New Alpha-Amylase Inhibitory Metabolites from Pericarps of Garcinia mangostana
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Experimental Procedures
2.2. Plant Material
2.3. Extraction and Separation
2.3.1. Garcimangophenone A (6)
2.3.2. Garcimangophenone B (7)
2.4. α-Amylase Inhibition
2.5. Molecular Docking and Dynamics Studies
2.5.1. Preparation of PDB Structures
2.5.2. Receptor Grids Generation and Docking
2.5.3. MD Simulation of Compound 7 and Acarbose in Complex with 5TD4
3. Results and Discussion
3.1. Purification of Compounds
3.2. Structural Assignment of Compounds 6 and 7
3.3. Identification of Known Metabolites
3.4. α-Amylase Inhibitory Activity
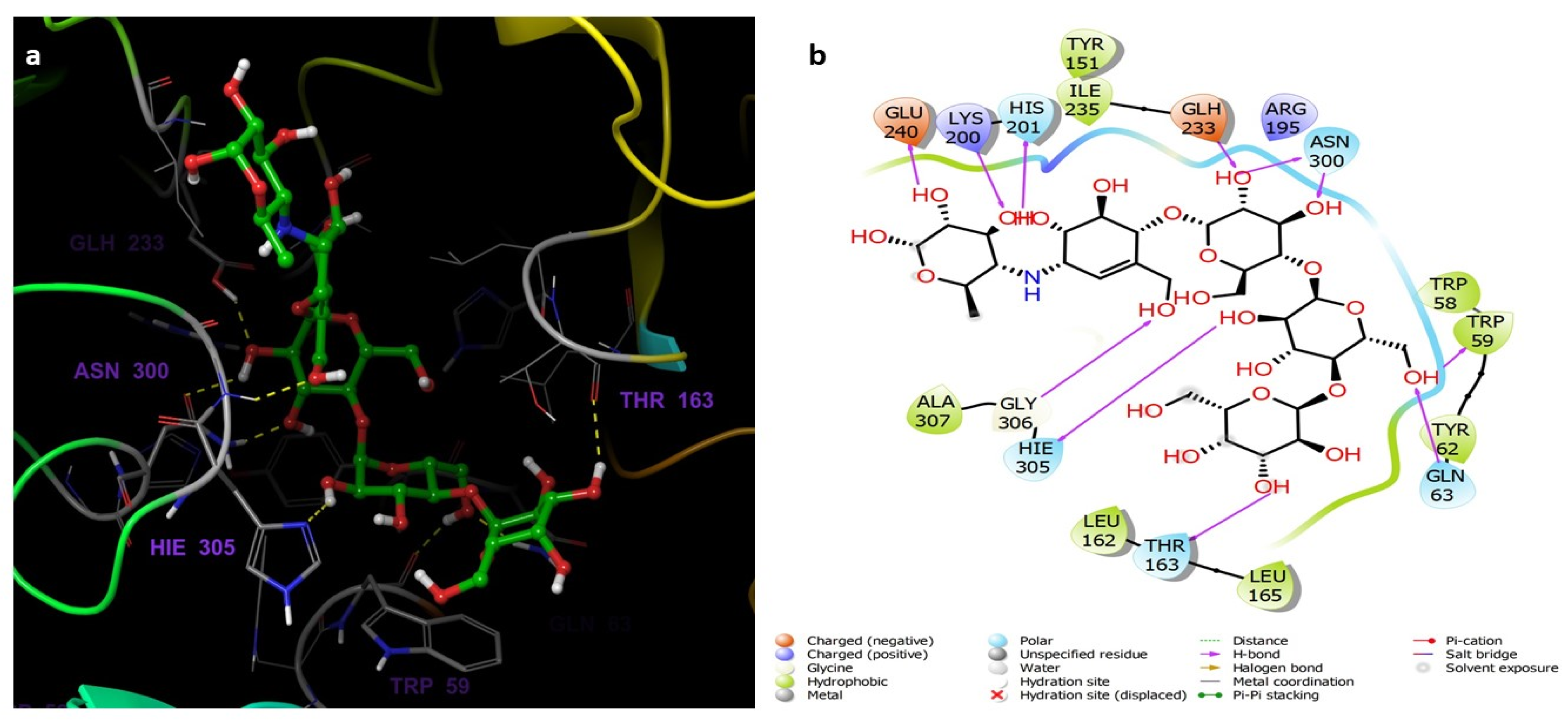
3.5. Molecular Docking and Dynamics Studies
3.5.1. Preparations of Ligands and Proteins
3.5.2. Molecular Docking Studies
3.5.3. Molecular Dynamic Simulation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ibrahim, S.R.M.; Mohamed, G.A.; Zayed, M.F.; Ross, S.A. 8-Hydroxyirilone 5-methyl ether and 8-hydroxyirilone, new antioxidant and α-amylase inhibitors isoflavonoids from Iris germanica rhizomes. Bioorg. Chem. 2017, 70, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.R.M.; Mohamed, G.A.; Khayat, M.T.; Ahmed, S.; Abo-Haded, H.; Alshali, K.Z. Mangostanaxanthone VIIII, a new xanthone from Garcinia mangostana pericarps, α-amylase inhibitory activity, and molecular docking studies. Rev. Bras. Farmacogn. 2019, 29, 206–212. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Mohamed, G.A.; Khayat, M.T.; Ahmed, S.; Abo-Haded, H. Garcixanthone D, a new xanthone, and other xanthone derivatives from Garcinia mangostana pericarps: Their α-amylase inhibitory potential and molecular docking studies. Starch-Stärke 2019, 71, 1800354. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Mohamed, G.A.; Khayat, M.T.; Ahmed, S.; Abo-Haded, H. α-Amylase inhibitors xanthones from Garcinia mangostana pericarps and its possible use for the treatment of diabetes with their molecular docking studies. J. Food. Biochem. 2019, 43, e12844. [Google Scholar] [CrossRef]
- Du, H.; Li, L.; Bennett, D.; Guo, Y.; Turnbull, I.; Yang, L.; Bragg, F.; Bian, Z.; Chen, Y.; Chen, J.; et al. Fresh fruit consumption in relation to incident diabetes and diabetic vascular complications: A 7-y prospective study of 0.5 million Chinese adults. PLoS Med. 2017, 14, e1002279. [Google Scholar] [CrossRef]
- Holman, R.R.; Paul, S.K.; Bethel, M.A.; Matthews, D.R.; Neil, H.A. 10-year follow-up of intensive glucose control in type 2 diabetes. N. Engl. J. Med. 2008, 359, 1577–1589. [Google Scholar] [CrossRef] [Green Version]
- Alothman, M.; Bhat, R.; Karim, A.A. Antioxidant capacity and phenolic content of selected tropical fruits from Malaysia, extracted with different solvents. Food Chem. 2009, 115, 785–788. [Google Scholar] [CrossRef]
- Muraki, I.; Imamura, F.; Manson, J.E.; Hu, F.B.; Willett, W.C.; van Dam, R.M.; Sun, Q. Fruit consumption and risk of type 2 diabetes: Results from three prospective longitudinal cohort studies. BMJ 2013, 347, f5001. [Google Scholar] [CrossRef] [Green Version]
- Ayala-Zavala, J.F.; Vega-Vega, V.; Rosas-Domínguez, C.; Palafox-Carlos, H.; Villa-Rodríguez, J.A.; Wasim Siddiqui, M.D.; González-Aguilar, G.A. Agro-industrial potential of exotic fruit by-products as a source of food additives. Food Res. Int. 2011, 44, 1866–1874. [Google Scholar] [CrossRef]
- Tundis, R.; Loizzo, M.R.; Menichini, F. Natural products as alpha-amylase and alpha-glucosidase inhibitors and their hypoglycaemic potential in the treatment of diabetes: An update. Mini-Rev. Med. Chem. 2010, 10, 315–331. [Google Scholar] [CrossRef]
- Sales, P.M.; Souza, P.M.; Simeoni, L.A.; Magalhães, P.O.; Silveira, D. α-Amylase inhibitors: A review of raw material and isolated compounds from plant source. J. Pharm. Pharmaceut. Sci. 2012, 15, 141–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, H.W.; Cho, J.K.; Curtis-Long, M.J.; Yuk, H.J.; Kim, Y.S.; Jung, S.; Kim, Y.S.; Lee, B.W.; Park, K.H. α-Glucosidase inhibition and antihyperglycemic activity of prenylated xanthones from Garcinia mangostana. Phytochemistry 2011, 72, 2148–2154. [Google Scholar] [CrossRef]
- Asgar, A. Anti-diabetic potential of phenolic compounds: A Review. Int. J. Food Prop. 2013, 16, 91–103. [Google Scholar] [CrossRef]
- Serina, J.J.C.; Castilho, P.C.M.F. Using polyphenols as a relevant therapy to diabetes and its complications, a review. Critical Rev. Food Sci. Nutr. 2021, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, G.A.; Ibrahim, S.R.M.; Shaaban, M.I.A.; Ross, S.A. Mangostanaxanthones I and II, new xanthones from the pericarp of Garcinia mangostana. Fitoterapia 2014, 98, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Palakawong, C.; Delaquis, P. Mangosteen processing: A review. J. Food Process. Preserv. 2018, 2018, e13744. [Google Scholar] [CrossRef]
- Abdallah, H.M.; El-Bassossy, H.; Mohamed, G.A.; El-Halawany, A.M.; Alshali, K.Z.; Banjar, Z.M. Phenolics from Garcinia mangostana inhibit advanced glycation endproducts formation: Effect on amadori products, cross-linked structures and protein thiols. Molecules 2016, 21, 251. [Google Scholar] [CrossRef] [Green Version]
- Abdallah, H.M.; El-Bassossy, H.; Mohamed, G.A.; El-Halawany, A.M.; Alshali, K.Z.; Banjar, Z.M. Phenolics from Garcinia mangostana alleviate exaggerated vasoconstriction in metabolic syndrome through direct vasodilatation and nitric oxide generation. BMC Complement. Altern. Med. 2016, 16, 359. [Google Scholar] [CrossRef]
- Abdallah, H.M.; El-Bassossy, H.; Mohamed, G.A.; El-Halawany, A.M.; Alshali, K.Z.; Banjar, Z.M. Mangostanaxanthones III and IV: Advanced glycation endproduct inhibitors from the pericarp of Garcinia mangostana. J. Nat. Med. 2017, 71, 216–226. [Google Scholar] [CrossRef]
- Muchtaridi, M.; Puteri, N.A.; Milanda, T.; Musfiroh, I. Validation analysis methods of α-mangostin, γ-mangostin and gartanin mixture in mangosteen (Garcinia mangostana L.) fruit rind extract from West Java with HPLC. J. App. Pharm. Sci. 2017, 7, 125–130. [Google Scholar]
- Jiang, H.Z.; Quan, X.F.; Tian, W.X.; Hu, J.M.; Wang, P.C.; Huang, S.Z.; Cheng, Z.Q.; Liang, W.J.; Zhou, J.; Ma, X.F.; et al. Fatty acid synthase inhibitors of phenolic constituents isolated from Garcinia mangostana. Bioorg. Med. Chem. Lett. 2010, 20, 6045–6047. [Google Scholar] [CrossRef] [PubMed]
- Zadernowski, R.; Czaplicki, S.; Naczk, M. Phenolic acid profiles of mangosteen fruits (Garcinia mangostana). Food Chem. 2009, 112, 685–689. [Google Scholar] [CrossRef]
- Mohamed, G.A.; Al-Abd, A.M.; El-Halawany, A.M.; Abdallaha, H.M.; Ibrahim, S.R.M. New xanthones and cytotoxic constituents from Garcinia mangostana fruit hulls against human hepatocellular, breast, and colorectal cancer cell lines. J. Ethnopharmacol. 2017, 198, 302–312. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Abdallah, H.M.; El-Halawany, A.M.; Radwan, M.F.; Shehata, I.A.; Al-Harshany, E.M.; Zayed, M.F.; Mohamed, G.A. Garcixanthones B and C, new xanthones from the pericarps of Garcinia mangostana and their cytotoxic activity. Phytochem, Lett. 2018, 25, 12–16. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Mohamed, G.A.; Elfaky, M.A.; Zayed, M.F.; El-Kholy, A.A.; Abdelmageed, O.H.; Ross, S.A. Mangostanaxanthone VII, a new cytotoxic xanthone from Garcinia mangostana. Z. Naturforsch. C 2018, 73, 185–189. [Google Scholar] [CrossRef]
- Mekseepralard, C.; Areebambud, C.; Suksamrarn, S.; Jariyapongskul, A. Effects of long-term alpha-mangostin supplementation on hyperglycemia and insulin resistance in type 2 diabetic rats induced by high fat diet and low dose streptozotocin. J. Med. Assoc. Thail. 2015, 98 (Suppl. S10), S23–S30. [Google Scholar]
- Taher, M.; Zakaria, T.M.F.S.T.; Susanti, D.; Zakaria, Z.A. Hypoglycaemic activity of ethanolic extract of Garcinia mangostana Linn. in normoglycaemic and streptozotocin-induced diabetic rats. BMC Complement. Altern. Med. 2016, 16, 135. [Google Scholar] [CrossRef] [Green Version]
- Adnyana, K.; Abuzaid, A.S.; Iskandar, E.Y.; Kurniati, N.F. Pancreatic lipase and α-amylase inhibitory potential of mangosteen (Garcinia mangostana linn.) pericarp extract. Int. J. Med. Res. Health Sci. 2016, 5, 23–28. [Google Scholar]
- Husen, S.A.; Kalqutny, S.H.; Ansori, A.N.M.; Susilo, R.J.K.; Alymahdy, A.D.; Winarni, D. Antioxidant and antidiabetic activity of Garcinia mangostana L. pericarp extract in streptozotocin-induced diabetic mice. Biosc. Res. 2017, 14, 1238–1245. [Google Scholar]
- Manaharan, T.; Palanisamy, U.D.; Ming, C.H. Tropical plant extracts as potential antihyperglycemic agents. Molecules 2012, 17, 5915–5923. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Abdallah, H.M.; El-Halawany, A.M.; Nafady, A.M.; Mohamed, G.A. Mangostanaxanthone VIII, a new xanthone from Garcinia mangostana and its cytotoxic activity. Nat. Prod. Res. 2019, 33, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Gangitano, E.; Francomano, D.; Addessi, E.; Toscano, R.; Costantini, D.; Tuccinardi, D.; Mariani, S.; Basciani, S.; Spera, G.; et al. Mangosteen extract shows a potent insulin sensitizing effect in obese female patients: A prospective randomized controlled pilot study. Nutrients 2018, 10, 586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibrahim, S.R.M.; El-Agamy, D.S.; Abdallah, H.M.; Ahmed, N.; Elkablawy, M.A.; Mohamed, G.A. Protective activity of tovophyllin A, a xanthone isolated from Garcinia mangostana pericarps, against acetaminophen-induced liver damage: Role of Nrf2 activation. Food Funct. 2018, 9, 3291–3300. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.R.M.; Mohamed, G.A.; Elfaky, M.A.; Al Haidari, R.A.; Zayed, M.F.; El-Kholy, A.A.; Ross, S.A. Garcixanthone A, a new cytotoxic xanthone from the pericarps of Garcinia mangostana. J. Asian Nat. Prod. Res. 2019, 21, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, G.A.; Alliuocide, G. A new flavonoid with potent α-amylase inhibitory activity from Allium cepa L. Arkivoc 2008, 11, 202–209. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://www.thermofisher.com/order/catalog/product/E33651?SID=srch-srp-E33651 (accessed on 15 November 2021).
- Qian, M.; Spinelli, S.; Driguez, H.; Payan, F. Structure of a pancreatic a-amylase bound to a substrate analogue at 2.03 a resolution. Protein Sci. 2008, 6, 2285–2296. [Google Scholar] [CrossRef]
- Proença, C.; Freitas, M.; Ribeiro, D.; Tomé, S.M.; Oliveira, E.F.T.; Viegas, M.F.; Araújo, A.N.; Ramos, M.J.; Silva, A.M.S.; Fernandes, P.A.; et al. Evaluation of a flavonoids library for inhibition of pancreatic α-amylase towards a structure-activity relationship. J. Enzyme Inhib. Med. Chem. 2019, 34, 577–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dandekar, P.D.; Kotmale, A.S.; Chavan, S.R.; Kadlag, P.P.; Sawant, S.V.; Dhavale, D.D.; RaviKumar, A. Insights into the inhibition mechanism of human pancreatic α-amylase, a type 2 diabetes target, by dehydrodieugenol B isolated from Ocimum tenuiflorum. ACS Omega 2021, 6, 1780–1786. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, LLC. Schrödinger Release 2021-4: LigPrep; Schrödinger, LLC.: New York, NY, USA, 2021; Available online: https://www.schrodinger.com/ligprep (accessed on 1 January 2022).
- Omar, A.M.; Mohamed, G.A.; Ibrahim, S.R.M. Chaetomugilins and chaetoviridins-promising natural metabolites: Structures, separation, characterization, biosynthesis, bioactivities, molecular docking, and molecular dynamics. J. Fungi 2022, 8, 127. [Google Scholar] [CrossRef]
- Schrödinger, LLC. Schrödinger Release 2021-4: Desmond Molecular Dynamics System; D.E. Shaw Research: New York, NY, USA, 2021; Available online: https://www.schrodinger.com/desmond (accessed on 1 January 2022).
- Ibrahim, S.R.M.; Omar, A.M.; Bagalagel, A.A.; Diri, R.M.; Noor, A.O.; Almasri, D.M.; Mohamed, S.G.A.; Mohamed, G.A. Thiophenes-naturally occurring plant metabolites: Biological activities and in silico evaluation of their potential as cathepsin D inhibitors. Plants 2022, 11, 539. [Google Scholar] [CrossRef]
- Ferrari, J.; Terreaux, C.; Sahpaz, S.; Msonthi, J.D.; Wolfender, J.; Hostettmann, K. Benzophenone glycosides from Gnidia involucrata. Phytochemistry 2000, 54, 883–889. [Google Scholar] [CrossRef]
- Kitanov, G.M.; Nedialkov, P.T. Benzophenone O-glucoside, a biogenic precursor of 1,3,7-trioxygenated xanthones in Hypericum annulatum. Phytochemistry 2001, 57, 1237–1243. [Google Scholar] [CrossRef]
- Agrawal, P.K. NMR spectroscopy in the structural elucidation of oligosaccharides and glycosides. Phytochemistry 1992, 31, 3307–3330. [Google Scholar] [CrossRef]
- Iwo, M.I.; Soemardji, A.A.; Hanafi, M. Sunscreen activity of α-mangostin from the pericarps of Garcinia mangostana Linn. J. Appl. Pharm. Sci. 2013, 3, 70–73. [Google Scholar]
- Sen, A.K.; Sarkar, K.K.; Mazumder, P.C.; Banerji, N.; Uusvuori, R.; Hase, T.A. The structures of garcinones A, B and C: Three new xanthones from Garcinia mangostana. Phytochemistry 1982, 21, 1747–1750. [Google Scholar] [CrossRef]
- Govindachari, T.R.; Kalyanaraman, P.S.; Muthukumaraswamy, N.; Pai, B.R. Xanthones of Garcinia mangostana Linn. Tetrahedron 1971, 27, 3919–3926. [Google Scholar] [CrossRef]
- Xu, Z.; Huang, L.; Chen, X.H.; Zhu, X.F.; Qian, X.J.; Feng, G.K.; Lan, W.J.; Li, H.J. Cytotoxic prenylated xanthones from the pericarps of Garcinia mangostana. Molecules 2014, 19, 1820–1827. [Google Scholar] [CrossRef] [Green Version]
- Osman, A.G.; Ali, Z.; Fantoukh, O.; Raman, V.; Kamdem, R.S.; Khan, I. Glycosides of ursane-type triterpenoid, benzophenone, and iridoid from Vangueria agrestis (Fadogia agrestis) and their anti-infective activities. Nat. Prod. Res. 2020, 34, 683–691. [Google Scholar] [CrossRef]
- Liu, B.; Chen, N.; Xu, Y.; Zhang, J.W.; Sun, Y.; Zhao, L.Z.; Ji, Y.B. A new benzophenone with biological activities from metabolites of butyrolactone I in rat faeces. Nat. Prod. Res. 2021, 35, 2489–2497. [Google Scholar] [CrossRef]
- Venditti, A.; Ukwueze, S.E. A possible glycosidic benzophenone with full substitution on B-ring from Psidium guajava leaves. Nat. Prod. Res. 2017, 31, 739–741. [Google Scholar] [CrossRef]
- Costa, J.S., Jr.; de Almeida, A.A.C.; Ferraz, A.D.B.F.; Rossatto, R.R.; Silva, T.G.; Silva, P.B.; Militão, G.C.; das Graças Lopes Citó, A.M.; Santana, L.C.; de Amorim Carvalho, F.A.; et al. Cytotoxic and leishmanicidal properties of garcinielliptone FC, a prenylated benzophenone from Platonia insignis. Nat. Prod. Res. 2013, 27, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Akoro, S.M.; Aiyelaagbe, O.O.; Onocha, P.A.; Gloer, J.B. Gakolanone: A new benzophenone derivative from Garcinia kola Heckel stem-bark. Nat. Prod. Res. 2020, 34, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Brayer, G.D.; Luo, Y.; Withers, S.G. The structure of human pancreatic alpha amylase at 1.8 A resolution and comparisons with related enzymes. Protein Sci. 1995, 4, 1730–1742. [Google Scholar] [CrossRef] [PubMed]
- Brayer, G.D.; Sidhu, G.; Maurus, R.; Rydberg, E.H.; Braun, C.; Wang, Y.; Nguyen, N.T.; Overall, C.M.; Withers, S.G. Subsite mapping of the human pancreatic alpha-amylase active site through structural, kinetic, and mutagenesis techniques. Biochemistry 2000, 39, 4778–4791. [Google Scholar] [CrossRef]
- Rydberg, E.H.; Sidhu, G.; Vo, H.C.; Hewitt, J.; Côte, H.C.; Wang, Y.; Numao, S.; MacGillivray, R.T.; Overall, C.M.; Brayer, G.D.; et al. Cloning, mutagenesis, and structural analysis of human pancreatic alpha-amylase expressed in Pichia pastoris. Protein Sci. 1999, 8, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, C.R.; Paidesetty, S.K.; Dehury, B.; Padhy, R.N. Molecular dynamics and computational study of Mannich-based coumarin derivatives: Potent tyrosine kinase inhibitor. J. Biomol. Struct. Dyn. 2020, 38, 5419–5428. [Google Scholar] [CrossRef] [PubMed]










| No. | 6 | 7 | ||||
|---|---|---|---|---|---|---|
| δH (m, J in Hz) | δC m | HMBC | δH (m, J in Hz) | δC m | HMBC | |
| 1 | - | 106.3 C | - | - | 109.4 C | - |
| 2 | - | 165.4 C | - | - | 163.8 C | - |
| 3 | 5.83 brs | 95.8 CH | 1, 2, 4, 5, 7 | 6.06 d (2.4) | 98.1 CH | 1, 2, 4, 5, 7 |
| 4 | - | 165.4 C | - | - | 161.7 C | - |
| 5 | 5.83 brs | 95.8 CH | 1, 3, 4, 6, 7 | 6.21 d (2.4) | 95.7 CH | 1, 3, 4, 7 |
| 6 | - | 165.5 C | - | - | 159.5 C | - |
| 7 | - | 200.6 C | - | - | 199.3 C | - |
| 1` | - | 123.9 C | - | - | 119.2 C | - |
| 2` | 6.52 d (2.4) | 107.4 CH | 4`, 6`, 1, 7 | 6.96 d (2.4) | 114.2 CH | 1`, 4`, 6`, 1, 7 |
| 3` | - | 153.8 C | - | - | 154.2 C | - |
| 4` | 6.36 d (2.4) | 112.1 CH | 2`, 5`, 6` | 7.18 brd (8.0) | 121.4 CH | 2`, 5`, 6` |
| 5` | - | 148.8 C | - | 7.22 d (8.0) | 120.4 CH | 1`, 3`, 4`, 6` |
| 6` | - | 145.7 C | - | - | 155.8 C | - |
| 1`` | - | - | - | 4.88 d (7.6) | 101.6 CH | 6 |
| 2`` | - | - | - | 2.92 m | 74.6 CH | 1`` |
| 3`` | - | - | - | 3.30 m | 77.8 CH | 1`` |
| 4`` | - | - | - | 3.29 m | 71.0 CH | 2``, 6`` |
| 5`` | - | - | - | 3.32 m | 78.2 CH | 3``, 6`` |
| 6`` | - | - | - | 3.86 dd (12.4, 2.0) 3.69 dd (12.4, 5.6) | 62.5 CH2 | 4``, 5`` |
| Compd | Docking Score | XP GScore | Glide GScore | Glide eModel |
|---|---|---|---|---|
| Native_ Inhibitor_5TD4 | −19.081 | −19.081 | −19.081 | −104.865 |
| Acarbose | −16.078 | −16.641 | −16.641 | −115.364 |
| 7 | −11.297 | −11.46 | −11.46 | −75.209 |
| 6 | −8.185 | −8.596 | −8.596 | −50.318 |
| 5 | −6.726 | −6.824 | −6.824 | −67.078 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhakamy, N.A.; Mohamed, G.A.; Fahmy, U.A.; Eid, B.G.; Ahmed, O.A.A.; Al-Rabia, M.W.; Khedr, A.I.M.; Nasrullah, M.Z.; Ibrahim, S.R.M. New Alpha-Amylase Inhibitory Metabolites from Pericarps of Garcinia mangostana. Life 2022, 12, 384. https://doi.org/10.3390/life12030384
Alhakamy NA, Mohamed GA, Fahmy UA, Eid BG, Ahmed OAA, Al-Rabia MW, Khedr AIM, Nasrullah MZ, Ibrahim SRM. New Alpha-Amylase Inhibitory Metabolites from Pericarps of Garcinia mangostana. Life. 2022; 12(3):384. https://doi.org/10.3390/life12030384
Chicago/Turabian StyleAlhakamy, Nabil Abdulhafiz, Gamal Abdallah Mohamed, Usama Ahmed Fahmy, Basma Ghazi Eid, Osama Abdelhakim Aly Ahmed, Mohammed Wanees Al-Rabia, Amgad Ibrahim Mansour Khedr, Mohammed Zahid Nasrullah, and Sabrin Ragab Mohamed Ibrahim. 2022. "New Alpha-Amylase Inhibitory Metabolites from Pericarps of Garcinia mangostana" Life 12, no. 3: 384. https://doi.org/10.3390/life12030384
APA StyleAlhakamy, N. A., Mohamed, G. A., Fahmy, U. A., Eid, B. G., Ahmed, O. A. A., Al-Rabia, M. W., Khedr, A. I. M., Nasrullah, M. Z., & Ibrahim, S. R. M. (2022). New Alpha-Amylase Inhibitory Metabolites from Pericarps of Garcinia mangostana. Life, 12(3), 384. https://doi.org/10.3390/life12030384








