Baseline Gene Expression Levels in Falkland-Malvinas Island Penguins: Towards a New Monitoring Paradigm
Abstract
:1. Introduction
2. Materials and Methods
2.1. Falkland-Malvinas Islands
2.2. Detroit Zoo
2.3. Blood Collection and RNA Extraction
2.4. Captive Penguins
2.5. Wild Penguins
2.6. cDNA Synthesis
2.7. Gene Selection
2.8. Polymerase Chain Reaction Primers
2.9. Real-Time Polymerase Chain Reaction
2.10. Statistical Methods
3. Results
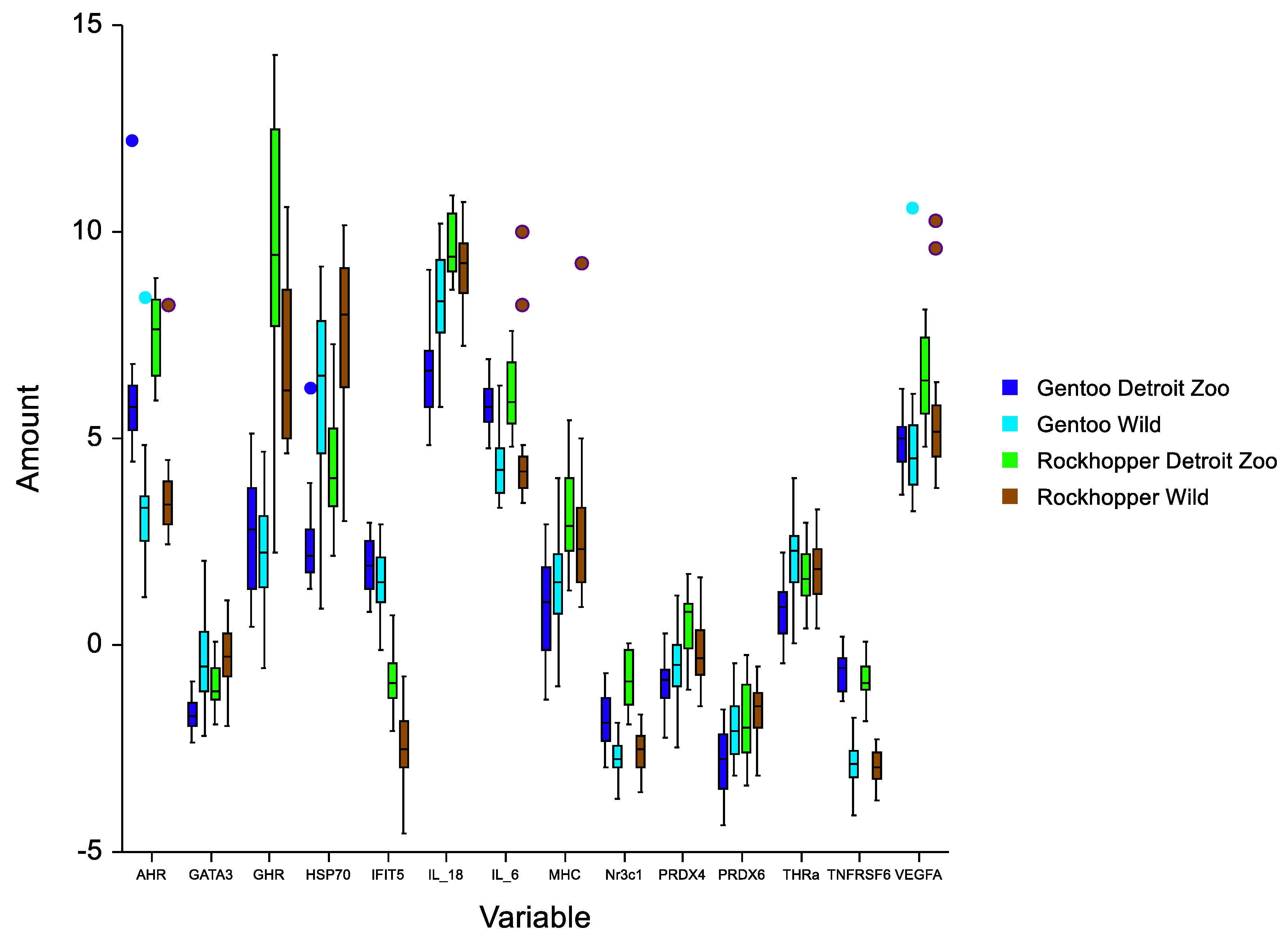
3.1. General Description
3.2. Falkland-Malvinas Islands
3.3. Detroit Zoo
4. Discussion
4.1. Gene Expression Differences by Sex
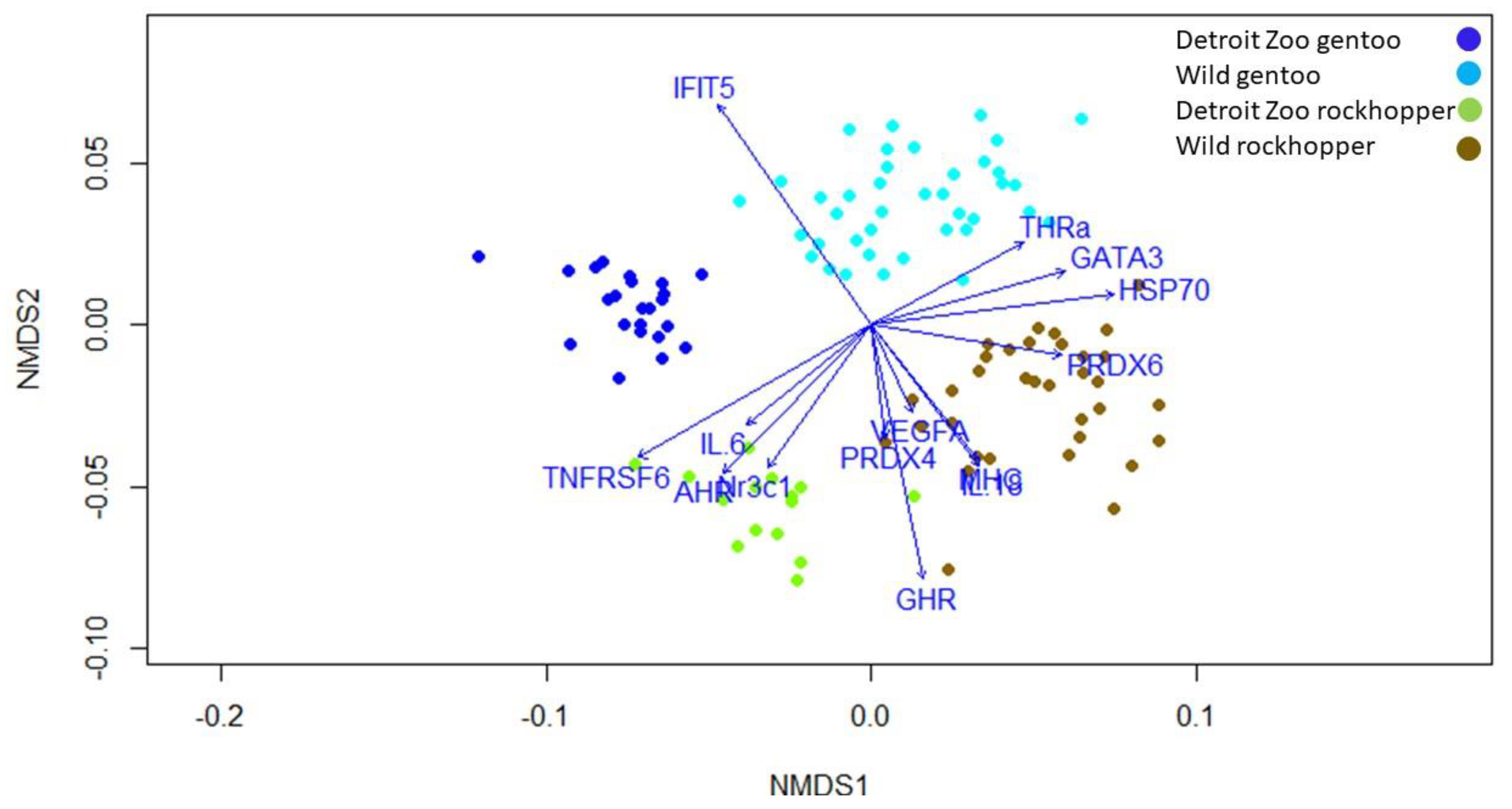
4.2. Gene Expression Differences by Location
4.3. Gene Expression Differences by Species
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Faulkner, C.B.; Simecka, J.W.; Davidson, M.K.; Davis, J.K.; Schoeb, T.R.; Lindsey, J.R.; Everson, M.P. Gene expression and production of tumor necrosis factor alpha, interleukin 1, interleukin 6, and gamma interferon in C3H/HeN and C57BL/6N mice in acute Mycoplasma pulmonis disease. Infect. Immun. 1995, 63, 4084–4090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acevedo-Whitehouse, K.; Duffus, A.L.J. Effects of Environmental Change on Wildlife Health. Philos. Trans. R. Soc. B 2009, 364, 3429–3438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLoughlin, K.; Turteltaub, K.; Bankaitis-Davis, D.; Gerren, R.; Siconolfi, L.; Storm, K.; Cheronis, J.; Trollinger, D.; Macejak, D.; Tryon, V.; et al. Limited dynamic range of immune response gene expression observed in healthy blood donors using RT-PCR. Mol. Med. 2006, 12, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, A.; Dubansky, B.; Bodinier, C.; Garcia, T.I.; Miles, S.; Pilley, C.; Raghunathan, V.; Roach, J.L.; Walker, N.; Walter, R.B.; et al. Genomic and physiological footprint of the Deepwater Horizon oil spill on resident marsh fishes. Proc. Natl. Acad. Sci. USA 2012, 109, 20298–20302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mancia, A.; Warr, G.W.; Champman, R.W. A transcriptomic analysis of the stress induced by capture–release health assessment studies in wild dolphins (Tursiops truncatus). Mol. Ecol. 2008, 17, 2581–2589. [Google Scholar] [CrossRef]
- Miller, K.M.; Li, S.; Kaukinen, K.H.; Ginther, N.; Hammill, E.; Curtis, J.M.R.; Patterson, D.A.; Sierocinski, T.; Donnison, L.; Pavlidis, P.; et al. Genomic signatures predict migration and spawning failure in wild Canadian salmon. Science 2011, 331, 214–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tinker, M.T.; Bodkin, J.L.; Bowen, L.; Ballachey, B.; Bentall, G.; Burdin, A.; Coletti, H.; Esslinger, G.; Hatfield, B.B.; Kenner, M.C.; et al. Sea otter population collapse in southwest Alaska: Assessing ecological covariates, consequences, and causal factors. Ecol. Monogr. 2021, 91, e01472. [Google Scholar] [CrossRef]
- Blanchong, J.A.; Robinson, S.J.; Samuel, M.D.; Foster, J.T. Application of Genetics and Genomics to Wildlife Epidemiology: Genetics and Wildlife Epidemiology. J. Wildl. Manag. 2016, 80, 593–608. [Google Scholar] [CrossRef] [Green Version]
- Signa, G.; Mazzola, A.; Vizzini, S. Seabird influence on ecological processes in coastal marine ecosystems: An overlooked role? A critical review. Estuar. Coast Shelf Sci. 2021, 250, 107164. [Google Scholar] [CrossRef]
- Sydeman, W.J.; Thompson, S.A.; Kitaysky, A. Seabirds and climate change: Roadmap for the future. Mar. Ecol. Prog. Ser. 2012, 454, 107–117. [Google Scholar] [CrossRef] [Green Version]
- Provencher, J.F.; Ammendolia, J.; Rochman, C.M.; Mallory, M.L. Assessing plastic debris in aquatic food webs: What we know and don’t know about uptake and trophic transfer. Environ. Rev. 2019, 27, 304–317. [Google Scholar] [CrossRef]
- Croxall, J.P.; Butchart, S.H.; Lascelles, B.E.; Stattersfield, A.J.; Sullivan, B.E.; Symes, A.; Taylor, P.H. Seabird conservation status, threats and priority actions: A global assessment. Bird Conserv. Int. 2012, 22, 1–34. [Google Scholar] [CrossRef] [Green Version]
- Trathan, P.N.; García-Borboroglu, P.; Boersma, D.; Bost, C.A.; Crawford, R.J.M.; Crossin, G.T.; Cuthbert, R.J.; Dann, P.; Davis, L.S.; De La Puente, S.; et al. Pollution, habitat loss, fishing, and climate change as critical threats to penguins. Conserv. Biol. 2015, 29, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Borboroglu, P.G.; Boersma, P.D. Penguins: Natural History and Conservation; University of Washington Press: Washington, DC, USA, 2015. [Google Scholar]
- Burfield, I.J.; Butchart, S.H.; Collar, N.J. BirdLife, conservation and taxonomy. Bird Conserv. Int. 2017, 27, 1–5. [Google Scholar] [CrossRef]
- Bowen, L.; Miles, A.K.; Murray, M.; Haulena, M.; Tuttle, J.; Van Bonn, W.; Adams, L.; Bodkin, J.L.; Ballachey, B.; Estes, J.; et al. Gene transcription in sea otters (Enhydra lutris); development of a diagnostic tool for sea otter and ecosystem health. Mol. Ecol. Resour. 2012, 12, 67–74. [Google Scholar] [CrossRef] [Green Version]
- Aarem, J.; Brunborg, G.; Aas, K.K.; Harbak, K.; Taipale, M.M.; Magnus, P.; Knudsen, G.P.; Duale, N. Comparison of blood RNA isolation methods from samples stabilized in Tempus tubes and stored at a large human biobank. BMC Res. 2016, 9, 430. [Google Scholar] [CrossRef] [Green Version]
- Bowen, L.; Miles, A.K.; Kolden, C.A.; Saarinen, J.A.; Bodkin, J.L.; Murray, M.J.; Tinker, M.T. Effects of wildfire on sea otter (Enhydra lutris) gene transcription profiles. Mar. Mamm. Sci. 2015, 31, 191–210. [Google Scholar] [CrossRef]
- Olias, P.; Adam, I.; Meyer, A.; Scharff, C.; Gruber, A.D. Reference genes for quantitative gene expression studies in multiple avian species. PLoS ONE 2014, 9, e99678. [Google Scholar] [CrossRef]
- Miller, K.M.; Günther, O.P.; Li, S.; Kaukinen, K.H.; Ming, T.J. Molecular indices of viral disease development in wild migrating salmon. Conserv. Physiol. 2017, 5, cox036. [Google Scholar] [CrossRef]
- Rohaim, M.A.; Santhakumar, D.; Naggar, R.F.E.; Iqbal, M.; Hussein, H.A.; Munir, M. Chickens Expressing IFIT5 Ameliorate Clinical Outcome and Pathology of Highly Pathogenic Avian Influenza and Velogenic Newcastle Disease Viruses. Front. Immunol. 2018, 9, 2025. [Google Scholar] [CrossRef]
- Castell, J.V.; Gómez-Lechón, M.J.; David, M.; Andus, T.; Geiger, T.; Trullenque, R.; Fabra, R.; Heinrich, P.C. Interleukin-6 is the major regulator of acute phase protein synthesis in adult human hepatocytes. FEBS Lett. 1989, 242, 237–239. [Google Scholar] [CrossRef] [Green Version]
- Gelain, M.E.; Bonsembiante, F. Acute Phase Proteins in Marine Mammals: State of Art, Perspectives and Challenges. Front. Immunol. 2019, 10, 1220. [Google Scholar] [CrossRef] [PubMed]
- Sallaberry-Pincheira, N.; González-Acuña, D.; Padilla, P.; Dantas, G.P.M.; Luna-Jorquera, G.; Frere, E.; Valdés-Velásquez, A.; Vianna, J.A. Contrasting patterns of selection between MHC I and II across populations of Humboldt and Magellanic penguins. Ecol. Evol. 2016, 6, 7498–7510. [Google Scholar] [CrossRef] [PubMed]
- Sheriff, M.J.; Dantzer, B.; Delehanty, B.; Palme, R.; Boonstra, R. Measuring stress in wildlife: Techniques for quantifying glucocorticoids. Oecologia 2011, 166, 869–887. [Google Scholar] [CrossRef]
- Wiens, G.D.; Glenney, G.W. Origin and evolution of TNF and TNF receptor superfamilies. Dev. Comp. Immuunol. 2011, 35, 1324–1335. [Google Scholar] [CrossRef]
- Seirafian, S.; Prod’homme, V.; Sugrue, D.; Davies, J.; Fielding, C.; Tomasec, P.; Wilkinson, G.W. Human cytomegalovirus suppresses Fas expression and function. J. Gen. Virol. 2014, 95, 933–939. [Google Scholar] [CrossRef] [Green Version]
- Oesch-Bartlomowicz, B.; Oesch, F. Phosphorylation of cytochromes P450: First discovery of a posttranslational modification of a drug-metabolizing enzyme. Biochem. Biophys. Resour. Commun. 2005, 338, 446–449. [Google Scholar] [CrossRef]
- Karchner, S.I.; Franks, D.G.; Kennedy, S.W.; Hahn, M.E. The molecular basis for differential dioxin sensitivity in birds: Role of the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 6252–6257. [Google Scholar] [CrossRef] [Green Version]
- Raccurt, M.; Baudimont, F.; Tirard, J.; Rey, B.; Moureaux, E.; Géloën, A.; Duchamp, C. Growing in Antarctica, a challenge for white adipose tissue development in Adélie penguin chicks (Pygoscelis adeliae). Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2008, 295, R1671–R1679. [Google Scholar] [CrossRef] [Green Version]
- Iwama, G.K.; Mathilakath, M.V.; Forsyth, R.B.; Ackerman, P.A. Heat shock proteins and physiological stress in fish. Am. Zool. 1999, 39, 901–909. [Google Scholar] [CrossRef] [Green Version]
- Tsan, M.F.; Gao, B. Cytokine function of heat shock proteins. Am. J. Physiol. Cell Physiol. 2004, 286, C739–C744. [Google Scholar] [CrossRef] [PubMed]
- De Maio, A. Heat shock proteins: Facts, thoughts, and dreams. Shock 1999, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bourgeon, S.; Martínez, J.; Criscuolo, F.; Maho, Y.L.; Raclot, T. Fasting-induced changes of immunological and stress indicators in breeding female eiders. Gen. Comp. Endocr. 2006, 147, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Krumm, B.; Meng, X.; Li, Y.; Xiang, Y.; Deng, J. Structural basis for antagonism of human interleukin 18 by poxvirus interleukin 18-binding protein. Proc. Natl. Acad. Sci. USA 2008, 105, 20711–20715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, K.; Puehler, F.; Baeuerle, D.; Elvers, S.; Staeheli, P.; Kaspers, B.; Weining, K.C. cDNA Cloning of Biologically Active Chicken Interleukin-18. J. Interf. Cytok. Res. 2000, 20, 879–883. [Google Scholar] [CrossRef] [Green Version]
- Parham, P. The Immune System, 4th ed.; Garland Science, Taylor and Francis Group, LLC: New York, NY, USA, 2014. [Google Scholar]
- Capilla-Lasheras, P.; Dominoni, D.M.; Babayan, S.A.; O’Shaughnessy, P.J.; Mladenova, M.; Woodford, L.; Pollock, C.J.; Barr, T.; Baldini, F.; Helm, B. Elevated immune gene expression is associated with poor reproductive success of urban blue tits. Front. Ecol. Evol. 2017, 5, 64. [Google Scholar] [CrossRef] [Green Version]
- Rey, B.; Degletagne, C.; Bodennec, J.; Monternier, P.A.; Mortz, M.; Roussel, D.; Romestaing, C.; Rouanet, J.L.; Tornos, J.; Duchamp, C. Hormetic response triggers multifaceted anti-oxidant strategies in immature king penguins (Aptenodytes patagonicus). Free Radic. Bio Med. 2016, 97, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Rey, B.; Dégletagne, C.; Duchamp, C. Transcriptomic data analysis and differential gene expression of antioxidant pathways in king penguin juveniles (Aptenodytes patagonicus) before and after acclimatization to marine life. Data Brief. 2016, 9, 549–555. [Google Scholar] [CrossRef]
- Yamada, S.; Guo, X. Peroxiredoxin 4 (PRDX4): Its critical in vivo roles in animal models of metabolic syndrome ranging from atherosclerosis to nonalcoholic fatty liver disease. Pathol. Int. 2018, 68, 91–101. [Google Scholar] [CrossRef]
- Dégletagne, C.; Roussel, D.; Rouanet, J.L.; Baudimont, F.; Moureaux, E.M.; Harvey, S.; Duchamp, C.; Le Maho, Y.; Raccurt, M.; Laudet, V. Growth prior to thermogenesis for a quick fledging of Adélie penguin chicks (Pygoscelis adeliae). PLoS ONE 2013, 8, e74154. [Google Scholar] [CrossRef] [Green Version]
- Dehkhoda, F.; Lee, C.M.M.; Medina, J.; Brooks, A.J. The growth hormone receptor: Mechanism of receptor activation, cell signaling, and physiological aspects. Front. Endocrinol. 2018, 9, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, L.; Zhang, W.M.; Lin, H.R.; Cheng, C.H. Effects of food deprivation on expression of growth hormone receptor and proximate composition in liver of black seabream Acanthopagrus schlegeli. Comp. Biochem. Physiol. 2004, 137, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, J.; Liu, B. Targeting VEGF/VEGFR to modulate antitumor immunity. Front. Immunol. 2018, 9, 978. [Google Scholar] [CrossRef] [Green Version]
- Bowen, L.; Riva, F.; Mohr, C.; Aldridge, B.; Schwartz, J.; Miles, A.K.; Stott, J.L. Differential gene expression induced by exposure of captive mink to fuel oil: A model for the sea otter. EcoHealth 2007, 4, 298–309. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Robinson, J.; Waller, M.; Parham, P.; Bodmer, J.G.; Marsh, S.G.E. IMGT/HLA Database—A sequence database for the human major histocompatibility complex. Nucl. Acids Res. 2001, 29, 210–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2012; Available online: http://www.R-project.org/ (accessed on 2 October 2020).
- Boersma, P.D.; Stokes, D.L.; Strange, I.J. Applying ecology to conservation: Tracking breeding penguins at New Island South reserve, Falkland Island. Aquat. Conserv. 2002, 12, 63–74. [Google Scholar] [CrossRef]
- Pedersen, A.B.; Babayan, S.A. Wild immunology. Mol. Ecol. 2011, 20, 872–880. [Google Scholar] [CrossRef]

| Gene | Gene Function | General Category |
|---|---|---|
| YWHAZ | Reference gene [19] | Reference |
| IFIT5 | Interferon Induced Protein With Tetratricopeptide Repeats 5 (IFIT5) is part of a novel class of IFN-effectors, known as IFN-induced proteins with tetratricopeptides repeats (IFITs). IFIT proteins are indicative of early response to virus [20,21]. | Inflammation |
| IL-6 | Interleukin 6 (IL-6) is a cytokine that stimulates the synthesis of the full spectrum of acute phase proteins as seen in inflammatory states [22]. The term “acute phase response” (APR) is referred to a nonspecific and complex reaction of an organism that occurs shortly after any tissue damage, such as infection, trauma, neoplasia, inflammation, and stress [23]. | Inflammation |
| MHC | Major histocompatibility complex class II beta (MHC) molecules play a key role in the adaptive immune responses of vertebrates. MHC class II beta has primarily been associated with extracellular infections (e.g., bacteria) [24]. | Targeted immunity |
| Nr3c1 | Nuclear Receptor Subfamily 3 Group C Member 1 (Nr3c1) is a glucocorticoid receptor expressed in response to stress [25]. | Stress response |
| TNFRSF6 | Tumor necrosis factor receptor super family 6 (TNFRSF6) is instrumental in a number of cellular signaling pathways involving inflammation, apoptosis, lymphocyte homeostasis, and tissue development [26]. TNFRSF6 also plays a prominent role in apoptotic clearance of virus-infected cells [27]. | Inflammation |
| AHR | The aryl hydrocarbon receptor (AHR) responds to classes of environmental toxicants including polycyclic aromatic hydrocarbons, polyhalogenated hydrocarbons, dibenzofurans, and dioxin [28]. Birds have been found to have different sensitivities to PHAHs and TCDD exposure in comparison to other species; this can be due to expression differences in AHR [29]. | Detoxification |
| THRa | Thyroid hormone receptor alpha (THRa) is associated with physiological stress and organic compound exposure [30]. | Stress response |
| HSP70 | The heat-shock protein 70 (HSP70) is produced in response to exposure to different kinds of environmental stress conditions, such as infection, inflammation, exercise, exposure of the cell to toxins, starvation, and thermal or other stress [31,32]. In addition to being expressed in response to a wide array of stressors, heat-shock proteins act as molecular chaperones [33]. In incubating female eiders, an increase in HSP70 resulted in a decrease of immunoglobulin [34]. | Stress response |
| IL-18 | Interleukin-18 (IL-18) plays an important role in inflammation and host defense against microbes. Induction of IL-18 initiates a TH1 immune response in chickens [35,36]. | Inflammation |
| Gata3 | Gata3 is a TH2-specific transcription factor that controls transcription of cytokines Interleukin (IL) IL-4, -5, and -13 [37]. Gata3 is involved in innate and adaptive immune responses to parasitic helminths [38]. Gata3 has also been shown to be involved in adipocyte development in Adelie penguin chicks [30]. | Innate and adaptive immune function |
| PRDX4 | Peroxiredoxin 4 (PRDX4) protects against oxidative damage by scavenging reactive oxygen species in both the intracellular (especially the endoplasmic reticulum) compartments and the extracellular space [39,40,41]. | Oxidative stress response |
| PRDX6 | Peroxiredoxin 6 (PRDX6) plays a role in redox regulation, phospholipid turnover, and protection against oxidative injury [39,40,41]. | Oxidative stress response |
| GHR | Growth hormone receptor (GHR) is associated with nutrition, growth, and is a regulator of aging and plays a significant role in cancer development [42,43]. GHR expression is decreased in association starvation in some species [44]. | Nutrition |
| VEGFA | Vascular endothelial growth factor A (VEGFA) is a cytokine involved in immune suppression [45]. | Immune suppression |
| Gene | Primer Name | FP1 | Primer Name | RP1 rc | Expected Amplicon (bp) |
|---|---|---|---|---|---|
| AHR | Sphen. AHR F1 | aggacgattaaagtttctccat | Sphen. AHR R1rc | gatagatggtggctgcagg | 111 |
| IL-18 | Sphen. IL18 F1 | tgttgtgagaaagaatgtggaa | Sphen. IL-18 R2rc | acttaaatgctctggagctac | 133 |
| GATA3 | Sphen. GATA3 F1 | ggtccatgacaaccttgaag | Sphen. GATA3 R2rc | tgcatcggtgtcggtgtag | 137 |
| PRDX6 | Sphen. PRDX6 F1 | aggacatcaatgcatacaacg | Sphen. PRDX6 R1rc | ccatccttgtcccgctcat | 126 |
| GHR | Sphen. GHR F1 | gatccaccaccaacagcag | Sphen. GHR R1rc | tggaactattgttgagagcct | 122 |
| VEGFA | Sphen. VEGFA F1 | gccttgctcagagaggaga | Sphen. VEGFA R1rc | cacatctgcaagtgcgctc | 127 |
| Nr3c1 | Sphen. Nr3c1 F1 | tgcatcgctctctcagcag | Sphen. Nr3c1 R1rc | aaggagctaacgtctcatcc | 118 |
| IFIT5 | Sphen. IFIT5 F2 | ttgccaggagaagtcttgtta | Sphen. IFIT5 R2rc | cttgaaagctttttgcagctg | 120 |
| THRa | Sphen. THRa F1 | ggcagccactggaagcag | Sphen. THRa R1rc | ctcgctgaacgcctccag | 107 |
| PRDX4 | Sphen. PRDX4 F1 | agcatggattaatactcctcg | Sphen. PRDX4 R1rc | cttggtcttccagatatacac | 115 |
| YWHAZ | Sphen. YWHAZ F1 | aaggagatgcagccaacaca | Sphen. YWHAZ R1rc | agttcagcaattgcttcatcaa | 136 |
| MHC | Sphen. MHC class II F | aacggcaccgagcgggtgaggt | Sphen. MHC class II R | cccgtagttgtgttggcag | 198 |
| IL-6 | Sphen. IL-6 F1 | cacctcatcctccgagact | Sphen. IL-6 R1rc | tgtaacaaaggattgtgcctg | 121 |
| TNFRSF6 | Sphen. TNFRSF6 F1 | aatgtcgggagagactggaa | Sphen. TNFRSF6 R1rc | gaagtgactgagccaactgt | 117 |
| HSP70 | Sphen. HSP70 F1 | gagcacaagcagaaagagct | Sphen. HSP70 R1rc | ttaatctacttcttcgatggtc | 119 |
| Gentoo | Rockhopper | |||
|---|---|---|---|---|
| Falkland-Malvinas Islands n = 39 | Detroit Zoo n = 23 | Falkland-Malvinas Islands n = 34 | Detroit Zoo n = 15 | |
| AHR | 3.23 (1.15) | 5.90 (1.54) | 3.58 (1.08) | 8.02 (2.04) |
| GATA3 | −0.39 (0.98) | −1.73 (0.44) | −0.30 (0.79) | −0.96 (0.52) |
| GHR | 2.16 (1.22) | 2.70 (1.46) | 6.80 (1.93) | 9.60 (3.14) |
| HSP70 | 6.07 (2.19) | 2.40 (1.04) | 7.49 (0.83) | 4.43 (1.58) |
| IFIT5 | 1.50 (0.85) | 1.86 (0.83) | −2.45 (0.82) | −0.79 (0.74) |
| IL-18 | 8.31 (1.10) | 6.52 (0.96) | 9.21 (0.93) | 9.71 (1.10) |
| IL-6 | 4.40 (0.84) | 5.78 (0.58) | 4.44 (1.27) | 6.02 (0.74) |
| MHC | 1.53 (1.22) | 0.89 (1.25) | 2.64 (1.52) | 3.13 (1.16) |
| Nr3c1 | −2.73 (0.50) | −1.83 (0.64) | −2.57 (0.59) | −0.84 (0.68) |
| PRDX4 | −0.54 (0.88) | −0.91 (0.69) | −0.18 (0.72) | 0.52 (0.74) |
| PRDX6 | −2.00 (0.74) | −2.82 (0.81) | −1.58 (0.62) | −1.91 (0.91) |
| THRa | 2.03 (0.93) | 0.87 (0.74) | 1.82 (0.76) | 1.67 (0.71) |
| TNFRSF6 | −2.91 (0.50) | −0.62 (0.48) | −2.92 (0.35) | −0.81 (0.46) |
| VEGFA | 5.00 (1.76) | 4.90 (0.67) | 5.52 (1.54) | 6.54 (1.00) |
| Gene | Sex | Location | Species |
|---|---|---|---|
| AHR | 1.45 × 10−5 | <2.2 × 10−16 | 5.9 × 10−3 |
| GATA3 | 5.98 × 10−10 | 1.0 × 10−1 | |
| GHR | 8.6 × 10−3 | 1.59 × 10−5 | <2.2 × 10−16 |
| HSP70 | 4.01 × 10−16 | 6.47 × 10−7 | |
| IFIT5 | 7.2 × 10−3 | 4.34 × 10−6 | <2.2 × 10−16 |
| IL-18 | 8.9 × 10−4 | 6.97 × 10−12 | |
| IL-6 | 7.7 × 10−3 | 3.11 × 10−11 | |
| MHC | 2.875 × 10−8 | ||
| Nr3c1 | 1.4 × 10−3 | <2.2 × 10−16 | 2.3 × 10−3 |
| PRDX4 | 1.17 × 10−5 | ||
| PRDX6 | 6.0 × 10−5 | 4.16 × 10−5 | |
| THRa | 1.49 × 10−5 | ||
| TNFRSF6 | 3.7 × 10−4 | <2.2 × 10−16 | 2.1 × 10−2 |
| VEGFA | 4.0 × 10−2 | 1.6 × 10−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bowen, L.; Waters, S.; Stott, J.L.; Duncan, A.; Meyerson, R.; Woodhouse, S. Baseline Gene Expression Levels in Falkland-Malvinas Island Penguins: Towards a New Monitoring Paradigm. Life 2022, 12, 258. https://doi.org/10.3390/life12020258
Bowen L, Waters S, Stott JL, Duncan A, Meyerson R, Woodhouse S. Baseline Gene Expression Levels in Falkland-Malvinas Island Penguins: Towards a New Monitoring Paradigm. Life. 2022; 12(2):258. https://doi.org/10.3390/life12020258
Chicago/Turabian StyleBowen, Lizabeth, Shannon Waters, Jeffrey L. Stott, Ann Duncan, Randi Meyerson, and Sarah Woodhouse. 2022. "Baseline Gene Expression Levels in Falkland-Malvinas Island Penguins: Towards a New Monitoring Paradigm" Life 12, no. 2: 258. https://doi.org/10.3390/life12020258
APA StyleBowen, L., Waters, S., Stott, J. L., Duncan, A., Meyerson, R., & Woodhouse, S. (2022). Baseline Gene Expression Levels in Falkland-Malvinas Island Penguins: Towards a New Monitoring Paradigm. Life, 12(2), 258. https://doi.org/10.3390/life12020258





