The Influence of Kerosene on Microbiomes of Diverse Soils
Abstract
1. Introduction
2. Materials and Methods
2.1. Pot Experiment
2.2. Field Experiment
2.3. Soil Sampling and Chemical Analysis
2.4. DNA Extraction, Amplification and Sequencing
2.5. 16S rRNA Sequencing Data Analysis
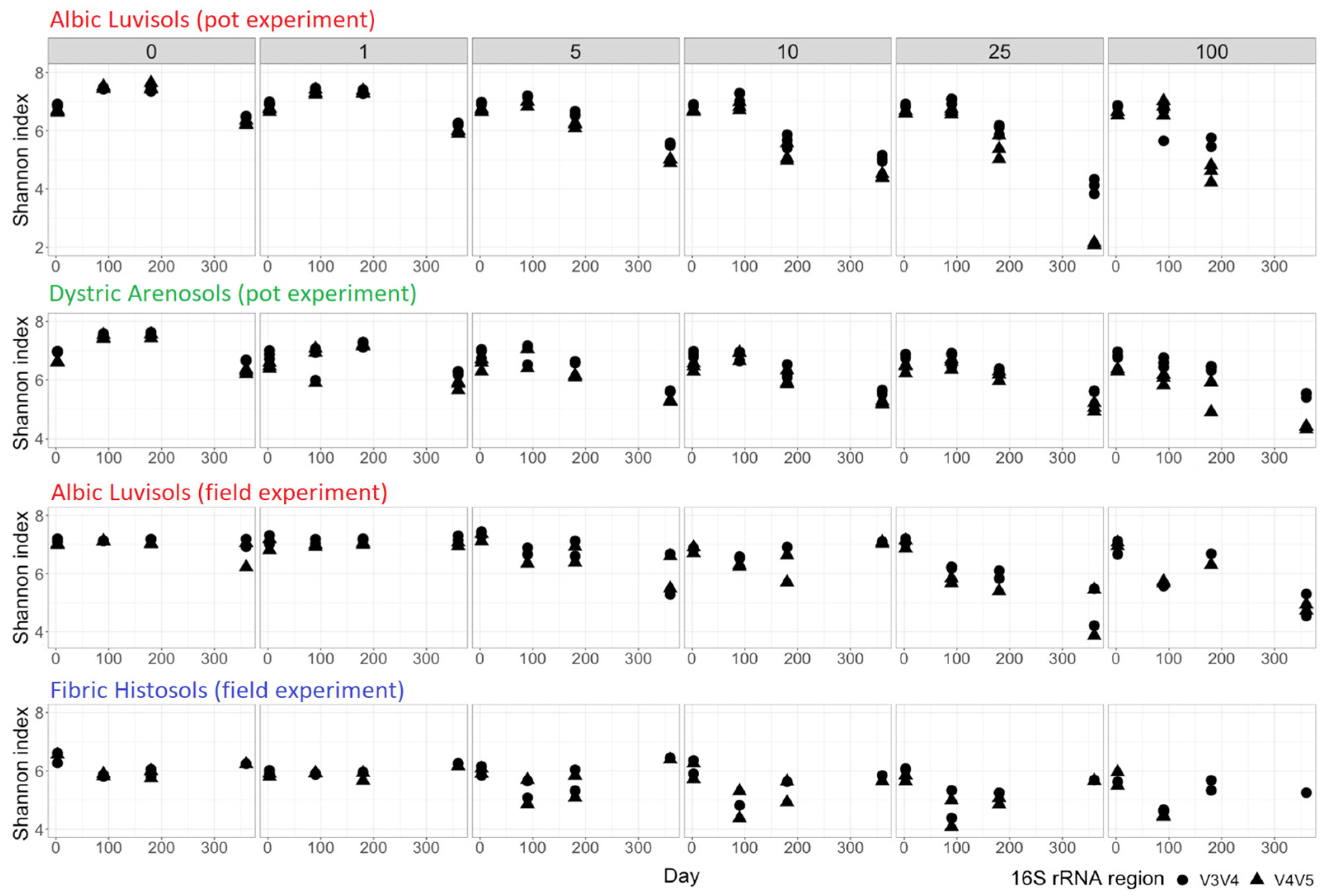
2.6. Assessment of Soil Microbial Community Diversity and Statistical Analysis
3. Results and Discussion
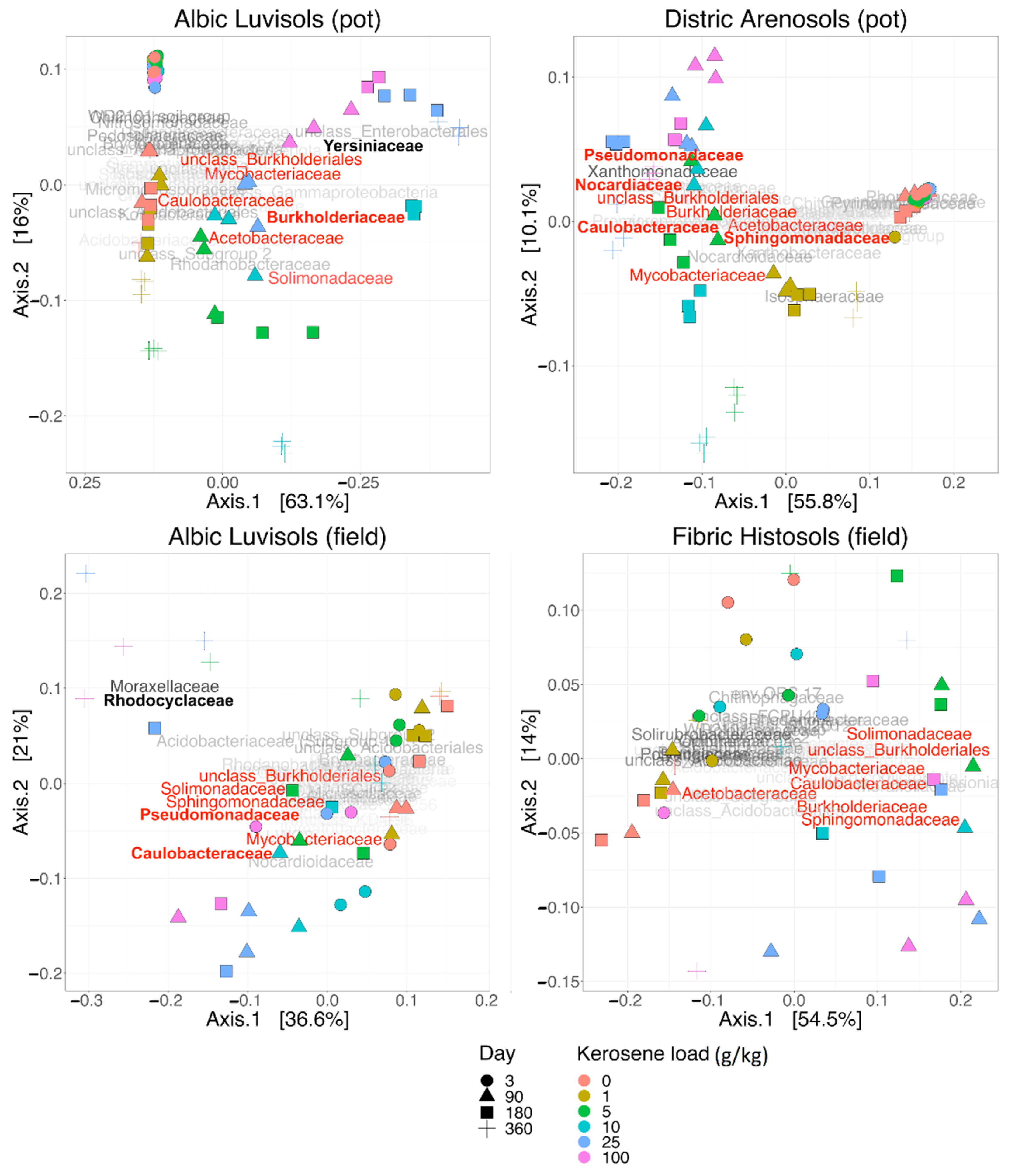
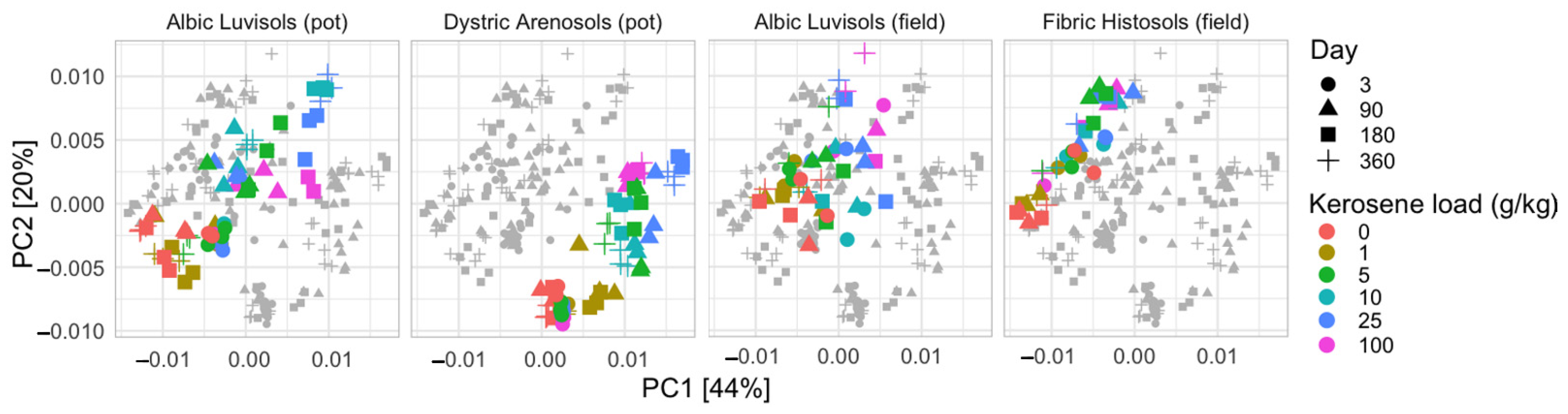
3.1. Soil Similarities and Differences in Physicochemical Properties and Microbiome Composition
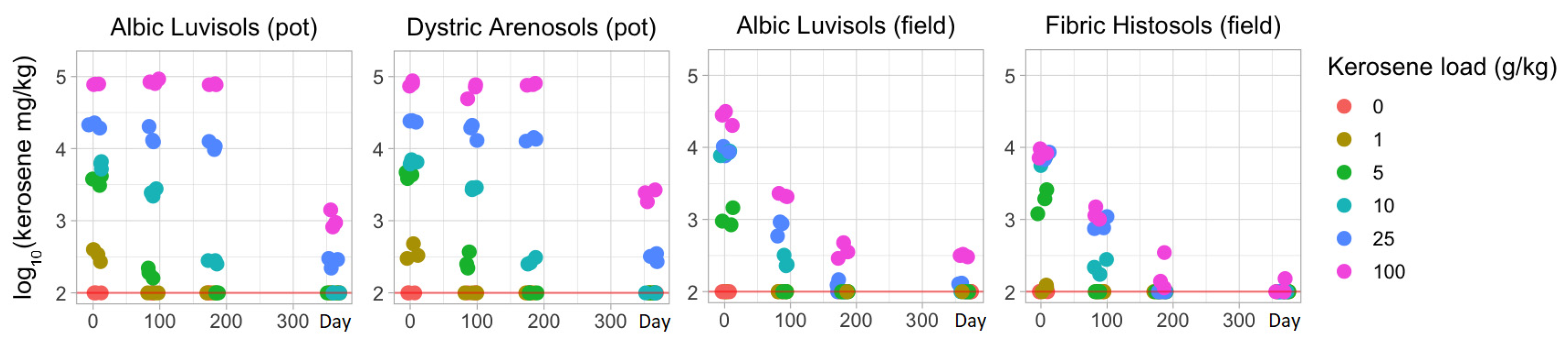
3.2. Temporal Changes in the Kerosene Concentration and the Physicochemical Soil Properties
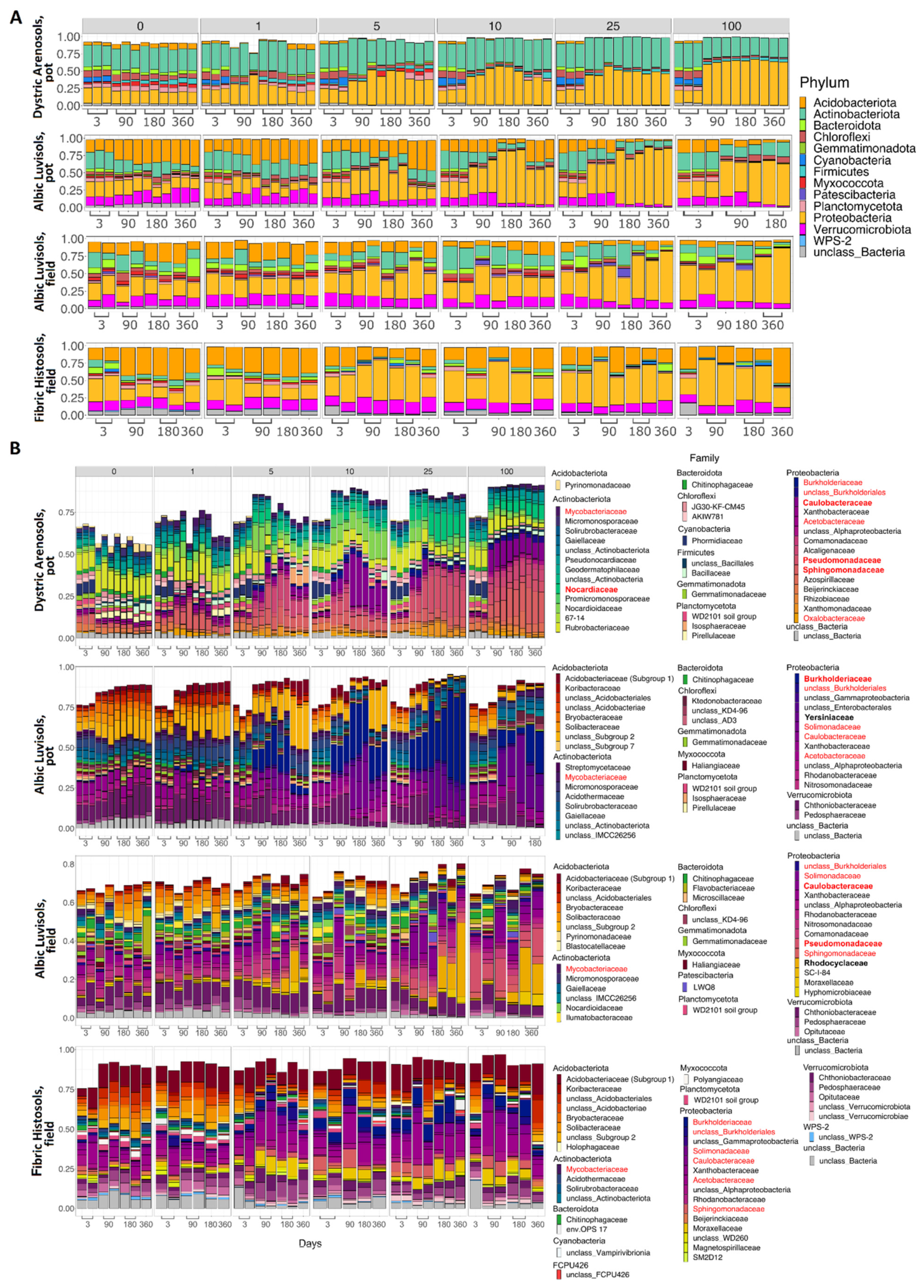
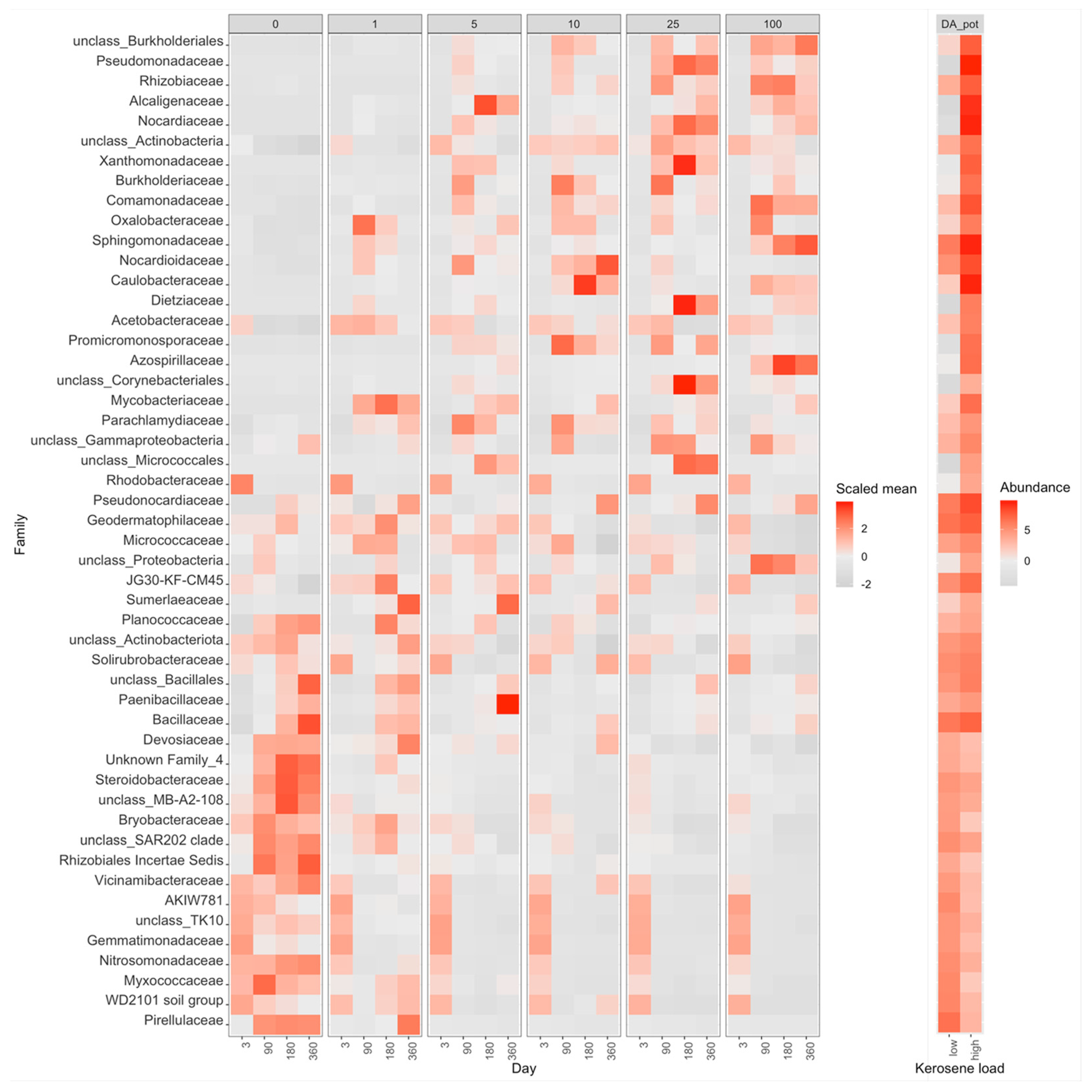
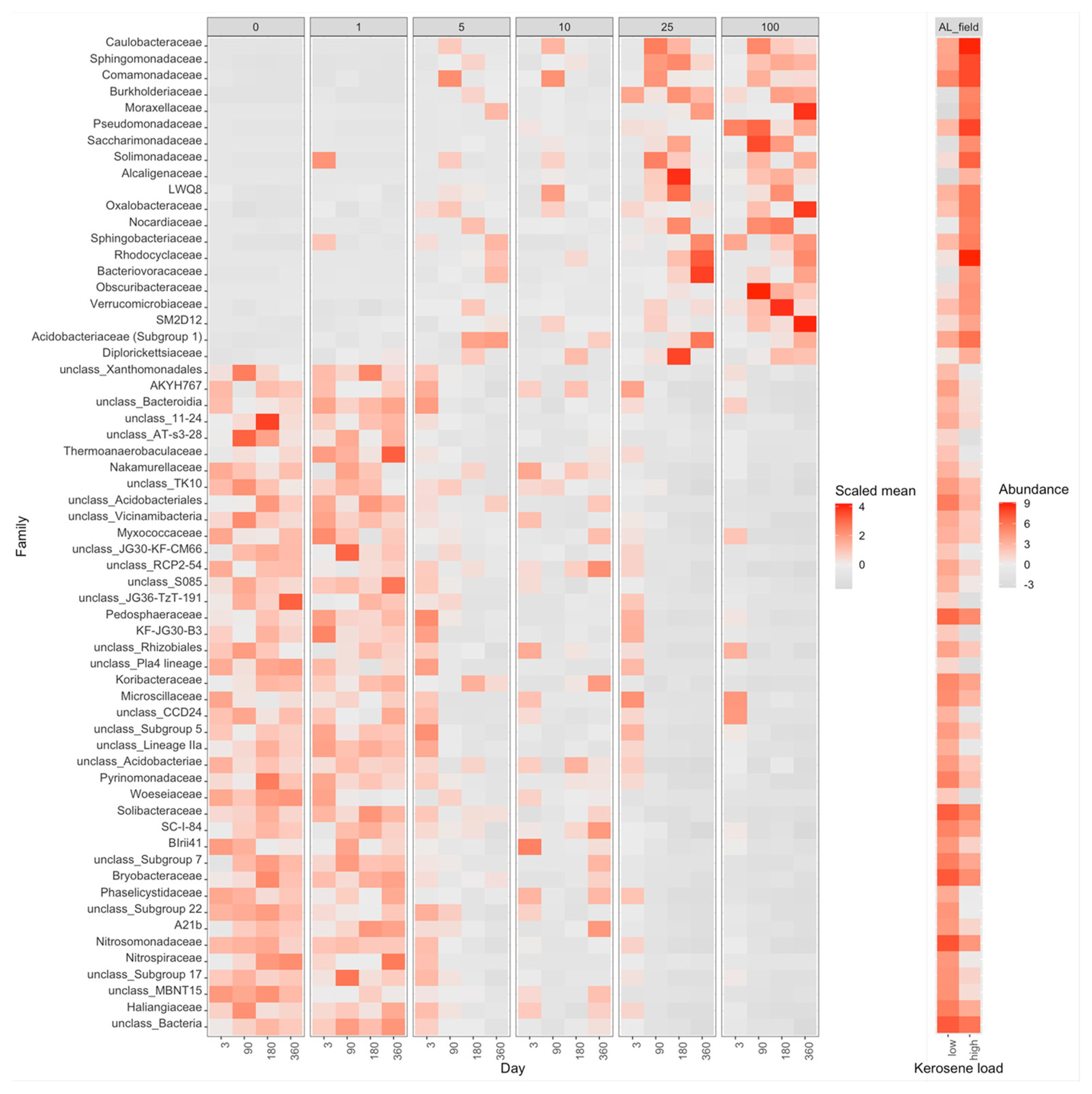
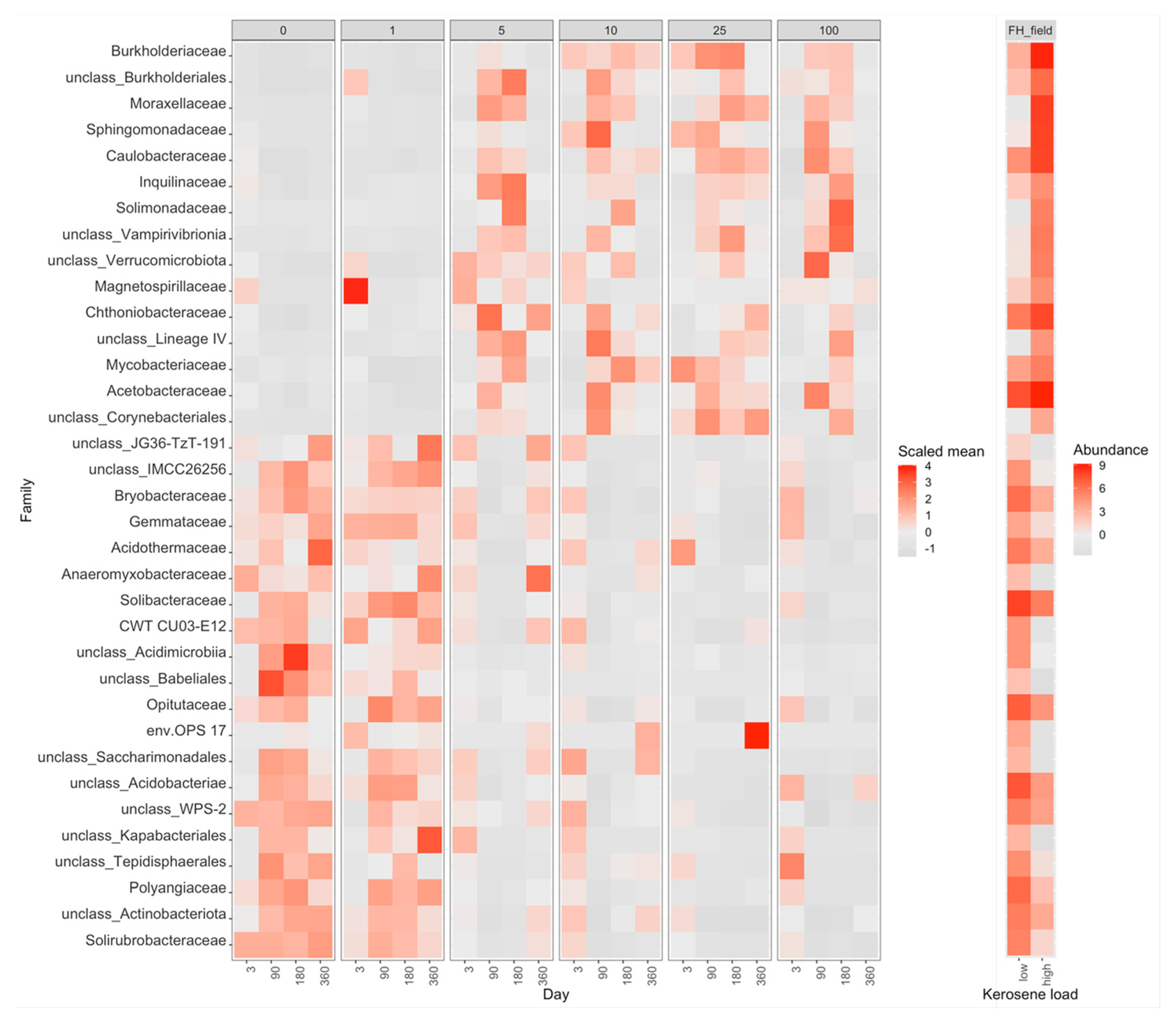
3.3. Taxonomic Composition of Soil Samples Treated with Kerosene
3.3.1. Albic Luvisols, the Pot Experiment
3.3.2. Dystric Arenosols, the Pot Experiment
3.3.3. Albic Luvisols, the Field Experiment
3.3.4. Fibric Histosols, the Field Experiment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bao, Y.J.; Xu, Z.; Li, Y.; Yao, Z.; Sun, J.; Song, H. High-throughput metagenomic analysis of petroleum-contaminated soil microbiome reveals the versatility in xenobiotic aromatics metabolism. J. Environ. Sci. 2017, 56, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Walker, D.A.; Webber, P.J.; Everett, K.R.; Brown, J. Effects of Crude and Diesel Oil Spills on Plant Communities at Prudhoe Bay, Alaska, and the Derivation of Oil Spill Sensitivity Maps. Arctic 1978, 31, 242–259. [Google Scholar] [CrossRef]
- Hutchinson, T.C.; Freedman, W. Effects of experimental crude oil spills on subarctic boreal forest vegetation near Norman Wells, N.W.T., Canada. Can. J. Bot. 1978, 56, 2424–2433. [Google Scholar] [CrossRef]
- Racine, C.H. Long-term recovery of vegetation on two experimental crude oil spills in interior Alaska black spruce taiga. Can. J. Bot. 1994, 72, 1171–1177. [Google Scholar] [CrossRef]
- Holt, S. The effects of crude and diesel oil spills on plant communities at Mesters Vig, northeast Greenland. Arct. Alp. Res. 1987, 19, 490–497. [Google Scholar] [CrossRef]
- Bay, C. Effects of Experimental Spills of Crude and Diesel Oil on Arctic Vegetation. A Long-Term Study on High Arctic Terrestrial Plant Communities in Jameson Land, Central East Greenland; NERI Technical Report; Ministry of Environment and Energy, National Environmental Research Institute: Aarhus C, Denmark, 1997.
- Koroleva, T.V.; Semenkov, I.N.; Sharapova, A.V.; Krechetov, P.P.; Lednev, S.A. Ecological consequences of space rocket accidents in Kazakhstan between 1999 and 2018. Environ. Pollut. 2021, 268, 115711. [Google Scholar] [CrossRef]
- Tremblay, J.; Yergeau, E.; Fortin, N.; Cobanli, S.; Elias, M.; King, T.L.; Lee, K.; Greer, C.W. Chemical dispersants enhance the activity of oil- and gas condensate-degrading marine bacteria. ISME J. 2017, 11, 2793–2808. [Google Scholar] [CrossRef]
- Khan, S.R.; Kumar, J.I.N.; Banker, M.; Kumar, R.N. Ex-Situ Studies on Biodegradation of Artificially Enriched Kerosene and Diesel Soils by Fungal Isolates. Soil Sediment Contam. 2015, 24, 796–810. [Google Scholar] [CrossRef]
- Bacosa, H.P.; Suto, K.; Inoue, C. Degradation potential and microbial community structure of heavy oil-enriched microbial consortia from mangrove sediments in Okinawa, Japan. J. Environ. Sci. Health Part A 2013, 48, 835–846. [Google Scholar] [CrossRef]
- Xu, X.; Liu, W.; Tian, S.; Wang, W.; Qi, Q.; Jiang, P.; Gao, X.; Li, F.; Li, H.; Yu, H. Petroleum Hydrocarbon-Degrading Bacteria for the Remediation of Oil Pollution Under Aerobic Conditions: A Perspective Analysis. Front. Microbiol. 2018, 9, 2885. [Google Scholar] [CrossRef]
- Karthikeyan, R.; Kulakow, P.A. Soil plant microbe interactions in phytoremediation. Adv. Biochem. Eng. Biotechnol. 2003, 78, 52–74. [Google Scholar] [CrossRef]
- Kim, K.D. Effects of diesel and kerosene on germination and growth of coastal wetland plant species. Bull. Environ. Contam. Toxicol. 2014, 93, 596–602. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mackenzie, J.S.; Jeggo, M. The one health approach-why is it so important? Trop. Med. Infect. Dis. 2019, 4, 88. [Google Scholar] [CrossRef]
- Koroleva, T.V.; Krechetov, P.P.; Semenkov, I.N.; Sharapova, A.V.; Lednev, S.A.; Karpachevskiy, A.M.; Kondratyev, A.D.; Kasimov, N.S. The environmental impact of space transport. Transp. Res. Part D Transp. Environ. 2018, 58, 54–69. [Google Scholar] [CrossRef]
- Lednev, S.A.; Koroleva, T.V.; Semenkov, I.N.; Klink, G.V.; Krechetov, P.P.; Sharapova, A.V.; Karpachevskiy, A.M.; Klink, G.V. The natural regeneration of desert ecosystem vegetation at the 2013 crash site of a Proton-M launch vehicle, Republic of Kazakhstan. Ecol. Indic. 2019, 101, 603–613. [Google Scholar] [CrossRef]
- Kottek, M.; Grieser, J.; Beck, C.; Rudolf, B.; Rubel, F. World Maps of Köppen-Geiger climate classification updated. Meteorol. Zeitschrift. 2006, 15, 259–263. [Google Scholar] [CrossRef]
- Lednev, S.A.; Semenkov, I.N.; Klink, G.V.; Krechetov, P.P.; Sharapova, A.V.; Koroleva, T.V. Impact of kerosene pollution on ground vegetation of southern taiga in the Amur Region, Russia. Sci. Total Environ. 2021, 772, 144965. [Google Scholar] [CrossRef]
- Dorokhova, M.F.; Krechetov, P.P.; Koroleva, T.V.; Sharapova, A.V. Algo-cyanobacterial communities as indicators of soil pollution with jet-fuel. Algae Cyanobacteria Nat. Agric. Ecosyst. In Proceedings of the II Int. Sci. Pract. Conf. Dedic. to 105th Anniv. Birth Profr. Emilia Adrianovna Shtina, Kirov, Russia, 19–23 October 2015; Vyatskaya GSHA: Kirov, Russia, 2015; pp. 118–122. [Google Scholar]
- Sharapova, A.V.; Semenkov, I.N.; Krechetov, P.P.; Lednev, S.A.; Koroleva, T.V. The Effect of Kerosene Pollution on the Cellulolytic Activity of Albic Retisols and Protic Arenosols: A Laboratory Experiment. Eurasian Soil Sci. Sci. 2022, 55, 233–239. [Google Scholar] [CrossRef]
- Bolotnik, T.A.; Timchenko, Y.V.; Plyushchenko, I.V.; Levkina, V.V.; Pirogov, A.V.; Smolenkov, A.D.; Popik, M.V.; Shpigun, O.A. Use of Chemometric Methods of Data Analysis for the Identification and Typification of Petroleum and Petroleum Products. J. Anal. Chem. 2019, 74, 1336–1340. [Google Scholar] [CrossRef]
- Andrews, S. A Quality Control Tool for High Throughput Sequence Data. 2019. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 16 December 2021).
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef] [PubMed]
- Murali, A.; Bhargava, A.; Wright, E.S. IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 2018, 6, 140. [Google Scholar] [CrossRef]
- Wright, E.S. Using DECIPHER v2.0 to analyze big biological sequence data in R. R J. 2016, 8, 352–359. [Google Scholar] [CrossRef]
- Kim, G.; Bae, J.; Kim, M.J.; Kwon, H.; Park, G.; Kim, S.J.; Choe, Y.H.; Kim, J.; Park, S.H.; Choe, B.H.; et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2019, 41, D590–D596. [Google Scholar]
- Wright, E.S. DECIPHER: Harnessing local sequence context to improve protein multiple sequence alignment. BMC Bioinform. 2015, 16, 322. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Gloor, G. ALDEx2: ANOVA-Like Differential Expression tool for compositional data. ALDEX Man. Modul. 2019, 20, 1–11. [Google Scholar]
- Abellan-Schneyder, I.; Matchado, M.S.; Reitmeier, S.; Sommer, A.; Sewald, Z.; Baumbach, J.; List, M.; Neuhaus, K. Primer, Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing. Msphere 2021, 6, e01202-20. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef]
- Janssen, P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef] [PubMed]
- Andronov, E.E.; Petrova, S.N.; Pinaev, A.G.; Pershina, E.V.; Rakhimgaliyeva, S.; Akhmedenov, K.M.; Gorobets, A.V.; Sergaliev, N.K. Analysis of the structure of microbial community in soils with different degrees of salinization using T-RFLP and real-time PCR techniques. Eurasian Soil Sci. 2012, 45, 147–156. [Google Scholar] [CrossRef]
- McHugh, T.A.; Compson, Z.; van Gestel, N.; Hayer, M.; Ballard, L.; Haverty, M.; Hines, J.; Irvine, N.; Krassner, D.; Lyons, T.; et al. Climate controls prokaryotic community composition in desert soils of the southwestern United States. FEMS Microbiol. Ecol. 2017, 93, fix116. [Google Scholar] [CrossRef] [PubMed]
- Dilly, O.; Bloem, J.; Vos, A.; Munch, J.C. Bacterial Diversity in Agricultural Soils during Litter Decomposition. Appl. Environ. Microbiol. 2004, 70, 468–474. [Google Scholar] [CrossRef]
- Naliukhin, A.N.; Khamitova, S.M.; Glinushkin, A.P.; Avdeev, Y.M.; Snetilova, V.S.; Laktionov, Y.V.; Surov, V.V.; Siluyanova, O.V.; Belozerov, D.A. Changes in the Metagenome of Prokaryotic Community as an Indicator of Fertility of Arable Soddy-Podzolic Soils upon Fertilizer Application. Eurasian Soil Sci. 2018, 51, 321–326. [Google Scholar] [CrossRef]
- Semenov, M.V.; Manucharova, N.A.; Krasnov, G.S.; Nikitin, D.A.; Stepanov, A.L. Biomass and Taxonomic Structure of Microbial Communities in Soils of the Right-Bank Basin of the Oka River. Eurasian Soil Sci. 2019, 52, 971–981. [Google Scholar] [CrossRef]
- Vilkiene, M.; Mockeviciene, I.; Karcauskiene, D.; Suproniene, S.; Doyeni, M.O.; Ambrazaitiene, D. Biological indicators of soil quality under different tillage systems in retisol. Sustainability 2021, 13, 9624. [Google Scholar] [CrossRef]
- Basiliko, N.; Henry, K.; Gupta, V.; Moore, T.R.; Driscoll, B.T.; Dunfield, P.F. Controls on bacterial and archaeal community structure and greenhouse gas production in natural, mined, and restored Canadian peatlands. Front. Microbiol. 2013, 4, 215. [Google Scholar] [CrossRef]
- Sun, H.; Terhonen, E.; Koskinen, K.; Paulin, L.; Kasanen, R.; Asiegbu, F.O. Bacterial diversity and community structure along different peat soils in boreal forest. Appl. Soil Ecol. 2014, 74, 37–45. [Google Scholar] [CrossRef]
- Grodnitskaya, I.D.; Trusova, M.Y.; Syrtsov, S.N.; Koroban, N.V. Structure of microbial communities of peat soils in two bogs in Siberian tundra and forest zones. Microbiology 2018, 87, 89–102. [Google Scholar] [CrossRef]
- Aksenov, A.S.; Shirokova, L.S.; Kisil, O.Y.; Kolesova, S.N.; Lim, A.G.; Kuzmina, D.; Pouillé, S.; Alexis, M.A.; Castrec-Rouelle, M.; Loiko, S.V.; et al. Bacterial number and genetic diversity in a permafrost peatland (Western Siberia): Testing a link with organic matter quality and elementary composition of a peat soil profile. Diversity 2021, 13, 328. [Google Scholar] [CrossRef]
- Qi, B.; Chen, Y.; Chen, D.; Chen, Y.; Ma, L.; Tian, X.; Li, Y.; Long, Y. Insight into Washing of Wet and Dry Crude Oil-Contaminated Soil, Clean—Soil, Air. Water 2021, 49, 2000440. [Google Scholar] [CrossRef]
- Yuan, L.; Habibi, A.; Dehghanpour, H. Liquid imbibition in tight rocks: The role of disjoining pressure, Colloids Surfaces A. Physicochem. Eng. Asp. 2021, 627, 127037. [Google Scholar] [CrossRef]
- Obire, O.; Nwaubeta, O.; Oofojekwu, P.C.; Ezenwaka, I.S.; Alegbeleye, W.O. Effects of Refined Petroleum Hydrocarbon on Soil Physicochemical and Bacteriological Characteristics. J. Appl. Sci. Environ. Manag. 2002, 6, 39–44. [Google Scholar] [CrossRef][Green Version]
- Fallah, M.; Shabanpor, M.; Ebrahimi, S. Evaluation of petroleum impacts on some properties of loamy sand soil with the main focus on hydraulic properties. Environ. Earth Sci. 2015, 74, 4751–4762. [Google Scholar] [CrossRef]
- Nseabasi, N.O.; Antai, S.P.; Bassey, I.U.; Iwatt, G.D.; Unimke, A.A. Chronic Kerosene Contamination and Variation in the Physicochemical and Heavy Metal Content of the Soil in Calabar, Cross River State, Nigeria. Imp. J. Interdiscip. Res. 2016, 2, 2094–2099. Available online: https://www.researchgate.net/publication/316668945_Chronic_Kerosene_Contamination_and_Variation_in_the_Physicochemical_and_Heavy_Metal_Content_of_the_Soil_in_Calabar_Cross_River_State_Nigeria (accessed on 16 December 2021).
- Beskrovnaya, P.; Fakih, D.; Morneau, I.; Hashimi, A.; Bello, D.G.; Xing, S.; Nanci, A.; Huan, T.; Tocheva, E.I. No Endospore Formation Confirmed in Members of the Phylum Proteobacteria. Appl. Environ. Microbiol. 2021, 87, 1–11. [Google Scholar] [CrossRef]
- Yang, S.; Wen, X.; Shi, Y.; Liebner, S.; Jin, H.; Perfumo, A. Hydrocarbon degraders establish at the costs of microbial richness, abundance and keystone taxa after crude oil contamination in permafrost environments. Sci. Rep. 2016, 6, 37473. [Google Scholar] [CrossRef]
- Shapiro, T.; Chekanov, K.; Alexandrova, A.; Dolnikova, G.; Ivanova, E.; Lobakova, E. Revealing of non-cultivable bacteria associated with the mycelium of fungi in the kerosene-degrading community isolated from the contaminated jet fuel. J. Fungi 2021, 7, 43. [Google Scholar] [CrossRef]
- Bacosa, H.; Suto, K.; Inoue, C. Preferential degradation of aromatic hydrocarbons in kerosene by a microbial consortium. Int. Biodeterior. Biodegrad. 2010, 64, 702–710. [Google Scholar] [CrossRef]
- Mitter, E.K.; Germida, J.J.; de Freitas, J.R. Impact of diesel and biodiesel contamination on soil microbial community activity and structure. Sci. Rep. 2021, 11, 10856. [Google Scholar] [CrossRef] [PubMed]
- Bacosa, H.P.; Steichen, J.; Kamalanathan, M.; Windham, R.; Lubguban, A.; Labonté, J.M.; Kaiser, K.; Hala, D.; Santschi, P.H.; Quigg, A. Polycyclic aromatic hydrocarbons (PAHs) and putative PAH-degrading bacteria in Galveston Bay, TX (USA), following Hurricane Harvey (2017). Environ. Sci. Pollut. Res. 2020, 27, 34987–34999. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, J.J.A.; Bacosa, H.P.; Chien, M.F.; Inoue, C. Enhanced degradation of polycyclic aromatic hydrocarbons (PAHs) in the rhizosphere of sudangrass (Sorghum × drummondii). Chemosphere 2019, 234, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Chikere, C.B.; Tekere, M.; Adeleke, R. Microbial communities in field-scale oil-polluted soil remediation using 16S rRNA amplicon sequencing. Int. J. Environ. Stud. 2021, 78, 410–426. [Google Scholar] [CrossRef]
- Shukor, M.Y.; Hassan, N.A.A.; Jusoh, A.Z.; Perumal, N.; Shamaan, N.A.; MacCormack, W.P.; Syed, M.A. Isolation and characterization of a Pseudomonas diesel-degrading strain from Antarctica. J. Environ. Biol. 2009, 30, 1–6. [Google Scholar]
- Ma, Y.; Wang, L.; Shao, Z. Pseudomonas, the dominant polycyclic aromatic hydrocarbon-degrading bacteria isolated from Antarctic soils and the role of large plasmids in horizontal gene transfer. Environ. Microbiol. 2006, 8, 455–465. [Google Scholar] [CrossRef]
- Yang, S.; Wen, X.; Zhao, L.; Shi, Y.; Jin, H. Crude oil treatment leads to shift of bacterial communities in soils from the deep active layer and upper permafrost along the China-Russia Crude Oil Pipeline route. PLoS ONE 2014, 9, e96552. [Google Scholar] [CrossRef]
- Byrne, E.R.; Roche, K.M.; Schaerer, L.G.; Techtmann, S.M. Temporal variation of crude and refined oil biodegradation rates and microbial community composition in freshwater systems. J. Great Lakes Res. 2021, 47, 1376–1385. [Google Scholar] [CrossRef]
- Bradford, L.M.; Vestergaard, G.; Táncsics, A.; Zhu, B.; Schloter, M.; Lueders, T. Transcriptome-Stable Isotope Probing Provides Targeted Functional and taxonomic insights into microaerobic pollutant-degrading aquifer microbiota. Front. Microbiol. 2018, 9, 2696. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, X.; Li, H.; Jia, J.; Liu, Y.; Ejenavi, O.; Ding, A.; Sun, Y.; Zhang, D. Separating and characterizing functional alkane degraders from crude-oil-contaminated sites via magnetic nanoparticle-mediated isolation. Res. Microbiol. 2016, 167, 731–744. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Juottonen, H.; Siivonen, P.; Lloret Quesada, C.; Tuomi, P.; Pulkkinen, P.; Yrjälä, K. Spatial patterns of microbial diversity and activity in an aged creosote-contaminated site. ISME J. 2014, 8, 2131–2142. [Google Scholar] [CrossRef]
- Zhang, S.; Hu, Z.; Wang, H. A retrospective review of microbiological methods applied in studies following the Deepwater Horizon oil spill. Front. Microbiol. 2018, 9, 520. [Google Scholar] [CrossRef] [PubMed]
- Makut, M.D.; Ishaya, P. Bacterial species associated with soils contaminated with used petroleum products in Keffi town, Nigeria. Afr. J. Microbiol. Res. 2010, 4, 1698–1702. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shelyakin, P.V.; Semenkov, I.N.; Tutukina, M.N.; Nikolaeva, D.D.; Sharapova, A.V.; Sarana, Y.V.; Lednev, S.A.; Smolenkov, A.D.; Gelfand, M.S.; Krechetov, P.P.; et al. The Influence of Kerosene on Microbiomes of Diverse Soils. Life 2022, 12, 221. https://doi.org/10.3390/life12020221
Shelyakin PV, Semenkov IN, Tutukina MN, Nikolaeva DD, Sharapova AV, Sarana YV, Lednev SA, Smolenkov AD, Gelfand MS, Krechetov PP, et al. The Influence of Kerosene on Microbiomes of Diverse Soils. Life. 2022; 12(2):221. https://doi.org/10.3390/life12020221
Chicago/Turabian StyleShelyakin, Pavel V., Ivan N. Semenkov, Maria N. Tutukina, Daria D. Nikolaeva, Anna V. Sharapova, Yulia V. Sarana, Sergey A. Lednev, Alexander D. Smolenkov, Mikhail S. Gelfand, Pavel P. Krechetov, and et al. 2022. "The Influence of Kerosene on Microbiomes of Diverse Soils" Life 12, no. 2: 221. https://doi.org/10.3390/life12020221
APA StyleShelyakin, P. V., Semenkov, I. N., Tutukina, M. N., Nikolaeva, D. D., Sharapova, A. V., Sarana, Y. V., Lednev, S. A., Smolenkov, A. D., Gelfand, M. S., Krechetov, P. P., & Koroleva, T. V. (2022). The Influence of Kerosene on Microbiomes of Diverse Soils. Life, 12(2), 221. https://doi.org/10.3390/life12020221







