance. PLoS Pathog. 2014, 10, e1003900. [Google Scholar] [CrossRef] [Green Version]
- Piao, W.; Shirey, K.A.; Ru, L.W.; Lai, W.; Szmacinski, H.; Snyder, G.A.; Sundberg, E.; Lakowicz, J.R.; Vogel, S.N.; Toshchakov, V.Y. A Decoy Peptide that Disrupts TIRAP Recruitment to TLRs Is Protective in a Murine Model of Influenza. Cell Rep. 2015, 11, 1941–1952. [Google Scholar] [CrossRef] [Green Version]
- Thanos, D.; Maniatis, T. Identification of the rel family members required for virus induction of the human beta interferon gene. Mol. Cell. Biol. 1995, 15, 152–164. [Google Scholar] [CrossRef] [Green Version]
- Traets, M.J.M.; Nijhuis, R.H.T.; Morré, S.A.; Ouburg, S.; Remijn, J.A.; Blok, B.A.; de Laat, B.; Jong, E.; Herder, G.J.M.; Fiolet, A.T.L.; et al. Association of genetic variations in ACE2, TIRAP and factor X with outcomes in COVID-19. PLoS ONE 2022, 17, e0260897. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Zhao, Z.; Yang, Z.; Meng, Q.; Tan, P.; Xie, W.; Qin, Y.; Wang, R.-F.; Cui, J. USP38 Inhibits Type I Interferon Signaling by Editing TBK1 Ubiquitination through NLRP4 Signalosome. Mol. Cell 2016, 64, 267–281. [Google Scholar] [CrossRef] [Green Version]
- Newby, B.N.; Brusko, T.M.; Zou, B.; Atkinson, M.A.; Clare-Salzler, M.; Mathews, C.E. Type 1 Interferons Potentiate Human CD8+ T-Cell Cytotoxicity Through a STAT4- and Granzyme B—Dependent Pathway. Diabetes 2017, 66, 3061–3071. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Liu, J.; Qian, L.; Feng, Q.; Wang, X.; Yuan, Y.; Zuo, Y.; Cheng, Q.; Miao, Y.; Guo, T.; et al. Induction of OTUD1 by RNA viruses potently inhibits innate immune responses by promoting degradation of the MAVS/TRAF3/TRAF6 signalosome. PLoS Pathog. 2018, 14, e1007067. [Google Scholar] [CrossRef] [Green Version]
- Carey, M.A.; Bradbury, J.A.; Rebolloso, Y.D.; Graves, J.P.; Zeldin, D.C.; Germolec, D.R. Pharmacologic Inhibition of COX-1 and COX-2 in Influenza A Viral Infection in Mice. PLoS ONE 2010, 5, e11610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carey, M.A.; Bradbury, J.A.; Seubert, J.M.; Langenbach, R.; Zeldin, D.; Germolec, D.R. Contrasting Effects of Cyclooxygenase-1 (COX-1) and COX-2 Deficiency on the Host Response to Influenza A Viral Infection. J. Immunol. 2005, 175, 6878–6884. [Google Scholar] [CrossRef] [PubMed]
- Staurengo-Ferrari, L.; Badaro-Garcia, S.; Hohmann, M.S.N.; Manchope, M.F.; Zaninelli, T.; Casagrande, R.; Verri, W. Contribution of Nrf2 Modulation to the Mechanism of Action of Analgesic and Anti-inflammatory Drugs in Pre-clinical and Clinical Stages. Front. Pharmacol. 2019, 9, 1536. [Google Scholar] [CrossRef] [Green Version]
- Bradley, L.M.; Douglass, M.F.; Chatterjee, D.; Akira, S.; Baaten, B.J.G. Matrix Metalloprotease 9 Mediates Neutrophil Migration into the Airways in Response to Influenza Virus-Induced Toll-Like Receptor Signaling. PLoS Pathog. 2012, 8, e1002641. [Google Scholar] [CrossRef] [Green Version]
- Sanders, C.; Thomas, P. MMP-7 promotes host recovery and lung function to influenza virus infection (VIR2P.1016). J. Immunol. 2014, 192, 75.5. [Google Scholar]
- Wang, S.; Le, T.Q.; Chida, J.; Cisse, Y.; Yano, M.; Kido, H. Mechanisms of matrix metalloproteinase-9 upregulation and tissue destruction in various organs in influenza A virus infection. J. Med. Investig. 2010, 57, 26–34. [Google Scholar] [CrossRef] [Green Version]
- Mendonca, P.; Soliman, K.F.A. Flavonoids Activation of the Transcription Factor Nrf2 as a Hypothesis Approach for the Prevention and Modulation of SARS-CoV-2 Infection Severity. Antioxidants 2020, 9, 659. [Google Scholar] [CrossRef] [PubMed]
- Desai, T.M.; Marin, M.; Chin, C.; Savidis, G.; Brass, A.L.; Melikyan, G.B. IFITM3 Restricts Influenza A Virus Entry by Blocking the Formation of Fusion Pores following Virus-Endosome Hemifusion. PLoS Pathog. 2014, 10, e1004048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, I.-C.; Bailey, C.C.; Weyer, J.L.; Radoshitzky, S.; Becker, M.M.; Chiang, J.J.; Brass, A.L.; Ahmed, A.A.; Chi, X.; Dong, L.; et al. Distinct Patterns of IFITM-Mediated Restriction of Filoviruses, SARS Coronavirus, and Influenza A Virus. PLoS Pathog. 2011, 7, e1001258. [Google Scholar] [CrossRef] [PubMed]
- Maiese, A.; Passaro, G.; De Matteis, A.; Fazio, V.; Raffaele, L.R.; Di Paolo, M. Thromboinflammatory response in SARS-CoV-2 sepsis. Med. -Leg. J. 2020, 88, 78–80. [Google Scholar] [CrossRef] [PubMed]
- Zanza, C.; Romenskaya, T.; Manetti, A.C.; Franceschi, F.; La Russa, R.; Bertozzi, G.; Maiese, A.; Savioli, G.; Volonnino, G.; Longhitano, Y. Cytokine Storm in COVID-19: Immunopathogenesis and Therapy. Medicina 2022, 58, 144. [Google Scholar] [CrossRef]
- Marro, B.S.; Hosking, M.P.; Lane, T.E. CXCR2 signaling and host defense following coronavirus-induced encephalomyelitis. Futur. Virol. 2012, 7, 349–359. [Google Scholar] [CrossRef] [Green Version]
- Simmons, G.; Zmora, P.; Gierer, S.; Heurich, A.; Pöhlmann, S. Proteolytic activation of the SARS-coronavirus spike protein: Cutting enzymes at the cutting edge of antiviral research. Antivir. Res. 2013, 100, 605–614. [Google Scholar] [CrossRef]
- Zhao, M.-M.; Yang, W.-L.; Yang, F.-Y.; Zhang, L.; Huang, W.-J.; Hou, W.; Fan, C.-F.; Jin, R.-H.; Feng, Y.-M.; Wang, Y.-C.; et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal. Transduct. Target. Ther. 2021, 6, 134. [Google Scholar] [CrossRef]
- Zhou, Y.; Vedantham, P.; Lu, K.; Agudelo, J.; Carrion, R., Jr.; Nunneley, J.W.; Barnard, D.; Pöhlmann, S.; McKerrow, J.H.; Renslo, A.R.; et al. Protease inhibitors targeting coronavirus and filovirus entry. Antivir. Res. 2015, 116, 76–84. [Google Scholar] [CrossRef]
- Martin, T.R.; Wurfel, M.M.; Zanoni, I.; Ulevitch, R. Targeting innate immunity by blocking CD14: Novel approach to control inflammation and organ dysfunction in COVID-19 illness. EBioMedicine 2020, 57, 102836. [Google Scholar] [CrossRef]
- Florin, L.; Lang, T. Tetraspanin Assemblies in Virus Infection. Front. Immunol. 2018, 9, 1140. [Google Scholar] [CrossRef] [PubMed]
- Messner, C.B.; Demichev, V.; Wendisch, D.; Michalick, L.; White, M.; Freiwald, A.; Textoris-Taube, K.; Vernardis, S.I.; Egger, A.S.; Kreidl, M.; et al. Clinical classifiers of COVID-19 infection from novel ultra-high-throughput proteomics. MedRxivr Prepr. 2020. [Google Scholar] [CrossRef]
- Cunningham, L.; Simmonds, P.; Kimber, I.; Basketter, D.A.; McFadden, J.P. Perforin and resistance to SARS coronavirus 2. J. Allergy Clin. Immunol. 2020, 146, 52–53. [Google Scholar] [CrossRef] [PubMed]
- Mariani, E. Perforins in human cytolytic cells: The effect of age. Mech. Ageing Dev. 1996, 92, 195–209. [Google Scholar] [CrossRef]
- Rukavina, D.; Laskarin, G.; Rubesa, G.; Strbo, N.; Bedenicki, I.; Manestar, D.; Glavas, M.; Christmas, S.E.; Podack, E.R. Age-Related Decline of Perforin Expression in Human Cytotoxic T Lymphocytes and Natural Killer Cells. Blood 1998, 92, 2410–2420. [Google Scholar] [CrossRef] [Green Version]
- Evoskoboinik, I.; Trapani, J.A. Perforinopathy: A Spectrum of Human Immune Disease Caused by Defective Perforin Delivery or Function. Front. Immunol. 2013, 4, 441. [Google Scholar] [CrossRef]
- Cortjens, B.; De Boer, O.J.; De Jong, R.; Antonis, A.F.; Sabogal Piñeros, Y.S.; Lutter, R.; Van Woensel, J.B.; Bem, R.A. Neutrophil extracellular traps cause airway obstruction during respiratory syncytial virus disease. J. Pathol. 2016, 238, 401–411. [Google Scholar] [CrossRef]
- Arisan, E.D.; Uysal-Onganer, P.; Lange, S. Putative Roles for Peptidylarginine Deiminases in COVID-19. Int. J. Mol. Sci. 2020, 21, 4662. [Google Scholar] [CrossRef]
- Liang, B.; Chen, J.; Li, T.; Wu, H.; Yang, W.; Li, Y.; Li, J.; Yu, C.; Nie, F.; Ma, Z.; et al. Clinical remission of a critically ill COVID-19 patient treated by human umbilical cord mesenchymal stem cells: A case report. Medicine 2020, 99, e21429. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Gene Symbol | Fold Change (logFC) | adjP |
|---|---|---|---|
| Annexin A2 | Anxa2 | 0.707804 | 0.019984 |
| Cd14 antigen | Cd14 | 1.154081 | 0.017954 |
| Dual specificity phosphatase 10 | Dusp10 | −1.41983 | 0.0107 |
| Formyl peptide receptor, related sequence 2 | Fpr-rs2 | 1.407792 | 0.019984 |
| Granzyme B | Gzmb | 1.211705 | 0.042207 |
| Macrophage galactose N-acetyl-galactosamine specific lectin 1 | Mgl1 | −1.41116 | 0.020827 |
| Matrix metallopeptidase 7 | Mmp7 | 1.146066 | 0.021632 |
| Matrix metallopeptidase 9 | Mmp9 | 2.031156 | 0.0107 |
| OTU domain containing 1 | Otud | −0.68072 | 0.021705 |
| Prostaglandin-endoperoxide synthase 1 | Ptgs1 | 0.84145 | 0.037095 |
| Prostaglandin-endoperoxide synthase 2 | Ptgs2 | 0.89212 | 0.026521 |
| Reticuloendotheliosis oncogene | Rel | −0.85632 | 0.048081 |
| Toll-interleukin 1 receptor (TIR) TIR domain-containing adaptor protein (Tirap) | Tirap | 0.777524 | 0.020839 |
| Triggering receptor expressed on myeloid cells 1 | Trem1 | 2.063924 | 0.023686 |
| Tumor necrosis factor (ligand) superfamily, member 13 | Tnfsf13 (April) | 0.880437 | 0.046223 |
| Ubiquitin specific peptidase 38 | Usp38 | −0.5067 | 0.035 |
| Gene Name | Gene Symbol | Fold Change | adjP |
|---|---|---|---|
| Cathepsin B | Cts B | 0.514308 | 0.040677 |
| Cd14 antigen | Cd14 | 1.154081 | 0.017954 |
| Cd151 antigen | Cd151 | 0.449516 | 0.047853 |
| Cd209d antigen | Cd209d | −1.78035 | 0.023051 |
| Interferon induced transmembrane protein 1 | Ifitm1 | 1.154081 | 0.021632 |
| Interferon induced transmembrane protein 2 | Ifitm2 | 1.174042 | 0.017954 |
| Interleukin 8 receptor, beta | Cxcr2 (Il8rb) | 2.650023 | 0.016262 |
| Oncostatin M | Osm | 0.748042 | 0.02052 |
| Peptidyl arginine deiminase, type IV | Padi4 | 1.156019 | 0.040966 |
| Perforin 1 | Prf1 | 0.69257 | 0.03873 |
| Protein phosphatase 1 regulatory subunit 3D | Ppp1r3d | 2.300879 | 0.020477 |
| Synapsin II | Syn2 | 1.057722 | 0.033471 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Németh, K.; Petschner, P.; Pálóczi, K.; Fekete, N.; Pállinger, É.; Buzás, E.I.; Tamási, V. Chronic Exposure to the Food Additive tBHQ Modulates Expression of Genes Related to SARS-CoV-2 and Influenza Viruses. Life 2022, 12, 642. https://doi.org/10.3390/life12050642
Németh K, Petschner P, Pálóczi K, Fekete N, Pállinger É, Buzás EI, Tamási V. Chronic Exposure to the Food Additive tBHQ Modulates Expression of Genes Related to SARS-CoV-2 and Influenza Viruses. Life. 2022; 12(5):642. https://doi.org/10.3390/life12050642
Chicago/Turabian StyleNémeth, Krisztina, Peter Petschner, Krisztina Pálóczi, Nóra Fekete, Éva Pállinger, Edit I. Buzás, and Viola Tamási. 2022. "Chronic Exposure to the Food Additive tBHQ Modulates Expression of Genes Related to SARS-CoV-2 and Influenza Viruses" Life 12, no. 5: 642. https://doi.org/10.3390/life12050642
APA StyleNémeth, K., Petschner, P., Pálóczi, K., Fekete, N., Pállinger, É., Buzás, E. I., & Tamási, V. (2022). Chronic Exposure to the Food Additive tBHQ Modulates Expression of Genes Related to SARS-CoV-2 and Influenza Viruses. Life, 12(5), 642. https://doi.org/10.3390/life12050642








