Frequency and Molecular Characterization of Staphylococcus aureus from Placenta of Mothers with Term and Preterm Deliveries
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Culturing on MSA Media
2.3. Antimicrobial Activity by Disc Diffusion Method
2.4. 16S rRNA Analysis of Isolates
2.5. Molecular Identification of Virulent Genes
2.6. Plasmid Profiling
3. Results
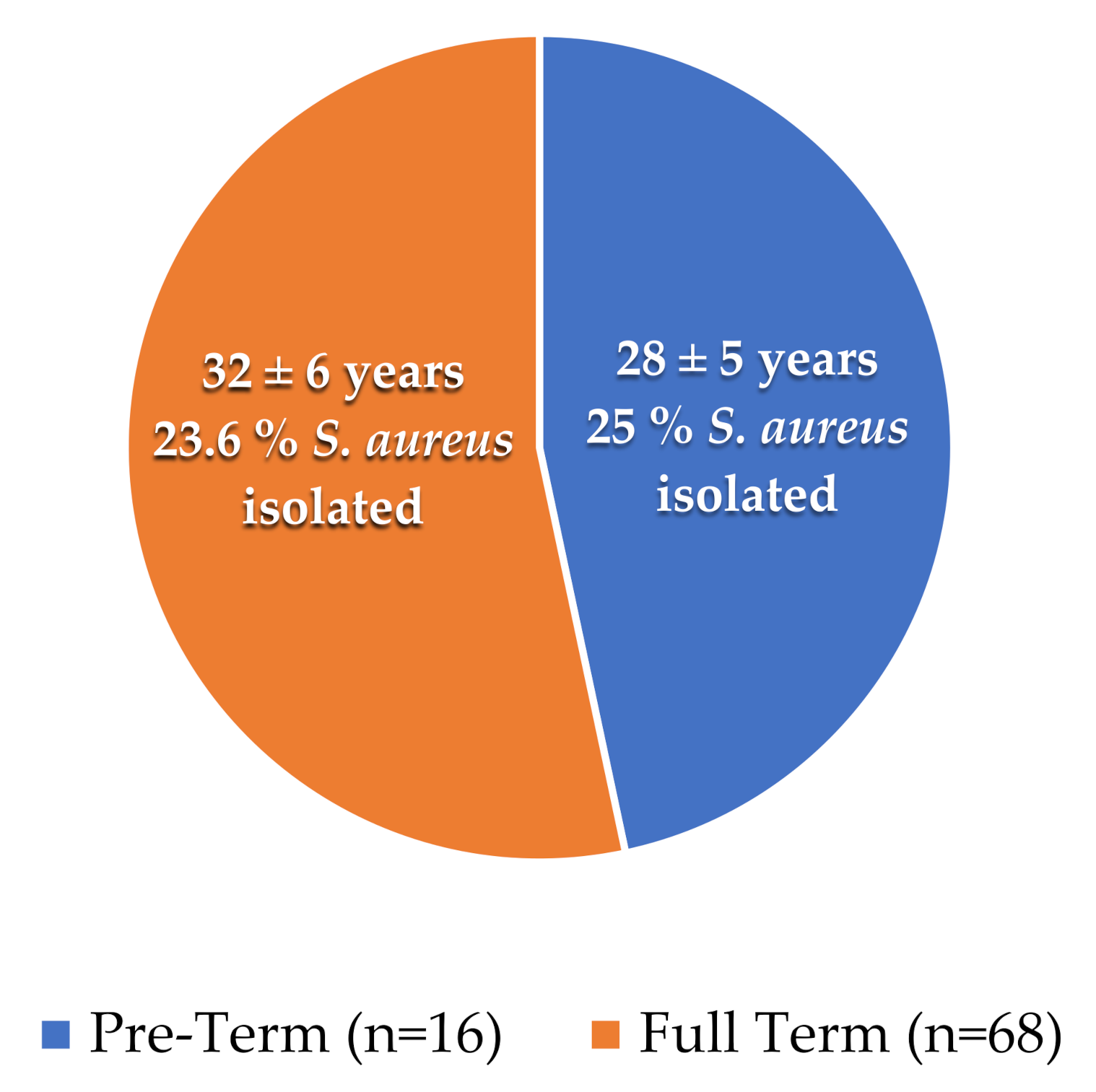
3.1. Distribution of Samples
3.2. S. aureus Culturing
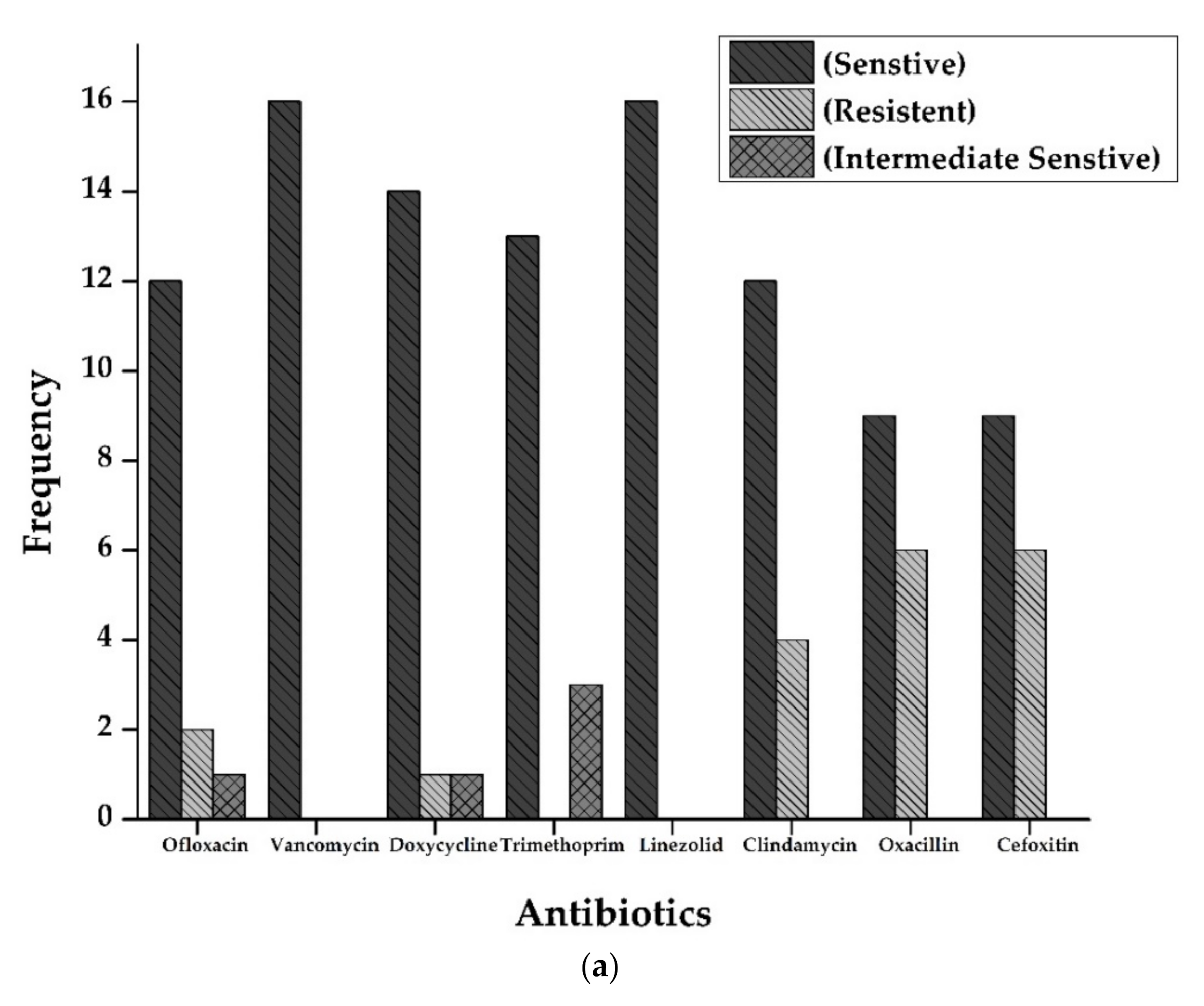
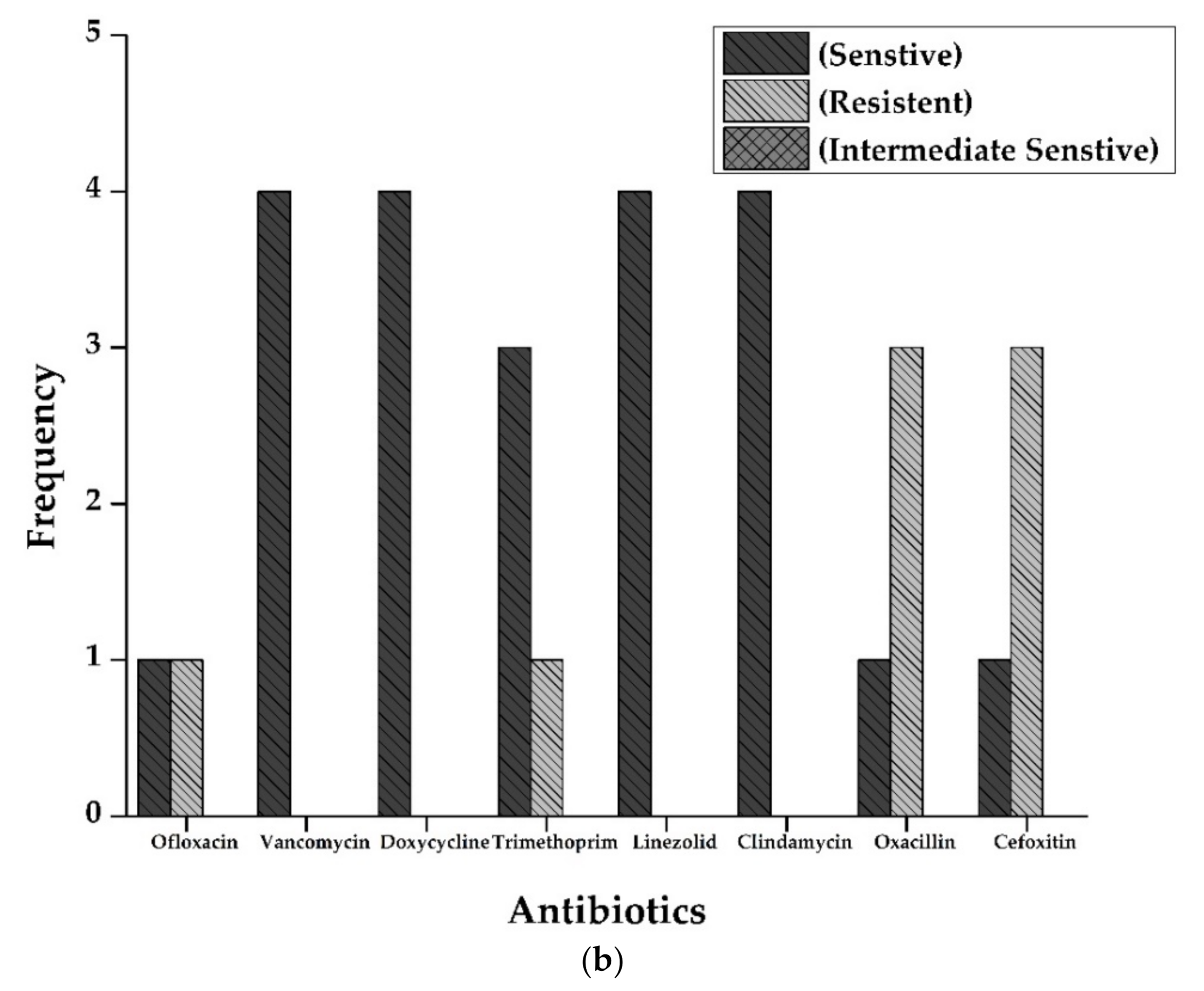
3.3. Antimicrobial Activity by Disc Diffusion Method
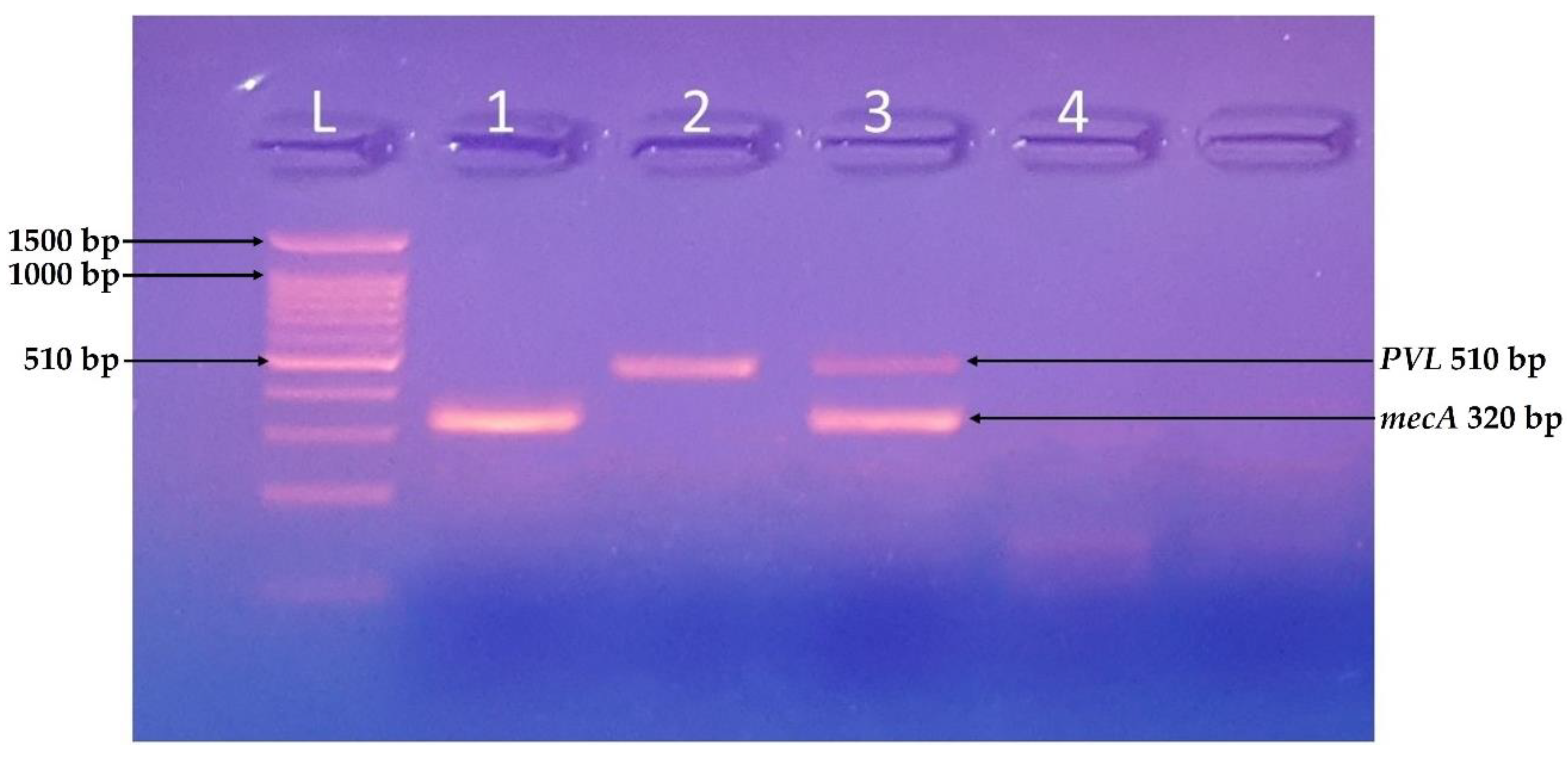
3.4. Molecular Identification of Virulent Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rajilic-Stojanovic, M.; Figueiredo, C.; Smet, A.; Hansen, R.; Kupcinskas, J.; Rokkas, T.; Andersen, L.; Machado, J.C.; Ianiro, G.; Gasbarrini, A.; et al. Systematic review: Gastric microbiota in health and disease. Aliment. Pharmacol. Ther. 2020, 51, 582–602. [Google Scholar] [CrossRef] [PubMed]
- Yasin, S.; Bhatti, T.; Farooqi, M.U.; Mateen, F. Immunologic aspect in diagnosis and treatment of SARS-COV-2 patients. J. Shifa Tameer-e-Millat Univ. 2020, 3, 113–121. [Google Scholar] [CrossRef]
- Koullali, B.; Van Zijl, M.D.; Kazemier, B.M.; Oudijk, M.A.; Mol, B.W.J.; Pajkrt, E.; Ravelli, A.C.J. The association between parity and spontaneous preterm birth: A population based study. BMC Pregnancy Childbirth 2020, 20, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dagklis, T.; Tsakiridis, I.; Mamopoulos, A.; Dardavessis, T.; Athanasiadis, A. Modifiable risk factors for spontaneous preterm birth in nulliparous women: A prospective study. J. Périnat. Med. 2020, 48, 96–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, E.; Romero, R.; Yoon, B.H.; Theis, K.R.; Gudicha, D.W.; Tarca, A.L.; Diaz-Primera, R.; Winters, A.D.; Gomez-Lopez, N.; Yeo, L.; et al. Bacteria in the amniotic fluid without inflammation: Early colonization vs. contamination. J. Périnat. Med. 2021. [Google Scholar] [CrossRef]
- Ciobanu, A.M.; Dumitru, A.E.; Gica, N.; Botezatu, R.; Peltecu, G.; Panaitescu, A.M. Benefits and Risks of IgG Transplacental Transfer. Diagnostics 2020, 10, 583. [Google Scholar] [CrossRef]
- Kumari, D.; Dash, D.R.K.; Behera, D.B.K.; Dash, D.D.K.; Mohanty, D.; Devi, M.; Srikant, A. Aetiological profile, risk factors, antibiotic sensitivity pattern and outcome of neonatal sepsis in tertiary care hospitals-a prospective observational study. Eur. J. Mol. Clin. Med. 2021, 8, 1073–1087. [Google Scholar]
- Bale, M.I.; Babatunde, S.K.; Awe, S. Prevalence of methicillin resistant Staphylococcus aureus bacteriuria among pregnant women attending secondary health hospitals in Ilorin, Nigeria. J. Adv. Microbiol. 2021, 21, 50–57. [Google Scholar] [CrossRef]
- Khan, S.; Siddiqui, S. Community-associated methicillin-resistant Staphylococcus aureus: Case report of acute sinusitis with orbital extension in a pregnant lady. Cureus 2020, 12, 12. [Google Scholar] [CrossRef]
- Jain, R.S.; Shirodkar, S.D. Associated comorbidities with obesity in pregnancy and its feto-maternal outcome. Int. J. Reprod. Contracept. Obstet. Gynecol. 2019, 8, 2785–2791. [Google Scholar] [CrossRef]
- Callahan, B.J.; DiGiulio, D.B.; Goltsman, D.S.A.; Sun, C.L.; Costello, E.K.; Jeganathan, P.; Biggio, J.R.; Wong, R.J.; Druzin, M.L.; Shaw, G.M.; et al. Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women. Proc. Natl. Acad. Sci. USA 2017, 114, 9966–9971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanni, D.; for the ELGAN Study Investigators; Korzeniewski, S.; Allred, E.N.; Fichorova, R.N.; O’Shea, T.M.; Kuban, K.; Dammann, O.; Leviton, A. Both antenatal and postnatal inflammation contribute information about the risk of brain damage in extremely preterm newborns. Pediatr. Res. 2017, 82, 691–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Dey, M.; Singh, S.; Sasidharan, S. Biochemical markers as predictor of preterm labor-their clinical relevance and the current status. Gy-Necology Obstet. Reprod. Med. 2021, 9, 1–8. [Google Scholar] [CrossRef]
- Hogan, P.G.; Mork, R.L.; Thompson, R.M.; Muenks, C.E.; Boyle, M.G.; Sullivan, M.L.; Morelli, J.J.; Williams, C.V.; Sanchez, N.; Hunstad, D.A.; et al. Environmental Methicillin-resistant Staphylococcus aureus Contamination, Persistent Colonization, and Subsequent Skin and Soft Tissue Infection. JAMA Pediatr. 2020, 174, 552. [Google Scholar] [CrossRef]
- Schuetz, C.R.; Hogan, P.G.; Reich, P.J.; Halili, S.; Wiseman, H.E.; Boyle, M.G.; Thompson, R.M.; Warner, B.B.; Fritz, S.A. Factors associated with progression to infection in methicillin-resistant Staphylococcus aureus-colonized, critically ill neonates. J. Perinatol. 2021, 41, 1285–1292. [Google Scholar] [CrossRef]
- Anafo, R.B.; Atiase, Y.; Kotey, F.C.N.; Dayie, N.T.K.D.; Tetteh-Quarcoo, P.B.; Duodu, S.; Osei, M.-M.; Alzahrani, K.J.; Donkor, E.S. Methicillin-resistant Staphylococcus aureus (MRSA) nasal carriage among patients with diabetes at the Korle Bu Teaching Hospital. PLoS ONE 2021, 16, e0257004. [Google Scholar] [CrossRef]
- Tsouklidis, N.; Kumar, R.; Heindl, S.E.; Soni, R.; Khan, S. Understanding the fight against resistance: Hospital-acquired methicillin-resistant Staphylococcus Aureus vs. community-acquired methicillin-resistant Staphylococcus aureus. Cureus 2020, 12, e8867. [Google Scholar] [CrossRef]
- Tumuhamye, J.; Steinsland, H.; Tumwine, J.K.; Namugga, O.; Mukunya, D.; Bwanga, F.; Sommerfelt, H.; Nankabirwa, V. Vaginal colonisation of women in labour with potentially pathogenic bacteria: A cross sectional study at three primary health care facilities in Central Uganda. BMC Infect. Dis. 2020, 20, 1–10. [Google Scholar] [CrossRef]
- Basset, P.; Feil, E.J.; Zanetti, G.; Blanc, D.S. The evolution and dynamics of methicillin-resistant Staphylococcus aureus. In Genetics and Evolution of Infectious Disease; Elsevier BV: Amsterdam, The Netherlands, 2011; pp. 669–688. [Google Scholar]
- Jevons, M. ‘Celebenin’-resistant staphylococci. BMJ 1961, 1, 124–125. [Google Scholar] [CrossRef]
- Noto, M.J.; Kreiswirth, B.N.; Monk, A.B.; Archer, G.L. Gene Acquisition at the Insertion Site for SCC mec, the Genomic Island Conferring Methicillin Resistance in Staphylococcus aureus. J. Bacteriol. 2008, 190, 1276–1283. [Google Scholar] [CrossRef] [Green Version]
- Parris, K.M.; Amabebe, E.; Cohen, M.C.; Anumba, D.O. Placental microbial–metabolite profiles and inflammatory mechanisms associated with preterm birth. J. Clin. Pathol. 2021, 74, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Burton, G.; Sebire, N.; Myatt, L.; Tannetta, D.; Wang, Y.-L.; Sadovsky, Y.; Staff, A.; Redman, C. Optimising sample collection for placental research. Placenta 2014, 35, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.N.; Bryant, J.E.; Madsen, E.L.; Ghiorse, W.C. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 1999, 65, 4715–4724. [Google Scholar] [CrossRef] [Green Version]
- Chen, F. GenElute™ five-minute plasmid miniprep kit: The fastest method for plasmid preparation. Vacuum 2013, 8, 6. [Google Scholar]
- Doster, R.S.; Kirk, L.A.; Tetz, L.M.; Rogers, L.M.; Aronoff, D.; Gaddy, J.A. Staphylococcus aureus infection of human gestational membranes induces bacterial biofilm formation and host production of cytokines. J. Infect. Dis. 2016, 215, 653–657. [Google Scholar] [CrossRef]
- Washam, M.; Woltmann, J.; Haberman, B.; Haslam, D.; Staat, M.A. Risk factors for methicillin-resistant Staphylococcus aureus colonization in the neonatal intensive care unit: A systematic review and meta-analysis. Am. J. Infect. Control 2017, 45, 1388–1393. [Google Scholar] [CrossRef] [PubMed]
- Gitman, M.R.; Alburquerque, B.; Chung, M.; van de Guchte, A.; Sullivan, M.J.; Obla, A.; Polanco, J.; Oussenko, I.; Smith, M.L.; Samaroo, F.; et al. Modified methicillin-resistant Staphylococcus aureus detected in neonatal intensive care patients. J. Antimicrob. Chemother. 2021, 76, 2774–2777. [Google Scholar] [CrossRef]
- Bauters, E.; Jonckheere, S.; Dehaene, I.; Vandecandelaere, P.; Argudín, M.A.; Page, G. Prevalence and clinical relevance of colonization with methicillin-resistant Staphylococcus aureus in the obstetric population. J. Matern. Neonatal Med. 2021, 1–6. [Google Scholar] [CrossRef]
- Atolagbe, C.T.; Tytler, B.A.; Jimoh, O.; Olayinka, A.T.; Olayinka, B. Prevalence and antimicrobial susceptibility pattern of coagulase-negative staphylococci obtained from nares of adult patients admitted to Ahmadu Bello University Teaching Hospital (ABUTH), Zaria. AROC Pharm. Biotechnol. 2021, 1, 34–43. [Google Scholar] [CrossRef]
- Skiba-Kurek, I.; Nowak, P.; Empel, J.; Tomczak, M.; Klepacka, J.; Sowa-Sierant, I.; Żak, I.; Pomierny, B.; Karczewska, E. Evaluation of biofilm formation and prevalence of multidrug-resistant strains of Staphylococcus epidermidis isolated from neonates with sepsis in Southern Poland. Pathogens 2021, 10, 877. [Google Scholar] [CrossRef]
| Source | Total Samples | Positive Samples |
|---|---|---|
| Full-Term Babies | 68 | 16 (23.5%) |
| Preterm Babies | 12 | 4 (33.3%) |
| Source | Total S. aureus | MRSA Positive |
|---|---|---|
| Full-Term | 16 | 7 (43.7%) |
| Preterm | 4 | 3 (75.0%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farooqi, H.M.U.; Kim, K.-H.; Kausar, F.; Muhammad, J.; Bukhari, H.; Choi, K.-H. Frequency and Molecular Characterization of Staphylococcus aureus from Placenta of Mothers with Term and Preterm Deliveries. Life 2022, 12, 257. https://doi.org/10.3390/life12020257
Farooqi HMU, Kim K-H, Kausar F, Muhammad J, Bukhari H, Choi K-H. Frequency and Molecular Characterization of Staphylococcus aureus from Placenta of Mothers with Term and Preterm Deliveries. Life. 2022; 12(2):257. https://doi.org/10.3390/life12020257
Chicago/Turabian StyleFarooqi, Hafiz Muhammad Umer, Kyung-Hwan Kim, Farzana Kausar, Javed Muhammad, Habib Bukhari, and Kyung-Hyun Choi. 2022. "Frequency and Molecular Characterization of Staphylococcus aureus from Placenta of Mothers with Term and Preterm Deliveries" Life 12, no. 2: 257. https://doi.org/10.3390/life12020257
APA StyleFarooqi, H. M. U., Kim, K.-H., Kausar, F., Muhammad, J., Bukhari, H., & Choi, K.-H. (2022). Frequency and Molecular Characterization of Staphylococcus aureus from Placenta of Mothers with Term and Preterm Deliveries. Life, 12(2), 257. https://doi.org/10.3390/life12020257






